Abstract
The phytohormone abscisic acid (ABA) plays an essential role in adaptive stress responses. The hormone regulates, among others, the expression of numerous stress-responsive genes. From various promoter analyses, ABA-responsive elements (ABREs) have been determined and a number of ABRE binding factors have been isolated, although their in vivo roles are not known. Here we report that the ABRE binding factors ABF3 and ABF4 function in ABA signaling. The constitutive overexpression of ABF3 or ABF4 in Arabidopsis resulted in ABA hypersensitivity and other ABA-associated phenotypes. In addition, the transgenic plants exhibited reduced transpiration and enhanced drought tolerance. At the molecular level, altered expression of ABA/stress-regulated genes was observed. Furthermore, the temporal and spatial expression patterns of ABF3 and ABF4 were consistent with their suggested roles. Thus, our results provide strong in vivo evidence that ABF3 and ABF4 mediate stress-responsive ABA signaling.
INTRODUCTION
Being sessile, plants have the capability to adapt to adverse environmental conditions such as drought, cold, and high salt. Under these stress conditions, the plant hormone abscisic acid (ABA) level increases in vegetative tissues, triggering adaptive responses that are essential for their survival and productivity (Zeevaart and Creelman, 1988; Leung and Giraudat, 1998). Under water deficit conditions, for example, ABA induces stomatal closure, minimizing water loss through transpiration. The ABA-controlled process is vital for plant survival, and ABA-deficient and ABA-responsive mutants are susceptible to water stress. On the other hand, high levels of ABA inhibit overall plant growth (Himmelbach et al., 1998).
Underlying the ABA-mediated stress responses is the transcriptional regulation of stress-responsive gene expression (Giraudat et al., 1994; Busk and Pages, 1998). Numerous genes have been reported that are upregulated under stress conditions in vegetative tissues (Ingram and Bartels, 1996; Shinozaki and Yamaguchi-Shinozaki, 1997). These include a class of genes known as LEA (for LATE EMBRYOGENESIS ABUNDANT) genes, which are expressed abundantly in developing seed under normal conditions, osmolyte biosynthetic genes, and genes of general cellular metabolism. In general, the gene products are considered to have protective or adaptive roles under stress conditions. In addition, the expression of many regulatory genes, including various kinase/phosphatase and transcription factor genes, also is induced by abiotic stresses. Not all stress-inducible genes are regulated by ABA. However, a large number of them also are responsive to exogenous ABA, and in many cases, their induction is impaired in ABA-deficient mutants. Meanwhile, the expression of some genes, such as rbcS and CAB genes, is suppressed by ABA and stress (Bartholomew et al., 1991; Wang et al., 1996; Weatherwax et al., 1996).
ABA-responsive elements (ABREs) that control ABA- and/or stress-responsive gene expression have been determined by numerous studies (Giraudat et al., 1994), and their putative cognate trans-acting factors have been isolated (Busk and Pages, 1998). Most ubiquitous among the cis elements is a group of sequences sharing the (C/T)ACGTGGC consensus. Many of these elements contain the G-box (CACGTG) sequence (Giuliano et al., 1988), which also is present in numerous genes regulated by other environmental cues (Menkens et al., 1995). Another group of ABREs, known as coupling element, hex3, or motif III (Busk and Pages, 1998), shares the CGCGTG core sequence. On the basis of their interactions with these two types of ABREs, which are referred to as ABREs hereafter, a number of putative trans-acting factors have been isolated (Busk and Pages, 1998). Also, their homologs and numerous other G-box binding factors, all belonging to the bZIP class proteins (Landschulz et al., 1988), are able to interact with the ABREs in vitro (Foster et al., 1994). However, the evidence showing that any of these biochemically identified factors play a role in ABA or stress signaling in planta is still lacking.
In an effort to identify transcription factors that control ABA-responsive gene expression during vegetative growth, we recently isolated four ABRE binding bZIP factors by yeast one-hybrid screening of an Arabidopsis cDNA expression library (Choi et al., 2000). Expression of the factors, referred to as ABF1 through ABF4 (ABRE Binding Factors 1 to 4), is ABA and stress inducible, and they can transactivate an ABRE-containing reporter gene in yeast. ABF2 and ABF4 also were reported by Uno et al. (2000), who named them AREB1 and AREB2, respectively, and showed that the factors can activate an ABA-responsive promoter in protoplasts. To investigate the in vivo functions of ABFs, we generated transgenic Arabidopsis plants constitutively overexpressing them. Here we show that ABF3 and ABF4 transgenic lines are hypersensitive to ABA and that they exhibit several other ABA/stress-associated phenotypes, including enhanced drought tolerance. The expression patterns of the two ABFs correlated well with their overexpression phenotypes.
RESULTS
Growth Phenotypes of ABF3 and ABF4 Overexpression Lines
To investigate the in vivo functions of ABF3 and ABF4, we used an overexpression approach. The coding region of ABF3 or ABF4 was fused to the 35S promoter of Cauliflower mosaic virus, and each construct was used to transform Arabidopsis (ecotype Landsberg erecta [Ler]) plants. Thirty-eight and 12 T3 homozygous lines were recovered from the 35S-ABF3 and 35S-ABF4 constructs, respectively, and after preliminary analysis, transgenic lines with higher ABF expression levels were selected for more detailed analysis.
Compared with wild-type plants, 35S-ABF3 transgenic plants exhibited mild growth retardation in the aerial parts: petioles were slightly shorter, and leaves were rounder in shape (Figure 1A). The degree of retardation was not severe, however, and overall growth patterns were similar to those of wild-type plants except that siliques were somewhat shorter and thicker (Figure 1A, inset). In contrast, 35S-ABF4 transgenic plants exhibited severe growth retardation (Figure 1A), which was dependent on the ABF4 expression level (Figure 1B). Petioles were shorter, leaves were smaller, flowering was delayed, and plants were shorter. Also, the germination of 35S-ABF3 plants was delayed several hours compared with that of wild-type plants in the absence of ABA (Figure 1C). 35S-ABF4 plants, on the other hand, germinated normally (data not shown); thus, the growth retardation observed with 35S-ABF4 plants was a postgermination process.
Figure 1.
Growth Phenotype of 35S-ABF3 and 35S-ABF4 Plants.
(A) Growth on soil. 35S-ABF3 (line A319) and 35S-ABF4 (line A405) transgenic plants were grown for 3 weeks on soil. The inset shows fully grown siliques, two each from Ler, 35S-ABF3, and 35S-ABF4 plants (left to right).
(B) Relationship between ABF4 expression level and the severity of the growth phenotype. Left, RNA gel blot analysis of ABF4 expression in Ler and the 35S-ABF4 transgenic lines A402, A405, and A406. The bottom panel shows ethidium bromide staining of the RNA gel. Each lane contained 25 μg of total RNA. Right, 35S-ABF4 transgenic lines with varying degrees of ABF4 expression. Only the aerial parts of the plants are shown for clarity.
