Abstract
Post-transcriptional gene silencing (PTGS) is a sequence-specific degradation mechanism that operates in almost all eukaryotic cells. In plants, double-stranded RNA triggers PTGS, generating 21- to 25-nucleotide guide RNAs responsible for specific degradation of cognate mRNA. The double stranded RNA intermediates of replicating plant viruses often induce PTGS, leading to symptom attenuation. Here we demonstrate the role of PTGS in defective interfering (DI) RNA–mediated symptom attenuation in plants infected with Cymbidium ringspot tombusvirus (CymRSV). Analysis of 21- to 25-nucleotide RNAs in Nicotiana benthamiana infected with CymRSV indicated that PTGS was not spread homogeneously along the viral genome. The 21- to 25-nucleotide RNAs derived mainly from plus-stranded RNA and likely arose from local basepaired structures. In contrast to helper viral RNA, short DI RNAs were not accessible to helper virus–induced RNA degradation guided by the 21- to 25-nucleotide RNAs. Our results suggest a model in which PTGS plays an important role in the selective accumulation and symptom attenuation mediated by DI RNAs. Because PTGS operates in a wide variety of different organisms, this model is applicable to DI RNA generation and accumulation in both plant and animal cells.
INTRODUCTION
Post-transcriptional gene silencing (PTGS), first identified in plants, is now thought to be an ancient self-defense mechanism acting against molecular parasites (Waterhouse et al., 2001). Introduction of double-stranded RNA (dsRNA) into plant cells triggers PTGS, resulting in the degradation of dsRNA and cognate mRNAs (Schweizer et al., 2000). A similar mechanism appears to operate in a wide variety of organisms, including filamentous fungi, nematodes, Drosophila, mice, and cultured HeLa cells, and generally is referred to as RNA interference (RNAi) (Cogoni and Macino, 1999a; Fire, 1999; Grant, 1999; Sharp and Zamore, 2000; Elbashir et al., 2001a; Svoboda et al., 2000). Recently, homologous genes required for PTGS were identified from different organisms, demonstrating the conservation of the gene-silencing machinery (Cogoni and Macino, 1999a, 1999b; Ketting et al., 1999; Tabara et al., 1999; Catalanotto et al., 2000; Dalmay et al., 2000, 2001; Domeier et al., 2000; Fagard et al., 2000; Mourrain et al., 2000; Smardon et al., 2000; Wu-Scharf et al., 2000). The accumulation of 21- to 25-nucleotide RNAs corresponding to both sense and antisense strands of target RNA occurs during PTGS in plant and animal cells (Hamilton and Baulcombe, 1999; Hammond et al., 2000; Parrish et al., 2000). These 21- to 25-nucleotide RNAs are generated by an RNase III–like enzyme (DICER) as the initiation step of RNAi, providing the specificity of a second RNase complex (RISC) that targets the cognate single-stranded (ss) RNAs (Bernstein et al., 2001).
In plants, PTGS has evolved as an antiviral system. PTGS is triggered efficiently by dsRNA intermediates of cytoplasmically replicating viruses. The RNA genome of the invading virus is targeted and eliminated specifically when this natural antiviral mechanism is activated (Waterhouse et al., 1998, 1999; Baulcombe, 1999; Smith et al., 2000; Zamore et al., 2000). In higher plants, PTGS is not limited to the cells in which it is activated, because mobile signals produced by PTGS can spread and confer sequence-specific RNA degradation in distant tissues (Palauqui et al., 1997; Voinnet and Baulcombe, 1997).
Consistent with the importance of PTGS as an antiviral response, many viruses encode gene-silencing suppressor proteins (Anandalakshmi et al., 1998; Beclin et al., 1998; Brigneti et al., 1998; Kasschau and Carrington, 1998; Voinnet et al., 1999, 2000). However, not all viruses are able to suppress PTGS, and some virus-infected plants recover after the development of the first systemic viral symptoms (e.g., nepovirus-infected tobacco plants; Ratcliff et al., 1997). Upper leaves of recovered plants lack symptoms (or show attenuated symptoms), and the virus content in these leaves is reduced significantly. This recovery phenomenon was shown to be the consequence of virus-induced gene silencing (Ratcliff et al., 1997).
Interestingly, a recovery-like phenotype also was observed in plants infected by tombusviruses that were associated with defective interfering (DI) RNAs (Russo et al., 1994; Havelda et al., 1998). DI RNAs are shortened forms of viral genomes that generally have lost essential viral genes for movement, replication, and encapsidation. They require the presence of a helper virus to multiply and accumulate, usually at the expense of the helper virus from which they originated. It is generally accepted that the formation of viral DI RNAs involves a series of replicase-mediated deletions. Interference with the helper virus frequently results in remarkable symptom modification associated with a substantial decrease in helper virus levels. It was assumed that the reduction in helper virus levels by DI RNAs was attributable to competition for replication components (Roux et al., 1991; Russo et al., 1994; White, 1996).
DI RNAs of Cymbidium ringspot tombusvirus (CymRSV) are among the most extensively studied DI RNA molecules. They are incapable of autonomous replication but contain all of the necessary cis-acting elements for replication (Havelda et al., 1995). During replication of CymRSV genomic RNA, a series of DI RNAs of ∼0.4 to 0.7 kb are generated de novo (Burgyán et al., 1991). The longest DI RNA (DI-13) is composed of three blocks of sequences (A, B, and C) derived entirely from the CymRSV genome (Burgyán et al., 1989, 1991). Short DI RNAs representing the final stage of DI RNA evolution have the same sequence blocks but with progressive deletions in block C, whereas blocks A and B are almost unaffected (Burgyán et al., 1991; Havelda et al., 1997). A highly basepaired structure in block C of the large DI-13 RNA directs the formation of the shorter DI RNA molecules (Havelda et al., 1997). The presence of DI RNAs in CymRSV-infected plants is associated with attenuated symptoms and significantly reduced helper virus levels. These similarities between PTGS-mediated-recovery plants and DI RNA–mediated symptom-attenuated plants prompted the analysis of a possible role of PTGS in DI RNA accumulation, reduction in helper virus levels, and the associated symptom attenuation effects.
In this article, we demonstrate that the replication of CymRSV in Nicotiana benthamiana plants triggers PTGS, resulting in the accumulation of 21- to 25-nucleotide RNAs corresponding to the CymRSV sequence. The evidence presented shows that the short DI RNAs are poorly accessible to RNA degradation by the RNase complex (RISC like) guided by the 21- to 25-nucleotide RNAs. In contrast, the same RNase complex efficiently targets the large DI-13 RNA and different helper virus sequences. We consistently identified a 235-nucleotide RNA sequence with a highly basepaired structure that was targeted by CymRSV-induced PTGS regardless of its presence in CymRSV large DI RNA or in a heterologous Potato virus X (PVX) vector. Analysis of the population of 21- to 25-nucleotide RNAs in CymRSV-infected N. benthamiana indicates that PTGS is not spread homogeneously along the viral genome. The 21- to 25-nucleotide RNAs were derived mainly from plus-stranded RNA thought to fold into basepaired structures. Our results also suggest that PTGS plays an important role in the evolution, accumulation, and symptom attenuation effects of DI RNAs.