(C) Left, germination of 35S-ABF3 transgenic seed on ABA-free medium. Ten-week-old seed of Ler, line A333, and line A319 were plated on ABA-free medium after 4 days of cold treatment, and germination (fully emerged radicle) was scored at various times. Each data point represents the mean of triplicate experiments (n = 50 each). Standard errors are smaller than the symbols. Right, RNA gel blot analysis of ABF3 expression in Ler and transgenic lines A333 and A319. The bottom panel shows ethidium bromide staining of the RNA gel. Each lane contained 25 μg of total RNA. The ABF3 expression levels in the A333 and A319 lines are comparable to the ABF4 expression levels in the A405 and A406 lines.
ABA Response of 35S-ABF Plants
To determine whether ABF3 or ABF4 overexpression affected ABA sensitivity, 35S-ABF3 and 35S-ABF4 transgenic plants were germinated and grown on media containing various concentrations of ABA. When ABA concentration was 0.5 μM or greater, the growth of 35S-ABF plants was arrested completely after radicles emerged; that is, cotyledon greening/expansion and root growth were inhibited severely, and none of the transgenic seedlings developed to have true leaves (Figure 2A). Under the same conditions, wild-type plants continued to grow and develop, although at slower rates than on ABA-free medium. The ABA hypersensitivity of 35S-ABF transgenic plants also was observed at 0.25 μM ABA (Figure 2B), although the transgenic seedlings eventually grew to have true leaves (data not shown).
Figure 2.
ABA Sensitivity of 35S-ABF3 and 35S-ABF4 Plants.
(A) Growth of transgenic plants on Murashige and Skoog (1962) medium (MS) containing 0.5 μM ABA. Seed were germinated and grown for 12 days.
(B) Growth of transgenic plants on MS medium containing 0.25 μM ABA. Seed were germinated on the medium for 3 days, and representative plants are shown.
(C) ABA dose response of germination. Seed, 6 months old after harvest and prechilled at 4°C for 4 days, were germinated on media containing various concentrations of ABA, and seedlings with fully emerged radicles were counted after 3 days. Experiments were performed in triplicate (n = 50 each), and the bars show standard errors.
(D) ABA dose response of root growth. Seed were germinated for 4 days on ABA-free medium, and the seedlings (n = 6) were transferred to media containing various concentrations of ABA. Root elongation was measured 5 days after the transfer. The experiments were performed more than four times, sometimes using different transgenic lines, and the results were consistent. The small bars represent standard errors. Transgenic lines A319 (ABF3) and A405 (ABF4) were used.
To determine the stage specificity of the ABA response, ABA dose response was examined during and after germination. As shown in Figure 2C, 35S-ABF transgenic plants were hypersensitive to ABA at the germination stage: 0.5 μM ABA was sufficient to inhibit their germination efficiencies to 8% (ABF3) or 28% (ABF4), whereas wild-type plants retained 80% germination under the same conditions. Likewise, root growth of the 35S-ABF transgenic lines was hypersensitive to ABA (Figure 2D). At 0.5 μM ABA, wild-type root growth was 84% of its control rate, whereas that of 35S-ABF4 and 35S-ABF3 plants was 35 and 51% of their control rates, respectively. At 1 μM ABA, root growth of the 35S-ABF4 plants was reduced to 6% and that of the 35S-ABF3 plants was reduced to 37%. In addition, lateral root and aerial part growth of 35S-ABF transgenic plants was inhibited significantly at this concentration (data not shown). Wild-type plants grew at 62% of the control rate at the same ABA concentration. Transgenic root growth was arrested almost completely when ABA concentration was >5 μM, whereas wild-type plants still continued to grow. These results indicate that both the germination and postgermination growth of 35S-ABF transgenic plants are hypersensitive to ABA.
Salt Response of 35S-ABF Plants
High concentrations of salts inhibit the germination of Arabidopsis (Werner and Finkelstein, 1995; Leon-Kloosterziel et al., 1996; Quesada et al., 2000; Zhu, 2000). Several studies show that ABA plays a role in the inhibition process. Although not all salt-insensitive mutants are ABA insensitive (Werner and Finkelstein, 1995), all ABA-deficient (aba) and ABA-insensitive (abi) mutants exhibit salt insensitivity during germination (Leon-Kloosterziel et al., 1996). This is probably because ABA, whose level increases under high salt conditions, promotes the inhibition process. Because the expression of both ABF3 and ABF4 is salt inducible (Choi et al., 2000), they may participate in salt response (in this case, salt-induced germination inhibition). To test this possibility, 35S-ABF transgenic plants were germinated on media containing various concentrations of NaCl. Figure 3A shows that the germination of wild-type and aba1, abi1, and abi2 mutant plants was not affected by NaCl <100 mM. In contrast, germination and growth of the 35S-ABF plants were affected significantly by 100 mM NaCl (Figures 3A and 3B): radicle emergence, root growth, and cotyledon opening/expansion were inhibited severely. In a parallel experiment, the transgenic plants responded to KCl in a similar manner (Figure 3B). On the other hand, their response to the same concentration (Figure 3B) or twice the concentration (data not shown) of mannitol, which gives the same osmotic pressure, was normal. Thus, in contrast to the salt-insensitive phenotype of ABA-deficient or ABA-insensitive mutants, both ABF3 and ABF4 overexpression resulted in salt hypersensitivity at the germination/young seedling stage, and the hypersensitivity appeared to be ionic rather than osmotic in nature.
Figure 3.
Salt and Glucose Sensitivity of 35S-ABF Plants.
(A) Salt effect on germination. Seed of Ler, aba1-1, abi1-1, abi2-1, 35S-ABF3 (line A319), and 35S-ABF4 (line A405) were plated after 4 days of cold treatment on media containing 50, 100, or 150 mM NaCl, and germination (fully open cotyledons) was scored after 4 days. Experiments were performed in triplicate (n = 50 each), and standard errors are smaller than the symbols.
(B) Salt effect on newly germinated seedling growth. Seed of the same transgenic lines were germinated for 4 days on MS medium containing 100 mM NaCl, KCl, or mannitol, and representative seedlings are shown.
(C) Glucose response of 35S-ABF transgenic plants. Seed were germinated and grown for 14 days on MS medium or the same medium supplemented with 3% glucose or mannitol in the vertical position. A319, A333, A405, and A406 indicate transgenic lines.
Sugar Response of 35S-ABF Plants
At higher concentrations, sugars inhibit the development of young seedlings; that is, they inhibit cotyledon greening/expansion and shoot growth (Jang et al., 1997). According to studies performed by other researchers, ABA plays an essential role in glucose or sucrose signal transduction (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000). For example, the ABA-deficient aba2 mutation is allelic to the sugar-insensitive sis4 mutation, and the glucose- or sugar-insensitive mutations gin6, sis5, and sun6 are allelic to the ABA-insensitive mutation abi4. Also, these studies show that other aba mutants, and to some degree abi5 mutants, are insensitive to glucose. Thus, ABF overexpression might have affected sugar sensitivity as well, if in fact ABF3 and ABF4 mediate ABA signaling. We addressed this possibility by examining their response to glucose, which exerts more severe growth inhibition than do other sugars (Jang et al., 1997). Under our experimental conditions, wild-type seedlings showed growth defects such as inhibition of cotyledon greening and true leaf development when glucose concentration was >4% (data not shown). The aerial part growth of 35S-ABF transgenic lines, on the other hand, was arrested completely at 3% glucose, at which level wild-type plants developed fully (Figure 3C). Thus, 35S-ABF transgenic plants were hypersensitive to glucose. This enhanced response of the transgenic plants was not observed with the same concentration of mannitol, which inhibited the growth of both wild-type and 35S-ABF transgenic plants significantly but similarly. Thus, the hypersensitivity was glucose specific rather than osmotic.
Epinasty and Obstacle-Touching Response of 35S-ABF4 Plants
Recent genetic studies show that ABA signaling pathways interact with those of ethylene. According to these studies, ABA-mediated inhibition of germination is regulated negatively by ethylene, whereas ethylene signaling components are required for the ABA inhibition of root growth (Beaudoin et al., 2000; Ghassemian et al., 2000). The involvement of auxin in ABA-dependent stress response also has been demonstrated (Vartanian et al., 1994). When exposed to progressive drought, new lateral roots take a short and tuberized form. This process, known as drought rhizogenesis, is impaired not only in the ABA-deficient aba-1 and ABA-insensitive abi1-1 mutants but also in the auxin-resistant axr1-3 mutant. To determine whether the overexpression of ABF3 or ABF4 affected ethylene or auxin sensitivity, we compared the effects of the ethylene precursor 1-aminocyclopropane-1-carboxylic acid and indole-3-acetic acid on the growth of 35S-ABF and wild-type plants. We did not see any differences in their responses. However, leaves of 35S-ABF4 plants have a tendency to have epinastic curvature when grown on plates (Figure 4A), which generally is attributed to ethylene action under stress conditions (Jackson, 1997). Also, roots of 35S-ABF4 transgenic plants exhibited abnormality in the obstacle-touching response (Okada and Shimura, 1990; Simmons et al., 1995). As shown in Figure 4B, roots of wild-type plants grow in a wavy pattern when grown on a hard agar surface at an inclined position. The wavy pattern of root growth, however, was diminished significantly in 35S-ABF4 plants. The obstacle-touching response is impaired in several auxin-resistant mutants, indicating that auxin signaling components are involved in the process (Okada and Shimura, 1992). Thus, this result implies that auxin signaling pathway(s) might have been perturbed in 35S-ABF4 plants. Together, our results suggest that ABF4 overexpression might have affected some aspects of ethylene and auxin responses.
Figure 4.
Epinasty and Obstacle-Touching Response of 35S-ABF4 Plants.
(A) Epinastic curvature of 35S-ABF4 transgenic leaves. Ler and transgenic plants were grown on MS plates for 2 weeks. ABF4 expression level is highest in the A406 line and lowest in the A402 line (Figure 1B).
(B) Obstacle-touching response of 35S-ABF4 plants. Plants (line A405) were grown on 1.5% agar plates for 4 days in the vertical position and then for 5 days at an angled (45°) position. Arrows indicate the root tip positions before changing to the slanted orientation.
Drought Tolerance of 35S-ABF Plants
One of the key ABA-controlled processes is stomatal closure under water stress conditions, which minimizes water loss through transpiration (Leung and Giraudat, 1998). ABA biosynthesis mutants and some of the ABA response mutants (i.e., abi1 and abi2), therefore, are very susceptible to drought because of the impaired stomatal aperture regulation (Leung and Giraudat, 1998; Schroeder et al., 2001). Thus, ABF3 and ABF4 overexpression lines are expected to exhibit an altered response to water deficit conditions if the factors are involved in ABA/stress signaling. To address this possibility, we examined the drought tolerance of 35S-ABF plants. As shown in Figure 5A, wild-type plants withered completely when withdrawn from water for 11 days, and only 16% of them survived to maturity when rewatered afterward. 35S-ABF3 plants, however, were not affected noticeably, and all survived the treatment to set seed. In a similar experiment, 35S-ABF4 plants also exhibited higher survival rates under water deficit conditions; all of them survived a 12-day drought treatment, whereas 33% of the wild-type plants survived to set seed (Figure 5B). Thus, both 35S-ABF3 and 35S-ABF4 plants survived the drought conditions better than did the wild-type plants.
Figure 5.
Drought Tolerance of 35S-ABF3 and 35S-ABF4 Plants.
(A) Drought tolerance of 35S-ABF3 transgenic plants (line A319). Transgenic and wild-type plants (n = 100 each) were grown on soil in the same container for 2 weeks, withheld from water for 11 days, and then rewatered. The photographs were taken 3 days after the rewatering.
(B) Drought tolerance of 35S-ABF4 transgenic plants (line A405). Plants at similar developmental stages (2-week-old wild-type plants and 3-week-old ABF4 plants) were withheld from water for 12 days and then rewatered. The photographs were taken 3 days after the rewatering.
(C) and (D) Transpiration rates of 35S-ABF3 and 35S-ABF4 transgenic plants, respectively. Leaves of similar developmental stages were excised and weighed at various times after the detachment. Each data point represents the mean of duplicate measurements (n = 9 each). Standard errors are smaller than the symbols.
(E) Stomatal aperture of ABF transgenic plants (lines A319 and A405). Stomatal guard cells were observed in the middle of the watering period. Arrows indicate guard cells, and the insets show representative stomata.
The enhanced drought tolerance of the transgenic plants could be attributed, at least in part, to their lower transpiration rates. When measured by the fresh weight loss of detached rosette leaves, the water loss rates of 35S-ABF3 and 35S-ABF4 transgenic lines were less than half and ∼70% of those of the wild-type plants, respectively (Figures 5C and 5D). Consistent with this result, the stomata of the transgenic plants had smaller openings than did the wild-type plants (Figure 5E). Under normal growth conditions, ∼85% (89 of 104) of the wild-type stomata were open, whereas ∼20% of them were open in 35S-ABF3 (15 of 71) and 35S-ABF4 (17 of 71) plants when observed in the middle of the watering period. Thus, constitutive overexpression of ABF3 or ABF4 resulted in partial stomatal closure, reduced transpiration, and enhanced drought tolerance.