RESULTS
CymRSV-Induced PTGS
Phenotypical similarities between recovery plants generated by PTGS and CymRSV+DI RNA–coinfected plants suggested a role for PTGS in DI RNA–mediated symptom attenuation. We assumed that DI RNAs could escape from helper virus–induced PTGS and accumulate at the expense of helper virus. An in vivo experimental system was developed to determine whether CymRSV could induce PTGS and whether DI RNAs could be targeted by helper-triggered gene silencing.
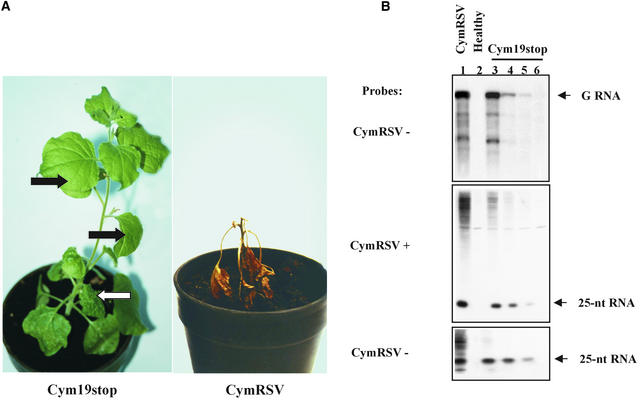
The system was based on a CymRSV mutant (Cym19stop) that is unable to express p19, a putative PTGS suppressor protein (Voinnet et al., 1999). Plants infected with the Cym19stop mutant showed a recovery-like phenotype (Figure 1A). The first virus symptoms developed at 5 to 7 days after inoculation (dai), similar to those in wild-type CymRSV-infected plants. In contrast to wild-type virus–infected plants that became necrotized rapidly (Figure 1A), new leaves of Cym19stop-infected plants that developed after the first symptomatic leaf were symptomless and contained very low or nondetectable levels of viral RNA (Figure 1B, lanes 3 to 6). To determine whether the recovery-like phenotype of Cym19stop-infected plants also was attributable to PTGS, the accumulation of 21- to 25-nucleotide RNAs characteristic of PTGS was tested. The 21- to 25-nucleotide RNAs accumulated in Cym19stop-infected leaves, and the amounts of these 21- to 25-nucleotide RNAs were related to the genomic RNA levels of mutant virus (Figure 1B, lanes 4 to 6). Moreover, 21- to 25-nucleotide RNAs accumulated to high levels in wild-type virus–infected cells (Figure 1B, lane 1). These results suggest that the replication of both Cym19stop and CymRSV triggered PTGS and that the gene-silencing machinery works similarly in wild-type virus–infected and Cym19stop-infected primary cells. However, the development of systemic PTGS differs significantly in wild-type and Cym19stop plants (see Discussion).
Figure 1.
Symptom Development and Viral RNA Accumulation in Virus-Infected N. benthamiana Plants.
(A) Symptoms of plants infected with Cym19stop and CymRSV. Photographs were taken 4 weeks after inoculation. White and black arrows show the first leaf with systemic symptoms and the recovered leaves used for challenge inoculation, respectively.
(B) RNA gel blot analysis of virus-specific RNAs extracted from the CymRSV-infected and Cym19stop-infected plants shown in (A). The first samples were taken at 7 dai from the first leaves with systemic symptoms (lanes 1 and 3). Lanes 4 to 6 contain total RNA extracted from leaves of Cym19stop-infected plants that developed after the first symptomatic leaf (black arrows in [A]). RNA gel blots were hybridized with 32P-labeled CymRSV genomic RNA. The strand polarities of the probes are indicated by + and −. The top gel shows high molecular weight RNAs separated on a 1.2% denaturing agarose gel. The middle and bottom gels show low molecular weight RNAs separated by denaturing PAGE. The positions of genomic (G) and 21- to 25-nucleotide (nt) virus-specific RNAs are indicated.
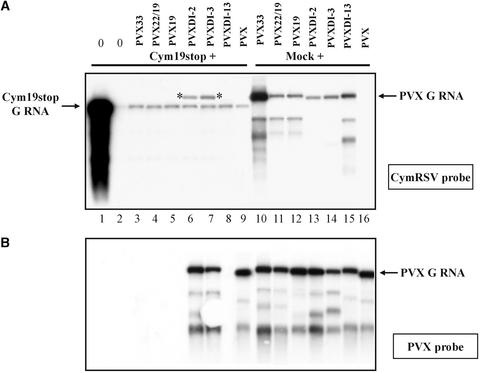
Resistance against a second infection of viruses containing sequences homologous with those of the first virus is a characteristic feature of PTGS-mediated-recovery plants (Ratcliff et al., 1997). To determine whether Cym19stop-infected recovery-like plants also are resistant to infection by a second related virus, symptomless upper leaves of Cym19stop-infected plants (Figure 1A) were challenge–inoculated with PVX and with PVX-Cym constructs carrying different coding sequences from the CymRSV genome (Figures 2A and 2B). The first systemic symptoms appeared at 7 to 8 dai on plants challenge inoculated with PVX. By 14 dai, newly developed leaves showed characteristic mosaic PVX symptoms (data not shown). RNA gel blot analysis of RNAs extracted from leaves with mosaic symptoms confirmed the accumulation of PVX RNAs (Figures 3A and 3B, lane 9). In contrast, plants challenge–inoculated with different PVX-Cym constructs (PVX33, PVX22, and PVX19) remained symptomless at least 4 weeks after inoculation (data not shown). PVX33, PVX22, and PVX19 accumulation was not detected in the challenge-inoculated leaves (data not shown) or newly developed leaves (Figures 3A and 3B, lanes 3 to 5). These results showing that Cym19stop-infected plants were resistant to further infection by PVX molecules carrying CymRSV sequences confirmed that the recovery phenotype of Cym19stop-infected plants was caused by the activation of PTGS.
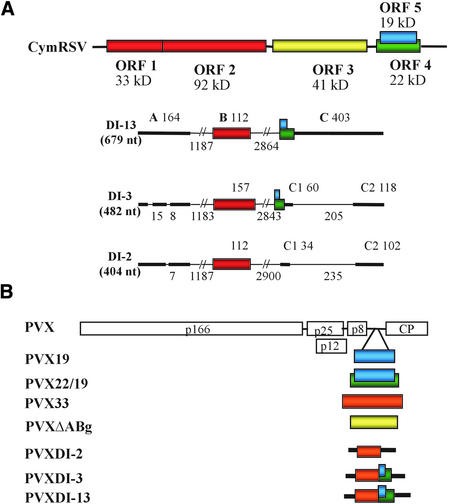
Figure 2.
Genome Organization of Wild-Type and Recombinant Viruses.
(A) Scheme of CymRSV and DI RNA genomes. CymRSV genomic RNA is shown at top, with ORFs and the molecular masses of encoded proteins indicated. Genomic RNA sequences conserved in DI RNAs are shown at bottom as colored blocks (derived from ORFs), thick lines (derived from noncoding sequences), or thin lines (deleted regions). Numbers above the shaded areas are the sizes (in nucleotides [nt]) of the RNA blocks (A, B, and C); the numbers of deleted bases are shown below the lines.
(B) Diagram of the PVX genome and recombinant derivatives expressing different ORFs and DI RNAs of CymRSV. The colored boxes indicate the corresponding CymRSV ORFs and DI RNA sequences; CP, coat protein.
Figure 3.
RNA Gel Blot Analysis of RNA Extracted from N. benthamiana Infected with Cym19stop and Challenge Inoculated with Different PVX Constructs.
Samples were taken from newly developed leaves at 14 days after the challenge inoculation. For hybridization, 32P-labeled RNA probes specific to CymRSV (A) or PVX (B) were used as indicated. Lanes 3 to 9 contain RNA extracted from plants preinoculated with Cym19stop, and lanes 10 to 16 contain RNA from mock (buffer)-preinoculated plants. Lanes 1 (the first leaf with systemic symptoms) and 2 (the first symptomless leaf) contain RNA from Cym19stop-infected nonchallenged plants. Asterisks indicate the replicating PVXDI-2 and -3 constructs. G RNA, genomic RNA.
Short DI RNAs Are Targeted Poorly by Helper Virus–Induced PTGS
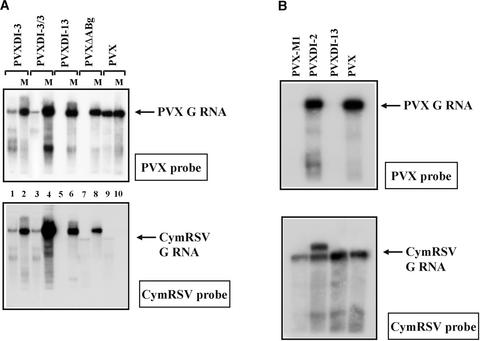
To determine whether DI RNAs could be targeted by helper virus–induced PTGS, three PVXcymDI constructs (PVXDI-2, PVXDI-3, and PVXDI-13) were generated (Figure 2B). The large DI-13 RNA represents an intermediate stage, whereas DI-2 and DI-3 represent the final stage of DI RNA evolution (Burgyán et al., 1991; Havelda et al., 1997). Transcripts synthesized from the three PVXcymDI constructs replicated efficiently in mock-inoculated N. benthamiana plants, confirming the viability of these recombinant viruses (Figures 3A and 3B, lanes 13 to 15). Simultaneously, the upper recovered leaves of Cym19stop-infected N. benthamiana plants were challenge inoculated with PVXDI-2, PVXDI-3, and PVXDI-13 transcripts. Like the PVX-inoculated control plants, the first systemic symptoms appeared by 7 to 8 dai on plants challenged inoculated with PVXDI-2 and PVXDI-3. RNA gel blot analysis also confirmed that PVX, PVXDI-2, and PVXDI-3 RNAs accumulated to detectable levels by 8 dai (data not shown). Two weeks after infection, the new leaves of the PVX–, PVXDI-2–, and PVXDI-3–challenged plants showed strong PVX symptoms (data not shown). Consistently, RNA gel blot analysis demonstrated efficient replication of PVXDI-2, PVXDI-3, and PVX constructs (Figures 3A and 3B, lanes 6, 7, and 9). Surprisingly, mosaic symptoms did not develop on plants challenge inoculated with PVXDI-13 transcript. Moreover, RNA gel blot analyses failed to detect PVXDI-13 RNA accumulation in inoculated (data not shown) or newly developed leaves of Cym19stop-infected plants (Figures 3A and 3B, lane 8). These results indicate that the analyzed DI-RNAs are targeted differently by helper virus–induced PTGS: the short DI-RNAs were not degraded, whereas the long DI-13 was targeted successfully (poor and good target activity, respectively).
Guide RNA–Mediated Degradation Is the Limiting Step in Short DI RNA Silencing
Short DI RNAs had poor target activity, whereas the long DI-13 RNA and different viral open reading frames (ORFs) could be targeted efficiently. The poor target activity of short DI RNAs could be the result of the limiting concentration of guide RNAs from regions within the small DI RNA in Cym19stop-inoculated plants. Alternatively, the accessibility of small DI RNAs for guide RNA–mediated degradation is the rate-limiting step in small DI RNA silencing. To determine whether target activity reflects the abundance of specific guide RNAs, the origin of the 21- to 25-nucleotide RNAs was determined. The viral genome was subjected to polymerase chain reaction (PCR) amplification in a series of ∼400-nucleotide-long fragments using appropriate primer pairs (Table 1, Figures 4A and 4C). Equal amounts (1 μg) of each DNA fragment were probed with ∼200 ng of 32P-labeled 21- to 25-nucleotide RNAs isolated from Cym19stop- or CymRSV-infected plants (Figure 4B). Guide RNA profiles (based on hybridization signals) of Cym19stop (data not shown) and CymRSV (Figure 4C) were similar. The results shown in Figure 4C indicate that the CymRSV genome was not represented homogeneously in the 21- to 25-nucleotide virus-specific RNA populations. No significant differences were observed among PCR fragments 3 to 14 in the labeled 21- to 25-nucleotide RNA population (Figure 4C). However, the 5′ terminal region (fragments 1 and 2) clearly was underrepresented. Because no obvious correlation could be found between genome representation in the guide RNAs and the poorer target activity of short DI RNAs compared with DI-13, the observed differences in target activity could not be determined by guide RNA levels.
Table 1.