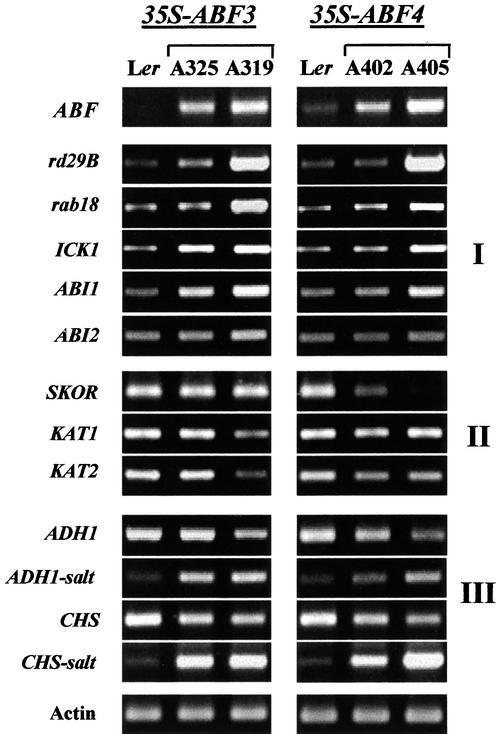
Expression of ABA-Responsive Genes in 35S-ABF Plants
To investigate the transcriptional regulatory roles of ABF3 and ABF4 in planta, the expression of various ABA/stress-responsive genes in 35S-ABF plants was determined. As shown in Figure 6, the transcript levels of a number of ABA-regulated genes (group I) were enhanced in 35S-ABF3 and 35S-ABF4 transgenic lines. These include LEA class genes rd29B (Yamaguchi-Shinozaki and Shinozaki, 1994) and rab18 (Lang and Palva, 1992), whose expression is induced by ABA and abiotic stresses. Expression of the ABA-inducible cell cycle regulator gene ICK1 (cyclin-dependent kinase inhibitor) (Wang et al., 1998) also was increased in the 35S-ABF transgenic lines. The ICK1 gene has been suggested to mediate cell division arrest by ABA. In the 35S-ABF3 lines, strong enhancement of ABI1 (Leung et al., 1994; Meyer et al., 1994) RNA level was observed, and ABI2 (Leung et al., 1997) transcript level was increased, although the degree of increase was lower. An increase in the ABI1 RNA level also was observed in the 35S-ABF4 line with higher ABF4 expression (A405). The expression of ABI1 and ABI2, which encode homologous protein phosphatase 2Cs and whose mutations result in defective stomatal closing and a wilty phenotype (Schroeder et al., 2001), is enhanced by ABA and water stress (Leung et al., 1997).
Figure 6.
Expression of ABA-Regulated Genes in 35S-ABF Transgenic Lines.
RNA levels of ABA-responsive genes were determined by coupled RT and PCR using total RNAs isolated from 2-week-old plants grown on MS plates. Lines A319 and A405 represent transgenic lines with higher ABF expression, whereas lines A325 and A402 represents transgenic lines with lower ABF expression.
Meanwhile, the RNA level of the ABA-repressible gene SKOR (Gaymard et al., 1998) was reduced significantly or was undetectable in 35S-ABF4 transgenic lines (group II). This gene encodes a root-specific K+ outward rectifying channel, and ABA repression of its expression has been suggested to be part of an adaptive water stress response. Similarly, guard cell ion channel genes KAT1 and KAT2 (Anderson et al., 1992; Pilot et al., 2001) were regulated negatively in the 35S-ABF3 line with higher ABF3 levels (A319). The two ion channels normally mediate K+ influx, enabling stomatal opening, but their activity is inhibited by ABA. Also, stress-responsive biosynthetic genes (group III), the chalcone synthase gene CHS (Feinbaum and Ausubel, 1988) and the alcohol dehydrogenase gene ADH1 (de Bruxelles et al., 1996), were downregulated. The transcript levels of these two genes, however, were higher in the 35S-ABF transgenic lines when plants were treated with high salt, suggesting that stress-induced post-translational modification of ABF3 and ABF4 may be required for the regulation of these genes. In summary, overexpression of ABF3 or ABF4 resulted in the modulation of ABA/stress-responsive gene expression, and the two factors played positive or negative roles depending on specific genes and environmental conditions.
Expression Patterns of ABF3 and ABF4
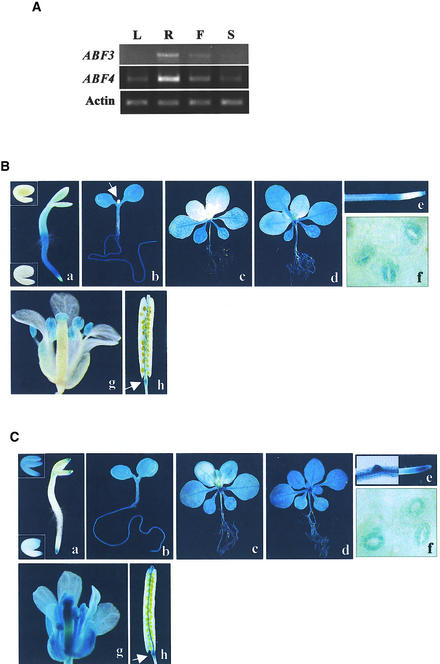
The phenotypes described so far indicated that the overexpression of ABF3 or ABF4 enhanced various aspects of ABA response. To assess the physiological relevance of the results and to obtain further clues about their functions, we investigated their temporal and spatial expression patterns. We first investigated the tissue specificity of their uninduced, basal expression. Because the basal expression levels of ABF3 and ABF4 are very low (Choi et al., 2000), we used coupled reverse transcription (RT) and polymerase chain reaction (PCR) for the analysis. As shown in Figure 7A, relatively higher expression of both ABF3 and ABF4 was detected in roots. Also, lower ABF3 expression was observed in flowers but not in leaves and siliques under the same conditions. On the other hand, weak ABF4 expression was detectable in leaves, flowers, and siliques.
Figure 7.
Expression Patterns of ABF3 and ABF4.
(A) Tissue specificity of ABF3 and ABF4 expression. RNA was isolated from various tissues of wild-type plants grown under normal conditions, and the expression of ABF3 and ABF4 was determined by coupled RT and PCR. L, leaves from 3-week-old plants; R, roots from 3-week-old plants; F, flowers; S, immature siliques.
(B) and (C) Histochemical GUS staining of ABF3 and ABF4 promoter activity, respectively. T3 homozygous plants were stained with 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for 6 hr ([e] and [f]) or 24 hr (other panels).
(a) Two-day-old seedlings. Insets show embryos from immature siliques (top) or dry seed (bottom).
(b) Five-day-old seedlings. The arrow in (B) shows the newly emerging shoot.
(c) Two-week-old seedlings.
(d) Two-week-old seedlings treated with 100 μM ABA (ABF3) or 200 mM NaCl (ABF4).
(e) Root tips. The left half of the image in (C) shows a lateral root primordium.
(f) Guard cells.
(g) Flowers.
(h) Siliques. Arrows indicate the silique abscission zone.
More detailed temporal and spatial expression patterns of ABF3 and ABF4 were determined by histochemical β-glucuronidase (GUS) staining of transgenic plants that harbored an ABF promoter–GUS reporter construct. With the ABF3 promoter (2.1 kb) construct, GUS activity was undetectable in embryos, but it was observed in the emerging radicles at the germination stage (Figure 7B, a) and in most of the vegetative tissues at later stages (Figure 7B, b and c). Roots were stained most strongly except the tip area, and petioles, leaf vascular tissues, and guard cells exhibited relatively strong GUS activity (Figure 7B, b to f). Emerging shoots and younger leaves (Figure 7B, b and c), on the other hand, exhibited GUS staining only after ABA, salt, or mannitol treatment (Figure 7B, d). In mature plants, GUS staining was detected also in anthers, stigma, and siliques (abscission zone, replum, and funiculi) (Figure 7B, g and h).
The ABF4 promoter was active in embryos from green siliques (Figure 7C, a), but its activity decreased as embryos became mature, and it was not detected in newly germinated seedlings except in some limited regions (radicle tip, shoot meristem region, and cotyledon tips) (Figure 7C, a). At later stages, starting from the stage of fully expanded cotyledons (Figure 7C, b), ABF4 promoter activity was observed in all vegetative tissues (Figure 7C, c to f) and also in floral organs and siliques (abscission zone, replum, and funiculi) (Figure 7C, g and h). The ABF4 promoter was most active in roots, especially in the growing regions (meristem, elongation zone, and lateral root primordia) (Figure 7C, e), suggesting its role in growth regulation. Also, it exhibited strong activity in petioles and guard cells (Figure 7C, c and f). Salt treatment of seedlings enhanced the ABF4 promoter activity somewhat (Figure 7C, d).
DISCUSSION
ABA-regulated gene expression plays a central role in ABA signaling, and numerous ABA/stress-responsive genes are regulated by the (C/T)ACGTGGC- or CGCGTG-containing ABREs. Thus, identifying relevant transcription factors is critical for the delineation of ABA signal transduction cascades. Many studies show that ABA signaling pathways are tissue specific (Giraudat et al., 1994; Leung and Giraudat, 1998), and several seed-specific ABA signaling components (ABI3, ABI4, and ABI5) have been identified by genetic screens. ABI3 and ABI4 encode transcription factors (Giraudat et al., 1992; Finkelstein et al., 1998) whose binding sites and immediate target genes are unknown. Recently, ABI5 has been shown to encode a bZIP factor that belongs to a seed-specific subfamily of ABF-related factors (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000), and its role in postgermination developmental arrest also has been demonstrated (Lopez-Molina et al., 2001). However, ABRE binding factors whose major function is to mediate ABA signaling during vegetative growth have not been reported, although numerous bZIP factors are known to interact with the ABREs in vitro (Foster et al., 1994).
ABFs are unique among the ABRE binding bZIP factors in that, unlike most of the other plant bZIP factors, they can interact with both the G-box type and the CGCGTG-containing ABREs (Choi et al., 2000). The broad binding specificity, together with the transactivation capability of an ABRE-containing reporter gene and the stress inducibility of their expression, suggested that ABFs have a potential to regulate a large number of ABA/stress-responsive genes and thus are likely to participate in stress-responsive ABA signaling. To address this question, we used an overexpression approach. Considering the potential functional redundancy of ABFs and numerous other bZIP factors interacting with ABREs (Foster et al., 1994), this approach would be better than loss-of-function approaches such as antisense, knockout, and RNA interference. The overexpression of ABFs (we estimate that ABF3 and ABF4 levels in the 35S-ABF transgenic lines used in our study range from approximately two- to 10-fold of their ABA-induced levels), however, might have caused unnatural conditions. For example, genes that are not normally regulated by ABFs might have been turned on or off. Also, it may have affected the functions of other ABFs or potentially other bZIP factors by titrating them out via nonnatural heterodimerization. Thus, the overexpression phenotypes need to be interpreted with caution, and their roles can be further confirmed by other experimental means. Nevertheless, our results show that ABF3 or ABF4 overexpression conferred several ABA-associated phenotypes, such as ABA hypersensitivity, sugar hypersensitivity, and enhanced drought tolerance, with altered expression of ABA/stress-responsive genes. Thus, our data provide a strong in vivo case for the involvement of ABF3 and ABF4 in stress-responsive ABA signaling.
Whereas the ABA hypersensitivity conferred by ABF3 or ABF4 overexpression was very distinct and observed at both the germination and later growth stages (Figure 2), the overexpression effects on growth in the absence of exogenous ABA were either moderate (ABF3) or developmental stage dependent (ABF4) (Figure 1). ABF3 exerted an inhibitory effect on both germination and seedling growth. However, the low degree of inhibition compared with that in the presence of exogenous ABA suggests that ABF3 alone is not sufficient for the inhibitory function. On the other hand, ABF4 overexpression had little effect on germination but had a severe effect on seedling growth, suggesting that ABF4 activity is modulated developmentally. Alternatively, this result may indicate that ABA inhibition of seedling growth is mediated by a mechanism that differs from the germination inhibition mechanism. Also, the developmental stage dependence of the ABF4 effects implies that the growth retardation of 35S-ABF4 plants probably results from the constitutive operation of part of the ABA signal transduction cascades rather than from the pleiotropic effects of ABF4 overexpression. Except for the varying degrees of growth retardation, neither 35S-ABF4 nor 35S-ABF3 plants showed any abnormality in general development. Thus, their overexpression affected growth rate but not developmental processes.
Other ABA- or stress-associated phenotypes of 35S-ABF3 and 35S-ABF4 transgenic plants include their hypersensitivities to salt and glucose, and 35S-ABF4 plants exhibited additional phenotypes (i.e., epinasty of leaves and abnormal obstacle-touching response) that can be related to altered ethylene or auxin response. The salt and glucose hypersensitivities may reflect the increased sensitivity to high osmolarity, because both high salt and high sugar accompany increases in osmolarity. However, 35S-ABF plants responded normally to mannitol, indicating that osmotic sensitivity was not affected. Thus, it appears that ABF3 and ABF4 are involved only in the nonosmotic branches of salt and glucose signaling pathways. The sugar-mediated developmental arrest is confined to a narrow window of developmental stages (∼2 days after germination) and is mediated by increased ABA level via an ABI4-dependent signaling cascade (Gazzarrini and McCourt, 2001). Also, it has been reported that ABA mediates developmental arrest at a similar stage and that the inhibition process requires ABI5 (Lopez-Molina et al., 2001), whose mutations result in weak sugar insensitivity. Thus, our results suggest that ABF3 and ABF4 have overlapping functions with ABI4 and ABI5 in mediating sugar- and ABA-induced developmental arrest. This is particularly so in the case of ABF3, because the onset of its expression coincides with the early developmental stage (Figure 7B, a).