Oligonucleotides Used for the Preparation of Short PCR Fragments Representing the CymRSV Genome
| No. of PCR Products |
Size of PCR Products (bp) |
Positions of Primers |
Orientation of Primers |
|---|---|---|---|
| 1 | 404 | 7 to 23 | S |
| 385 to 404 | A | ||
| 2 | 409 | 161 to 184 | S |
| 543 to 564 | A | ||
| 3 | 403 | 525 to 546 | S |
| 908 to 929 | A | ||
| 4 | 408 | 929 to 952 | S |
| 1318 to 1338 | A | ||
| 5 | 410 | 1051 to 1071 | S |
| 1441 to 1462 | A | ||
| 6 | 403 | 1463 to 1484 | S |
| 1846 to 1867 | A | ||
| 7 | 403 | 1811 to 1831 | S |
| 2195 to 2215 | A | ||
| 8 | 411 | 2215 to 2236 | S |
| 2595 to 2615 | A | ||
| 9 | 411 | 2629 to 2652 | S |
| 3012 to 3033 | A | ||
| 10 | 403 | 3175 to 3196 | S |
| 3555 to 3579 | A | ||
| 11 | 413 | 3367 to 3388 | S |
| 3749 to 3773 | A | ||
| 12 | 377 | 3844 to 3864 | S |
| 4196 to 4217 | A | ||
| 13 | 404 | 4230 to 4253 | S |
| 4614 to 4635 | A | ||
| 14 | 404 | 4331 to 4552 | S |
| 4715 to 4733 | A |
S and A indicate the sense and antisense orientation, respectively, of oligonucleotides.
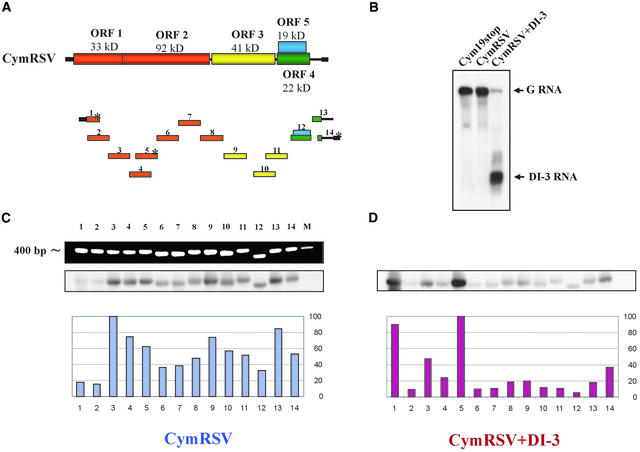
Figure 4.
Identification of the Origin of Small 21- to 25-Nucleotide Guide RNAs Derived from CymRSV–Infected or CymRSV+DI RNA–Infected N. benthamiana Plants.
(A) Scheme of the CymRSV genome and PCR-amplified ∼400-nucleotide DNA fragments 1 through 14. The colors of the PCR fragments indicate the corresponding ORFs. Asterisks indicate the PCR fragments that contained DI RNA sequences.
(B) RNA gel blot analysis of RNA extracts used for isolation of the 21- to 25-nucleotide guide RNAs. G RNA, genomic RNA.
(C) and (D) DNA gel blot analysis of the PCR fragment separated in an ethidium bromide–stained 1.2% agarose gel (top) and hybridized subsequently with 5′-labeled small 21- to 25-nucleotide guide RNAs derived from CymRSV-infected (C) or CymRSV+DI-3 RNA–infected (D) N. benthamiana isolated from the corresponding samples shown in (B) (bottom). Diagrams below the DNA gel blots represent the intensity of the obtained signals expressed as values of 0 to 100 where the highest signal was scored as 100. Numbers below the diagrams indicate the PCR-amplified DNA fragments. M indicates the 400-bp DNA size marker.
In cells coinfected with DI RNA and helper virus, guide RNAs generated from replicating DI RNAs also could influence DI RNA and helper virus accumulation. To determine whether guide RNAs could be produced from DI RNA molecules (as a result of the DICER-like enzyme activity), we defined the 21- to 25-nucleotide RNA profile for CymRSV+DI-3 RNA–infected plants (Figures 4B and 4D). The presence of high levels of DI RNA in coinfected cells (Figure 4B) caused a dramatic change in the composition of 21- to 25-nucleotide RNAs (cf. Figures 4C and 4D). In the coinfected cells, the majority of the guide RNAs (46%) were derived from two regions (fragments 1 [22%] and 5 [24%]) contained in the DI RNA (Figures 4A and 4D). In contrast, those fragments (2 and 4) that partially overlapped fragments 1 and 5 but did not contain DI RNA sequence hybridized weakly with the labeled guide RNAs (Figures 4A and 4D). Surprisingly, the presence of 3′ terminal sequence in the guide RNA population still was low.
These findings suggest that guide RNAs can be generated efficiently from DI RNAs present at high molar excess. Therefore, in natural DI RNA–infected and helper virus–infected plants, the 21- to 25-nucleotide RNAs corresponding to DI RNA molecules should be present in significantly higher concentrations than in plants infected only with DI RNA–free inoculum. To determine whether short DI RNAs can be targeted and degraded (by the RISC-like enzyme) when guide RNAs containing DI RNA sequences are abundant, we used Cym19stop+DI-3 transcripts for preinoculation in another experiment. After confirming that coinfection of N. benthamiana plants with Cym19stop+DI-3 also resulted in a recovery phenotype (data not shown), symptomless upper leaves of recovered plants were challenge inoculated with PVX and PVXDI-3. PVX infection was identical using Cym19stop- and Cym19stop+DI-3 RNA–preinoculated plants, suggesting that the addition of DI-3 RNA to Cym19stop did not influence PVX replication.
The first mosaic symptoms appeared at 8 dai, and virus replication also was detected by RNA gel blot analysis at this time (data not shown). Interestingly, PVXDI-3 symptom development differed only slightly on Cym19stop- and Cym19stop+DI-3 RNA–preinoculated plants. By 8 dai, mosaic symptoms appeared on Cym19stop-inoculated plants, whereas on Cym19stop+DI-3 RNA–preinoculated plants, the first symptoms developed 2 days later (data not shown). Consistently, PVXDI-3 RNA was first detected at 10 dai by RNA gel blot analysis in Cym19stop+DI-3 RNA–preinoculated plants (Figure 5A, lane 1). Moreover, PVXDI-3 RNA accumulation was reduced only when DI RNA was added to Cym19stop preinoculation transcripts (Figure 5A, lane 1). Therefore, we conclude that poor target activity of short DI RNA molecules is the consequence of inefficient guide RNA–mediated targeting of ssDI RNAs by the RISC-like degradation complex.
Figure 5.
RNA Gel Blot Analysis of RNA Extracted from Challenge-Inoculated N. benthamiana.
Plants were preinoculated with Cym19stop+DI-3 RNA (A) or Cym19stop only (B). The constructs used for challenge inoculation are indicated above the lanes. The challenge inoculum indicated above the lanes marked M indicates samples that were extracted from mock-treated plants. Samples were taken 10 days after the challenge inoculation. 32P-labeled probe specific to CymRSV or PVX was used for hybridization as indicated. G RNA, genomic RNA.