Stomatal closure is a key ABA-controlled process in coping with water deficit conditions. Our data indicate that ABF3 and ABF4 are involved in this process. Their overexpression resulted in lower transpiration and enhanced drought tolerance (Figure 5), which are reminiscent of the phenotypes of the ABA-hypersensitive mutant era1 (Pei et al., 1998). Furthermore, the stomatal openings of 35S-ABF transgenic plants were smaller than those of wild-type plants, and altered expression of several genes involved in stomatal aperture regulation has been observed in transgenic plants (Figure 6). Among the genes we investigated, ABI1 was the most strongly affected, especially in ABF3 overexpression lines. ABI1 is known to be a negative regulator of ABA signaling (Gosti et al., 1999), although its expression is enhanced by ABA and high osmolarity and reduced in the aba1 and abi1 mutants (Leung et al., 1997). It is not clear, though, whether the increased ABI1 level played a positive or a negative role in the stomatal closing. Whatever ABI1's role might be, our results indicate that ABI1 expression is subject to ABF3 regulation and that ABF3 and ABF4 overexpression affected the expression of genes involved in stomatal movement and/or guard cell ABA signaling (Schroeder et al., 2001), the net result of which was enhanced stomatal closure.
The transcript level changes of ABA-responsive genes in 35S-ABF transgenic lines demonstrate that ABF3 and ABF4 function as transcriptional regulators in planta. As shown in Figure 6, both positive and negative changes were observed depending on specific genes. This result suggests that different subsets of ABF3 and ABF4 target genes are regulated by different mechanisms. Also, the negative regulation of some genes under normal growth conditions and the positive regulation of the same genes after salt treatment (Figure 6, group III) suggest that stress-induced modification of ABF activities is required for the regulation of these genes. The modifying activity may be limiting under normal conditions. Thus, the negative regulation of some genes can be explained, for example, by the binding of transcriptionally inactive, unmodified ABFs, whose proportion increases with higher ABF levels. The modification may involve phosphorylation of ABFs. The involvement of kinase/phosphatases in ABA/stress signaling is well known (Leung and Giraudat, 1998), and several phosphorylation sites are highly conserved among ABFs (Choi et al., 2000). More recently, Uno et al. (2000) reported an ABA-activated kinase activity in cultured cells that phosphorylates AREB1 (ABF2) and AREB2 (ABF4). Alternatively, the modification may involve interaction with other regulatory proteins.
The expression patterns of ABF3 and ABF4 were consistent with the functions suggested by their overexpression phenotypes. Spatially, both promoters were most active in roots and guard cells, consistent with their roles during water stress response. Also, ABF3, which exerted only minor growth inhibition, was expressed weakly in the growing tissues (root tips, new shoots, and new leaves), whereas ABF4 was expressed strongly in the growing regions of roots (meristem, elongation zone, and lateral root primordia). Temporally, strong ABF3 promoter activity was observed in the newly germinated seedlings, consistent with its more pronounced effect on germination (Figures 1C and 2C). On the other hand, the ABF4 promoter exhibited major activity at the onset of seedling growth, in agreement with its severe effect on seedling growth. Our results also show that both ABF3 and ABF4 might function during reproductive stages and seed abscission. Both promoters exhibited strong activity in the abscission zone, replum, and funiculi of siliques, and relatively strong activity was detected in stigma and anthers.
The isolation of multiple factors with similar binding activities but different expression patterns suggested that the ABA/stress signaling involving the ABREs is likely to be mediated by multiple factors (Choi et al., 2000). Our current results further support this observation. Although their overexpression phenotypes and expression patterns were similar, ABF3 and ABF4 were different from each other in several respects. The details of their temporal and spatial expression patterns differed (Figure 7). Growth retardation, root growth inhibition, and impaired stimulus-touching response were more prominent in ABF4 transgenic lines. Also, minor differences were observed at the molecular level. ABI1 and ABI2 expression levels were higher in 35S-ABF3 transgenic lines, whereas downregulation of SKOR expression was observed only in 35S-ABF4 plants. The functions of other ABFs (ABF1 and ABF2) remain to be determined, but their roles appear to be quite different from each other and from those of ABF3 and ABF4, according to our preliminary data. Thus, ABRE-dependent ABA/stress signaling in vegetative tissues appears to be mediated by multiple factors with overlapping but distinct functions.
As mentioned above, extensive genetic studies have been performed and a number of ABA signaling components have been identified (Leung and Giraudat, 1998; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000). However, none of ABFs has been isolated in these genetic screens, although they are highly homologous with ABI5, and our results strongly suggest that ABF3 and ABF4 are involved in ABA signaling. This is probably because of the selection criteria used in the mutant screenings. In most of the genetic studies, mutants were selected based on their altered sensitivities to ABA during germination. Thus, mutants that specifically affect vegetative ABA signaling might have been bypassed. Our observations that ABF3 and ABF4 are expressed mainly in vegetative tissues and that their overexpression effects on germination are moderate or insignificant support this hypothesis. Functional redundancy among ABFs (see above) also may have contributed to the negative results.
METHODS
Arabidopsis Growth
Arabidopsis thaliana ecotype Landsberg erecta (Ler) was used in this study. aba1-1, abi1-1, and abi2-1 seed (Koornneef et al., 1982, 1984) were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus), and their phenotypes were confirmed before use.
Plants were grown at 22°C under long day conditions (16-hr-light/8-hr-dark cycle) aseptically or on soil. For soil growth, seed were sown on a 1:1:1 mixture of vermiculite, perlite, and peat moss irrigated with 0.1% Hyponex (Hyponex Co., Marysville, OH) placed at 4°C for 4 days in the dark to break residual dormancy, and transferred to normal growth conditions. Unless stated otherwise, the plants were watered once per week. For aseptic growth, seed were treated with 70% ethanol for 5 min and then with 30% household bleach for 5 min, washed five times with sterile water, and plated on MS medium (Murashige and Skoog, 1962) solidified with 0.8% phytoagar. The MS medium was supplemented with 1% sucrose and, as described in Results, with abscisic acid, salts, glucose, or mannitol as needed. For the germination test, seed collected at the same or similar times were used. For root growth measurements, plants were germinated and grown in the vertical position.
Constructs and Arabidopsis Transformation
The 35S promoter–ABF coding region constructs (35S-ABF3 and 35S-ABF4) were prepared by replacing the β-glucuronidase (GUS) coding region of pBI121 (Jefferson et al., 1987) with the coding region of ABF3 or ABF4. The GUS sequence was removed after BamHI–SacI digestion, and after T4 DNA polymerase treatment to remove the 3′ overhang, the remaining portion of pBI121 was ligated with the ABF3 or ABF4 coding region, which was prepared by polymerase chain reaction (PCR) followed by BamHI digestion. The ABF coding regions included their entire coding regions with the stop codons, and a BamHI linker sequence was attached in front of the initiation codons for cloning. The ABF promoter–GUS reporter fusions were prepared by inserting 2.1 kb (ABF3) or 1.2 kb (ABF4) of their 5′ flanking sequences from the initiation codons in front of the GUS reporter gene of pBI101.2 (Jefferson et al., 1987).