Specific Sequences/Structures Rather Than the Length of Target Molecules Are Responsible for Target Activity
It is likely that the observed differences in target activity between DI-13 and the shorter DI RNAs is determined by the different efficiencies of the guide RNA–mediated targeting step. If the length of the target molecule correlates with the efficiency of the targeting step, the large DI-13 RNA, which contains 235 nucleotides of extra sequence compared with DI-2 RNA (Figure 2A), should be degraded more efficiently. Alternatively, the extra 235-nucleotide RNA segment may contain specific sequences that are responsible for good target activity. To distinguish between these possibilities, symptomless upper leaves of Cym19stop+DI-3 RNA–preinoculated N. benthamiana plants were challenged with PVXDI-13, PVXΔABg (Figures 2A and 2B) containing a 400-nucleotide-long fragment from the coat protein ORF (Szittya and Burgyán, 2001), or PVXDI-3/3, which contains a head-to-tail dimer of DI-3 RNA (Dalmay et al., 1995).
All constructs replicated efficiently in mock-inoculated control plants (Figure 5A, lanes 4, 6, and 8). At 8 dai, none of these challenge-inoculated plants showed symptoms, in accordance with the failure of RNA gel blot analysis to detect virus accumulation (data not shown). However, by 10 dai, the first mosaic symptoms appeared on PVXDI-3/3 challenge-inoculated plants (data not shown). Consistently, virus RNA accumulation could be detected in RNA samples taken from these plants (Figure 5A, lane 3). In contrast, the PVXDI-13 and PVXΔABg constructs failed to accumulate (up to 4 weeks after inoculation) in plants preinoculated with Cym19stop+ DI-3 RNA (Figure 5A, lanes 5 and 7, respectively). These observations that the short PVXΔABg was well targeted but the PVXDI-3/3 containing a longer DI RNA insert than PVXDI-13 was not degraded demonstrate that target activity was not defined by the length of DI RNA.
These results also suggest that the extra 235-nucleotide sequence (motif 1 region, referred to as M1) (Havelda et al., 1997) present in DI-13 and absent in the short DI RNAs is responsible for the good target activity of DI-13 RNA. To confirm this possibility, the M1 region was inserted into the PVX vector (PVX-M1) and in vitro transcripts of PVX-M1 were used to inoculate symptomless leaves of N. benthamiana plants infected previously with the Cym19stop virus. PVX, PVXDI-2, and PVXDI-13 transcripts were used as controls for challenge inoculations. As demonstrated previously, the PVX and PVXDI-2 positive controls replicated efficiently, whereas the negative control PVXDI-13 failed to accumulate to detectable levels in Cym19stop-preinoculated plants (Figure 5B). PVX-M1 challenge-inoculated plants were similar to PVXDI-13 controls, remaining symptomless (data not shown). Consistently, RNA gel blot analysis could not detect PVX-M1 viral RNA accumulation in Cym19stop-preinoculated plants (Figure 5B), even though PVX-M1 replicated efficiently in mock-preinoculated plants. These experiments demonstrate that M1 was responsible for the good target activity of DI-13 RNA. Moreover, the target activity of M1 was independent of the DI-13 context.
A Highly Basepaired Structure in Single-Stranded Viral RNA Can Trigger PTGS
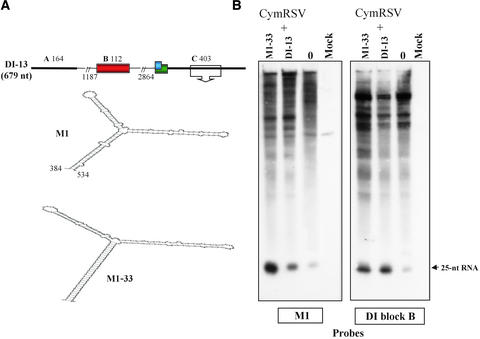
We have shown that a highly base-paired structure in the 3′ region of the longer DI RNAs directed the formation of the smaller class of molecules (Havelda et al., 1997). Mutations that increased the stability of the highly structured 3′ region (M1 stem) of the longest DI-13 RNA of CymRSV significantly enhanced the generation and accumulation of the smaller derivatives. Therefore, it was concluded that locally occurring secondary structures play an important role in DI RNA evolution (Havelda et al., 1997). We also reported that the extension of the M1 stem (18 bp) to 33 bp in the DI-13 mutant RNA (M1-33) (Figure 6) accelerated dramatically the transition of DI-13 RNA to shorter molecules (Havelda et al., 1997). To determine whether PTGS might play a role in the accelerated elimination of M1-33, we compared the composition of 21- to 25-nucleotide RNAs of plants infected with CymRSV+DI-13 and CymRSV+M1-33.
Figure 6.
Accumulation of 21- to 25-Nucleotide RNAs Derived from DI-13 RNA with Different Basepaired Structures in Block C.
(A) Scheme of the DI-13 RNA genome with the previously identified basepaired structures (M1) in the wild-type and M1-33 mutant molecules. The open box with the arrow indicates the position of the wild-type and mutant basepaired structures depicted below. nt, nucleotides.
(B) RNA gel blots of low molecular weight RNAs show the accumulation and the origin of virus-specific guide RNAs in CymRSV+0–, CymRSV+DI-13–, and CymRSV+M1-33–infected plants. Samples were taken at 7 to 8 dai from the first leaves with systemic symptoms, and 32P-labeled motif 1 (M1) and block B–specific probes with negative polarity were used for hybridization. The arrow shows the position of the 21- to 25-nucleotide guide RNAs.
Both DI-13 and M1-33 RNAs replicated efficiently in the presence of the helper virus (data not shown). Gel blots of RNAs extracted from CymRSV+DI-13 RNA–infected and CymRSV+M1-33 RNA–infected plants were hybridized with labeled block B of DI-13 RNA and a 5′-labeled 36-nucleotide-long oligonucleotide containing the complementary sequence of the DI-13 RNA M1 stem between positions 505 and 541 (Figure 6; Havelda et al., 1997). The accumulation of 21- to 25-nucleotide RNAs derived from block B was very similar in both infections (Figure 6). This result suggests that guide RNAs from the common regions of DI-13 and M1-33 RNAs are generated at similar efficiency. However, M1-specific 21- to 25-nucleotide RNAs accumulated seven times higher in CymRSV+M1-33 RNA–infected than in CymRSV+ DI-13 RNA–infected plants (Figure 6). This result indicates that the 33-bp stem structure is a more efficient inducer of guide RNA generation than is the wild-type 18-bp-long stem of M1.
Asymmetric Accumulation of Virus-Derived Plus- and Minus-Stranded 21- to 25-Nucleotide RNAs
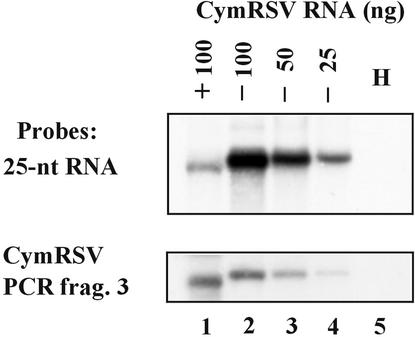
The experiment described above suggests that the 21- to 25-nucleotide RNAs may be generated from cleavage of the highly basepaired structures on single-stranded viral RNA molecules. If this is true, the majority of guide RNAs should be in the plus orientation because of the high molar excess of plus-stranded over minus-stranded viral RNAs. To test the polarity of the virus-derived 21- to 25-nucleotide RNAs, the CymRSV genomic RNA was transcribed in both positive and negative polarity and the transcripts were transferred to membranes and then probed with 32P-labeled PCR fragment 3 (Figure 4A) as a loading control and 5′-labeled 21- to 25-nucleotide RNAs extracted from CymRSV-infected plants to investigate the ratio of plus- and minus-stranded virus-derived molecules in the guide RNA population. The results, shown in Figure 7, indicate that ∼80% of the labeled guide RNAs hybridized with negative transcripts; thus, they were derived from viral RNA with positive polarity (Figure 7, cf. lanes 1 and 4). Therefore, it was concluded that the majority of small 21- to 25-nucleotide RNAs were derived from the plus strand of viral RNA.
Figure 7.
Identification of the Polarities of 21- to 25-Nucleotide RNAs Accumulating in CymRSV-Infected Plants.
Known amounts of plus and minus strands of CymRSV were transcribed in vitro, and transcripts were transferred to membranes and then hybridized with 5′-labeled 21- to 25-nucleotide guide RNAs derived from CymRSV (top) or 32P-labeled, PCR-amplified DNA fragment (frag.) 3 as a loading control (bottom). Numbers above the lanes indicate the quantity of loaded RNA in nanograms; + and − indicate the polarities of the loaded CymRSV RNA; H indicates control RNA extracted from a healthy plant.
DISCUSSION
Host defense responses that target differentially parasite organisms and molecules are decisive factors of their competition (Read and Taylor, 2001). In this study, we analyzed experimentally the effect of PTGS, a sequence-specific host defense system, on DI RNA–helper virus competition. Here we report that PTGS has a significant effect on the generation, accumulation, and symptom attenuation effects of DI RNAs.
Recovery Phenotype of Cym19stop-Infected Plants Is a Result of PTGS
An experimental system based on a CymRSV mutant (Cym19stop) unable to express the viral p19 protein was developed to analyze the consequence of virus-induced PTGS in tombusvirus-infected plants. Many lines of evidence support the finding that the recovery phenotype of Cym19stop-infected N. benthamiana plants is a consequence of virus-induced PTGS, similar to the recovery phenotype of nepovirus-inoculated plants (Ratcliff et al., 1997). The upper leaves of Cym19stop-infected plants were symptomless and contained very low levels of virus RNAs. Furthermore, the symptomless leaves were susceptible to PVX infection but resistant to PVX-Cym constructs that contained sequences homologous with Cym19stop that were silencing competent. Finally, 21- to 25-nucleotide virus-specific RNAs, the hallmarks of virus-induced PTGS (Hamilton and Baulcombe, 1999), accumulated to high levels in the first systemic leaves of Cym19stop plants.
The cellular mechanism of PTGS is considered to be highly conserved in eukaryotes. At least two RNases are involved in PTGS in Drosophila cultured cells. The first complex (DICER) cleaves the dsRNAs into 21- to 25-nucleotide molecules by an RNase III–like enzyme, after which the 21- to 25-nucleotide RNAs guide RISC complexes to degrade the homologous mRNAs (Bernstein et al., 2001). The specific advantage of the Cym19stop-PVX challenge inoculation system is that these two steps, the generation of guide RNAs and the guide RNA–mediated targeting of ssRNAs, can be studied separately. In this system, guide RNAs always are generated identically by Cym19stop; therefore, PTGS efficiency against the challenging PVX-Cym constructs depends only on the target activity of the CymRSV segment carried by the PVX vector. The induction and guide RNA generation step of CymRSV-triggered PTGS can be investigated separately by examining the 21- to 25-nucleotide RNA profiles of virus-infected plants.
Conclusions drawn about PTGS from the Cym19stop system also are valid for wild-type virus–infected cells, because profiles of the 21- to 25-nucleotide RNAs extracted from CymRSV-infected and Cym19stop-infected plants are very similar. However, wild-type virus infection leads to lethal necrosis, whereas Cym19stop inoculation results in a recovery phenotype. We interpreted these observations to indicate that PTGS is active but incomplete in the presence of p19. This conclusion is consistent with the previous finding that the p19 protein of Tomato bushy stunt virus is a weak PTGS suppressor (Voinnet et al., 1999). In addition, whereas the p19 protein of CymRSV inhibited the spread of systemic silencing, it did not interfere with virus-induced gene silencing in protoplasts (D. Silhavy, unpublished results). These results suggest that the initial cellular processes of PTGS are identical in primary infected cells of CymRSV-infected and Cym19stop-infected plants, although the spread of PTGS and consequently the phenotypes of infected plants are different.
PTGS Selects for DI RNAs Containing Poor PTGS Target Regions
Comparing the target activity of different DI RNAs and viral genomic regions that are not included in DI RNA molecules, we found that the target activity of different regions of the viral genome can vary significantly. Our finding that short DI RNAs were targeted poorly by the helper virus–induced PTGS suggested that short DI RNAs accumulate in virus-infected plants because they are poor targets of PTGS. We also demonstrated that the poor target activity of short DI RNAs was not caused by either the low level of the DI RNA–specific guide RNAs or the size of the target molecule. Short DI RNAs, which contain almost exclusively cis-acting sequences required for replication (Havelda et al., 1995), may bind efficiently to the replicase complex and thus are not accessible to the RISC degradation complex of PTGS. Alternatively, the folding of DI RNAs may prevent PTGS-mediated degradation. The target activity of endogenous genes also might vary. The capacity of distinct regions from the phytoene desaturase (PDS) gene expressed by PVX to confer virus-induced gene silencing differed significantly (Thomas et al., 2001). Furthermore, certain genes were resistant to RNAi inactivation in Caenorhabditis elegans (Fraser et al., 2000) and in HeLa cells (Elbashir et al., 2001a), even though guide RNAs were abundant in both systems. It is likely that the low target activity of these endogenous genes contributed to their resistance against RNAi-mediated inactivation.
In contrast to the short DI RNAs, the large DI-13 RNA was a good target for PTGS induced by the helper virus. We demonstrated that the 235-nucleotide M1, the only region present in DI-13 and absent in short DI RNAs, was responsible for the good target activity of DI-13 RNA. Previously, we reported that M1 was deleted preferentially during the evolution of CymRSV DI RNAs (Havelda et al., 1997). These coinciding results suggest that PTGS provides selective pressure on DI RNA accumulation by eliminating DI RNA molecules containing PTGS target sequences. We also tested the target activity of different coding regions of CymRSV and found that all of these sequences, like the M1 region, were good PTGS targets. These observations suggest that good target activity is the general feature of viral RNAs. Consequently, short DI RNAs are exceptional molecules, being weak targets of PTGS. Therefore, we suggest that, in addition to a size-related replication advantage (Roux et al., 1991; White, 1996), PTGS-mediated selection also plays a decisive role in the selective accumulation of short DI RNAs.