The promoter fragments were prepared by PCR using Arabidopsis (ecotype Columbia) genomic DNA as a template and the primer sets 5′-caaacttaccctgttgttgcaact-3′ and 5′-ctagtctagaaggatcaagcttctgga-tatttac-3′ for ABF3 and 5′-gatcaatttgaatttttgatatacatc-3′ and 5′-cta-gtctagattcaatgaaaacaaagcatccaag-3′ for ABF4. The PCR fragments were digested with XbaI and ligated with pBI101.2, which was prepared by HindIII digestion followed by Klenow treatment and XbaI digestion. DNA manipulation was according to standard procedures (Sambrook et al., 1989; Ausubel et al., 1994), and the intactness of the ABF coding regions and the junction sequences was confirmed by DNA sequencing.
Transformation of Arabidopsis was according to the vacuum infiltration method (Bechtold and Pelletier, 1998) using Agrobacterium tumefaciens strain GV3101. For the phenotypic investigation, T3 or T4 homozygous lines were used. GUS staining patterns were confirmed by observing at least five different transgenic lines, and T3 homozygous lines were used for detailed analysis.
RNA Isolation, Coupled Reverse Transcription and PCR, and RNA Gel Blot Analysis
RNA was isolated by the method of Chomczynski and Mackey (1995), with a minor modification (Choi et al., 2000). RNA gel blot analysis and coupled reverse transcription (RT)-PCR were performed as described (Choi et al., 2000) with the following modifications. For RNA gel blot analysis, hybridization was performed at 65°C in the Rapid-hyb buffer from Amersham Pharmacia Biotech. Exposure time was 6 hr (ABF3) or 20 hr (ABF4). RT-PCR was performed using the Access RT-PCR System from Promega or the Superscript One-Step RT-PCR System from Gibco BRL. Each RT-PCR result was confirmed by several independent reactions, and RNA preparations were confirmed to be free of contaminating genomic DNA using primer sets spanning introns whenever possible. Primers used in the RT-PCR reactions are listed in Table 1.
Table 1.
Coupled RT and PCR Primers
| Gene Name | Sequence (5′ to 3′) |
|---|---|
| Actin | F: cat cag gaa gga ctt gta cgg R: gat gga cct gac tcg tca tac |
| ABF3 | F: aga acc tca acc ggt gga gag tg R: gga gtc aga tca ggt gac atc tgg |
| ABF4 | F: aac tgt gtt caa cag atg ggt cag R: ggt tcc tcc gta act agc taa tcc |
| rd29B | F: gtg aag atg act atc tcg gtg gtc R: gcc taa ctc tcc ggt gta acc tag |
| rab18 | F: atg acg agt acg gaa atc cga tgg R: tat gta tac acg att gtt cga agc |
| ICK1 | F: acg cac acg taa cct aaa tcg R: gca tct ccg tca tca att tcg |
| ABI1 | F: tca aga ttc cga gaa cgg aga tc R: gag gat caa acc gac cat cta ac |
| ABI2 | F: gtt ctt gtt ctg gcg acg gag c R: cca tta gtg act cga cca tca ag |
| rd29A | F: gat aac gtt gga gga aga gtc ggc R: cag ctc agc tcc tga ttc act acc |
| SKOR | F: atg gga ggt agt agc ggc ggc R: gat tct ctg gta atc ccc tga ag |
| KAT1 | F: ttc tgc gtc gag gaa tac aat ata g R: ctt agg gtc aac tag aag ata g |
| KAT2 | F: aca caa gac caa tgt caa tct ctt g R: gtc gac tag aag ata tga gtg gc |
| ADH1 | F: tcc acg tat ctt cgg cca tg R: tag cac ctt ctg cag cgc c |
| CHS | F: tca cca aca gtg aac aca tga cc R: gag tca agg tgg gtg tca gag g |
F, forward primer; R, reverse primer.
Histochemical GUS Staining
In situ assay of GUS activity was performed as described by Jefferson et al. (1987). Whole plants were immersed in 1 mM 5-bromo-4-chloro- 3-indolyl-β-glucuronic acid solution in 100 mM sodium phosphate, pH 7.0, 0.1 mM EDTA, 0.5 mM ferricyanide, 0.5 mM ferrocyanide, and 0.1% Triton X-100, and after applying vacuum for 5 min, they were incubated at 37°C for the indicated times (see Figure 7). Chlorophyll was cleared from the plant tissues by immersing them in 70% ethanol.
Drought Treatment and Measurement of Transpiration Rate
For drought treatment, 3-week-old soil-grown plants were withheld completely from water for the specified times. To minimize experimental variations, the same numbers of plants was grown on the same tray. With 35S-ABF4 plants, which show growth retardation, two batches of plants, one of the same age and the other of similar developmental stages (i.e., wild-type seed were sown 7 days later so that they were at similar developmental stages at the end of the treatment) were tested, and similar results were obtained. The entire test was repeated at least four times, sometimes using different arrangements of plants (i.e., test plants on different containers, etc.), and the results were consistent. The transpiration rate of detached leaves was measured by weighing freshly harvested leaves placed abaxial side up on open Petri dishes on the laboratory bench. Leaves of similar developmental stages (third to fifth true rosette leaves) from 3-week-old soil-grown plants were used.
Guard Cells
To examine guard cells, leaves were excised from 3-week-old soil-grown plants in the middle of the watering (3 days after watering) and light periods. Leaves of similar developmental stages (third to sixth true rosette leaves) from 20 different plants of wild-type and transgenic lines were placed on slides abaxial side up immediately after excision, and photographs were taken. The number of guard cells then was counted in the randomly chosen fields, usually six to seven.
Acknowledgments
We thank Drs. Pill-Soon Song, Jungmook Kim, and Moon Soo Soh for critical reading of the manuscript and Dr. Moon Soo Soh for his technical advice and helpful discussions throughout this work. This work was supported in part by a grant from the Agricultural Plant Stress Research Center funded by the Science Research Center Program of the Korea Science and Engineering Foundation. This paper is Kumho Life and Environmental Science Laboratory publication No. 51.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010362.
References
- Anderson, J.A., Huprikar, S.S., Kochian, L.V., Lucas, W.J., and Gaber, R.F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89, 3736–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Huertero, F., Arroyo, A., Zhou, L., Sheen, J., and Leon, P. (2000). Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev. 14, 2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. (New York: Green Publishing Associates/Wiley Interscience).
- Bartholomew, D.M., Bartley, G.E., and Scolnik, P.A. (1991). Abscisic acid control of rbcS and cab transcription in tomato leaves. Plant Physiol. 96, 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12, 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82, 259–266. [DOI] [PubMed] [Google Scholar]
- Busk, P.K., and Pages, M. (1998). Regulation of abscisic acid-induced transcription. Plant Mol. Biol. 37, 425–435. [DOI] [PubMed] [Google Scholar]
- Choi, H., Hong, J., Ha, J., Kang, J., and Kim, S.Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 21, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Mackey, K. (1995). Modification of the TRI reagent procedure for isolation of RNA from polysaccharide- and proteoglycan-rich sources. Biotechniques 19, 942–945. [PubMed] [Google Scholar]
- de Bruxelles, G.L., Peacock, W.J., Dennis, E.S., and Dolferus, R. (1996). Abscisic acid induces the alcohol dehydrogenase gene in Arabidopsis. Plant Physiol. 111, 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinbaum, R.L., and Ausubel, F.M. (1988). Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 8, 1985–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., and Lynch, T.J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R., Wang, M.L., Lynch, T.J., Rao, S., and Goodman, H.M. (1998). The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, R., Izawa, T., and Chua, N.-H. (1994). Plant bZIP proteins gather at ACGT elements. FASEB J. 8, 192–200. [DOI] [PubMed] [Google Scholar]
- Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., Michaux Ferriere, N., Thibaud, J.B., and Sentenac, H. (1998). Identification and disruption of a plant shaker-like outward channel involved in K+ release into the xylem sap. Cell 94, 647–655. [DOI] [PubMed] [Google Scholar]
- Gazzarrini, S., and McCourt, P. (2001). Genetic interactions be-tween ABA, ethylene and sugar signaling pathways. Curr. Opin. Plant Biol. 4, 387–391. [DOI] [PubMed] [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12, 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Hauge, B.M., Valon, C., Smalle, J., Parcy, F., and Goodman, H.M. (1992). Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4, 1251–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudat, J., Parcy, F., Bertauche, N., Gosti, F., and Leung, J. (1994). Current advances in abscisic acid action and signaling. Plant Mol. Biol. 26, 1557–1577. [DOI] [PubMed] [Google Scholar]
- Giuliano, G., Pichersky, E., Malik, V.S., Timko, M.P., Scolnik, P.A., and Cashmore, A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85, 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Beaudoin, N., Serizet, C., Webb, A.A., Vartanian, N., and Giraudat, J. (1999). ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11, 1897–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelbach, A., Iten, M., and Grill, E. (1998). Signalling of abscisic acid to regulate plant growth. Philos. Trans. R. Soc. Lond. B 353, 1439–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser, C., Kortstee, A., Pego, J., Weisbeek, P., Wisman, E., and Smeekens, S. (2000). The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: Involvement of abscisic acid in sugar responses. Plant J. 23, 577–585. [DOI] [PubMed] [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. [DOI] [PubMed] [Google Scholar]
- Jackson, M. (1997). Hormones from roots as signals for the roots of stressed plants. Trends Plant Sci. 2, 22–28. [Google Scholar]
- Jang, J.-C., Leon, P., Zhou, L., and Sheen, J. (1997). Hexokinase as a sugar sensor in higher plants. Plant Cell 9, 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 20, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Jorna, M.L., Brinkhorst-van-der Swan, D.L.C., and Karssen, C.M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid–insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61, 377–383. [Google Scholar]
- Laby, R.J., Kincaid, M.S., Kim, D., and Gibson, S.I. (2000). The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J. 23, 587–596. [DOI] [PubMed] [Google Scholar]
- Landschulz, W.H., Johnson, P.F., and McKnight, S.L. (1988). The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- Lang, V., and Palva, E.T. (1992). The expression of a rab-related gene, rab18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heynh. Plant Mol. Biol. 20, 951–962. [DOI] [PubMed] [Google Scholar]
- Leon-Kloosterziel, K.M., Gil, M.A., Ruijs, G.J., Jacobsen, S.E., Olszewski, N.E., Schwartz, S.H., Zeevaart, J.A., and Koornneef, M. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. [DOI] [PubMed] [Google Scholar]
- Leung, J., Bouvier-Durand, M., Morris, P.C., Guerrier, D., Chefdor, F., and Giraudat, J. (1994). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Science 264, 1448–1452. [DOI] [PubMed] [Google Scholar]
- Leung, J., Merlot, S., and Giraudat, J. (1997). Arabidopsis ABA response gene ABI1: Features of a calcium-modulated protein phosphatase. Plant Cell 9, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina, L., and Chua, N.-H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina, L., Mongrand, S., and Chua, N.-H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–512. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Leube, M.P., and Grill, E. (1994). A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science 264, 1452–1455. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Okada, K., and Shimura, Y. (1990). Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250, 274–276. [DOI] [PubMed] [Google Scholar]
- Okada, K., and Shimura, Y. (1992). Aspects of recent development in mutational studies of plant signaling pathways. Cell 70, 369–372. [DOI] [PubMed] [Google Scholar]
- Pei, Z.M., Ghassemian, M., Kwak, C.M., McCourt, P., and Schroeder, J.I. (1998). Role of farnesyltransferase in ABA regulation of guard cell anion channels and plant water loss. Science 282, 287–290. [DOI] [PubMed] [Google Scholar]
- Pilot, G., Lacombe, B., Gaymard, F., Cherel, I., Boucherez, J., Thibaud, J.B., and Sentenac, H. (2001). Guard cell inward K+ channel activity in Arabidopsis involves expression of the twin channel subunits KAT1 and KAT2. J. Biol. Chem. 276, 3215–3221. [DOI] [PubMed] [Google Scholar]
- Quesada, V., Ponce, M.R., and Micol, J.L. (2000). Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics 154, 421–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Schroeder, J.I., Kwak, J.M., and Allen, G.J. (2001). Guard cell abscisic acid signaling and engineering drought hardiness in plants. Nature 410, 327–330. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons, C., Soll, D., and Migliaccio, F. (1995). Circumnutation and gravitropism cause root waving in Arabidopsis thaliana. J. Exp. Bot. 46, 143–150. [Google Scholar]
- Uno, Y., Furihata, T., Abe, H., Yoshida, R., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid–dependent signal transduction pathway under drought and high-salinity. Proc. Natl. Acad. Sci. USA 97, 11632–11637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian, N., Marcotte, L., and Giraudat, J. (1994). Drought rhizogenesis in Arabidopsis thaliana. Plant Physiol. 104, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana, interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15, 501–510. [DOI] [PubMed] [Google Scholar]
- Wang, S.M.., Lue, W.L., Eimert, K., and Chen, J. (1996). Phytohormone-regulated β-amylase gene expression in rice. Plant Mol. Biol. 31, 975–982. [DOI] [PubMed] [Google Scholar]
- Weatherwax, S.C., Ong, M.S., Degenhardt, J., Bray, E.A., and Tobin, E.M. (1996). The interaction of light and abscisic acid in the regulation of plant gene expression. Plant Physiol. 111, 363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, J.E., and Finkelstein, R.R. (1995). Arabidopsis mutants with reduced response to NaCl and osmotic stress. Physiol. Plant. 93, 659–666. [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salinity stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart, J.A.D., and Creelman, R.A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473. [Google Scholar]
- Zhu, J.-K. (2000). Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 124, 941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]