PTGS Eliminates DI RNA Molecules with Strong Secondary Structures
Previously, we reported that extension of the 18-nucleotide stem structure of M1 to 33 nucleotides accelerated the evolution of large DI RNAs to short ones (Havelda et al., 1997). We have found that the extended basepaired structure of M1-33 resulted in a specific increase of 21- to 25-nucleotide RNAs, suggesting that highly basepaired structures within a ssRNA also can induce PTGS. The role of such basepaired structures in PTGS induction in other systems is not clear. In Drosophila embryo lysates, single-stranded regions flanking short dsRNA sequences suppressed the generation of guide RNAs, suggesting that highly basepaired structures in an otherwise ssRNA did not induce PTGS (Elbashir et al., 2001b). In contrast, a recent model of higher plant PTGS hypothesized the involvement of dsRNA segments in PTGS activation and in systemic signaling (Carrington, 2000). Our finding that M1-33 triggers PTGS experimentally supports the notion that a short double-stranded region within a ssRNA can induce PTGS. Because ssRNA viral genomes generally are highly structured, dsRNA regions in ssRNAs could be important inducers of virus-triggered PTGS. The nonhomogeneous representation of the viral genome in guide RNA populations also could reflect the formation of double-stranded structures along the viral RNA. On the basis of the results reported here, we propose that PTGS also plays an important role in eliminating highly basepaired segments from large DI RNAs because these structures trigger effective PTGS.
Asymmetric Accumulation of 21- to 25-Nucleotide Guide RNAs
In CymRSV-infected plants, guide RNAs accumulated asym-metrically, with the majority of 21- to 25-nucleotide RNAs being derived from plus-stranded viral RNA (Figure 7). This observation is in agreement with highly structured regions within ssRNAs being involved in PTGS induction. Alternatively, the differential stability of plus- and minus-stranded guide molecules might explain the dominance of plus-oriented 21- to 25-nucleotide RNAs. However, no experimental data are available to support this alternative explanation. Asymmetric guide RNA accumulation also might occur in other virus-induced PTGS systems. Antisense-oriented short PDS sequences were more effective than sense inserts in PVX-PDS–induced gene-silencing assays (Thomas et al., 2001). It is conceivable that the majority of 21- to 25-nucleotide RNAs derived from PVX-PDS also were of positive orientation; therefore, higher levels of guide RNAs complementary to PDS mRNA were generated when antisense inserts were expressed. Asymmetric accumulation of 21-nucleotide small temporal RNAs controlling the developmental timing in C. elegans and Drosophila also was observed, and the small temporal RNAs were produced by a PTGS-like mechanism (Grishok et al., 2001; Hutvágner et al., 2001).
Model of PTGS-Mediated DI RNA Evolution and Symptom Attenuation
We suggest a new model, which integrates PTGS in the evolution and symptom attenuation effects of tombusvirus DI RNAs. Defective RNAs are generated from the viral genome randomly by replicase error. These molecules are selected for by the presence of cis elements required for replication and by the absence of sequences that are targeted by PTGS. Therefore, short DI RNAs that are the final products of DI RNA evolution can accumulate to high levels because they are poor targets of PTGS. Abundant DI RNAs could reduce helper virus titer in two ways, by using the majority of limiting host and viral factors (e.g., viral replicase) and by inducing PTGS targeting the helper virus genome. Therefore, we suggest that, in addition to the competition of DI RNAs and helper virus for limiting host and viral factors, DI RNA–induced PTGS also contributes significantly to DI-RNA–mediated symptom attenuation by reducing the level of helper RNAs. Because PTGS operates in wide variety of cells, including cultured animal cells, our model may apply to DI RNA generation and accumulation in both plant and animal systems.
METHODS
Plasmid Constructs
The infectious cDNA clone of Cymbidium ringspot tombusvirus (CymRSV) was described previously (Dalmay et al., 1993). Site-directed mutagenesis (Kunkel et al., 1985) was used to construct the Cym19stop mutant. The full-length cDNA clone of CymRSV was mutagenized by introducing two stop codons in the third and sixth codon positions of open reading frame 5 (ORF 5), which encodes the p19 protein. These modifications did not alter the amino acid sequence of the p22 movement protein encoded by ORF 4. The accuracy of mutagenesis was controlled by sequencing of the region of interest.
The Potato virus X (PVX) vector (pP2C2S) used to express CymRSV and defective interfering (DI) RNA sequences has been described (Chapman et al., 1992; Baulcombe et al., 1995). The CymRSV cDNA fragments encoding p33, p22/19, and p19 were amplified by polymerase chain reaction (PCR) from the CymRSV plasmid (Dalmay et al., 1993) using oligonucleotides homologous with the first 22 nucleotides and complementary to the last 21 nucleotides of ORF 1, ORF 4, and ORF 5, respectively. The PCR-amplified DNA fragments were cloned individually into the EcoRV-linearized pP2C2S. Similarly, PVXcymDI constructs were prepared as follows. CymRSV DI-13 (679 nucleotides), DI-3 (482 nucleotides), and DI-2 (404 nucleotides) sequences were amplified by PCR with oligonucleotides homologous with the first 22 nucleotides and complementary to the last 21 nucleotides of CymRSV using the appropriate cDNA clones of CymRSV DI RNAs (Burgyán et al., 1991). The PCR-amplified fragments were cloned into EcoRV-linearized pP2C2S. The recombinant constructs obtained were designated PVXDI-2, -3, and -13, respectively.
In Vitro RNA Transcription and Plant Inoculation
In vitro transcription of CymRSV and Cym19stop RNAs from linearized template plasmids and inoculation of RNA transcripts onto Nicotiana benthamiana plants were performed as described previously (Dalmay et al., 1993). PVX-derived plasmids were linearized with SpeI, and in vitro RNA transcripts were capped using a cap analog (New England Biolabs, Beverly, MA) (Chapman et al., 1992).
RNA Extraction and Analysis
Total RNA was extracted from 100 mg of leaf tissue (White and Kaper, 1989). Briefly, the homogenized plant materials were resuspended in 600 μL of extraction buffer (0.1 M glycine-NaOH, pH 9.0, 100 mM NaCl, 10 mM EDTA, 2% SDS, and 1% sodium lauroylsarcosine) and mixed with an equal volume of phenol. The aqueous phase was treated with equal volumes of phenol and chloroform, precipitated with ethanol, and resuspended in sterile water. RNA gel blot analysis of higher molecular weight RNAs was performed as described previously (Dalmay et al., 1993).
Detection, Isolation, and Labeling of 21- to 25-Nucleotide RNAs
RNA gel blot analysis of 21- to 25-nucleotide RNAs was performed as follows. Approximately 1 to 5 μg of total RNA was separated by 8% PAGE with 8.3 M urea and 1 × Tris-borate-EDTA. The gel was soaked in 20 × SSC solution (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) for 15 min, and then RNA was blotted onto Hybond-N membranes and fixed by oven baking at 80°C for 2 hr. Strand-specific RNA probes were generated by in vitro RNA transcription from the indicated cDNA clones (see Figures) in the presence of 32P-α-UTP. Hybridization was performed in 50% formamide, 5 × SSPE (1 × SSPE is 0.115 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA, pH 7.4), 5 × Denhardt's solution (1 × Denhardt's solution is 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% BSA), and 0.5% SDS with competitor DNA. After overnight incubation at 40°C, the filter was washed twice in 2 × SSC and 0.1% SDS for 10 min at 40°C.
For preparation of 21- to 25-nucleotide RNA, 15 to 20 μg of total RNA was subjected to electrophoresis through an 8% denaturing polyacrylamide gel followed by staining in 1 × Tris-borate-EDTA and 0.5 μg/mL ethidium bromide solution for 20 min. The 21- to 25-nucleotide RNAs were visualized by UV light and excised from the gel. The gel slice was crushed, covered with 2 volumes of elution buffer (80% formamide, 40 mM Pipes, pH 6.4, 1 mM EDTA, and 400 mM NaCl), and incubated overnight. The gel residues were pelleted by centrifugation, and the supernatant was precipitated with ethanol. The 21- to 25-nucleotide RNAs (∼1 μg) were dephosphorylated and labeled subsequently in a 10-μL reaction in the presence of γ-32P-ATP and RNasin with 8 units of T4 polynucleotide kinase. The labeled CymRSV- specific 21- to 25-nucleotide RNAs were used for hybridization as described above.
Acknowledgments
We are grateful to Anne Simon for her comments and invaluable help during the preparation of the manuscript. We thank David Baulcombe for generously supplying the PVX vector. This research was supported by grants from the Hungarian Orszagos Tudomanyos Kutatasi Alapok (No. 31929) and the Ministry of Education (No. FKFP0442/1999).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010366.
References
- Anandalakshmi, R., Pruss, G.J., Ge, X., Marathe, R., Mallory, A.C., Smith, T.H., and Vance, V.B. (1998). A viral suppressor of gene silencing in plants. Proc. Natl. Acad. Sci. USA 95, 13079–13084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C., Chapman, S., and Santa Cruz, S. (1995). Jellyfish green fluorescent protein as a reporter for virus infections. Plant J. 7, 1045–1053. [DOI] [PubMed] [Google Scholar]
- Beclin, C., Berthome, R., Palauqui, J.C., Tepfer, M., and Vaucheret, H. (1998). Infection of tobacco or Arabidopsis plants by CMV counteracts systemic post-transcriptional silencing of nonviral (trans)genes. Virology 252, 313–317. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Brigneti, G., Voinnet, O., Li, W.X., Ji, L.H., Ding, S.W., and Baulcombe, D.C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17, 6739–6746. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Burgyán, J., Grieco, F., and Russo, M. (1989). A defective interfering RNA in cymbidium ringspot virus infections. J. Gen. Virol. 70, 235–239. [Google Scholar]
- Burgyán, J., Rubino, L., and Russo, M. (1991). De novo generation of cymbidium ringspot virus defective interfering RNA. J. Gen. Virol. 72, 505–509. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. (2000). RNA silencing: Moving targets. Nature 408, 150–151. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., Azzalin, G., Macino, G., and Cogoni, C. (2000). Gene silencing in worms and fungi. Nature 404, 245. [DOI] [PubMed] [Google Scholar]
- Chapman, S., Kavanagh, T., and Baulcombe, D.C. (1992). Potato virus X as a vector for gene expression in plants. Plant J. 2, 549–557. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. a). Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399, 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and Macino, G. (1999. b). Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286, 2342–2344. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Rubino, L., Burgyán, J., Kollár, Á., and Russo, M. (1993). Functional analysis of cymbidium ringspot virus genome. Virology 194, 697–704. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Szittya, G., and Burgyán, J. (1995). Generation of defective interfering RNA dimers of cymbidium ringspot tombusvirus. Virology 207, 510–517. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101, 543–553. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Horsefield, R., Braunstein, T.H., and Baulcombe, D.C. (2001). SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 20, 2069–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domeier, M.E., Morse, D.P., Knight, S.W., Portereiko, M., Bass, B.L., and Mango, S.E. (2000). A link between RNA interference and nonsense-mediated decay in Caenorhabditis elegans. Science 289, 1928–1931. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001. a). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001. b). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15, 358–363. [DOI] [PubMed] [Google Scholar]
- Fraser, A.G., Kamath, R.S., Zipperlen, P., Martinez-Campos, M., Sohrmann, M., and Ahringer, J. (2000). Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408, 325–330. [DOI] [PubMed] [Google Scholar]
- Grant, S.R. (1999). Dissecting the mechanisms of posttranscriptional gene silencing: Divide and conquer. Cell 96, 303–306. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A novel species of small antisense RNA in post-transcriptional gene silencing. Science 286, 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S.M., Bernstein, E., Beach, D., and Hannon, G.J. (2000). An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404, 293–296. [DOI] [PubMed] [Google Scholar]
- Havelda, Z., Dalmay, T., and Burgyán, J. (1995). Localization of cis-acting sequences essential for cymbidium ringspot tombusvirus defective interfering RNA replication. J. Gen. Virol. 76, 2311–2316. [DOI] [PubMed] [Google Scholar]
- Havelda, Z., Dalmay, T., and Burgyán, J. (1997). Secondary structure-dependent evolution of Cymbidium ringspot virus defective interfering RNA. J. Gen. Virol. 78, 1227–1234. [DOI] [PubMed] [Google Scholar]
- Havelda, Z., Szittya, G., and Burgyán, J. (1998). Characterization of the molecular mechanism of defective interfering RNA mediated symptom attenuation in tombusvirus-infected plants. J. Virol. 72, 6251–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Pasquinelli, A.E., Bálint, E., Tuschl, T., and Zamore, P.D. (2001). A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293, 834–838. [DOI] [PubMed] [Google Scholar]
- Kasschau, K.D., and Carrington, J.C. (1998). A counterdefensive strategy of plant viruses: Suppression of posttranscriptional gene silencing. Cell 95, 461–470. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Haverkamp, T.H., van Luenen, H.G., and Plasterk, R.H. (1999). Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNase D. Cell 99, 133–141. [DOI] [PubMed] [Google Scholar]
- Kunkel, T.A., Roberts, J.D., and Zakour, R.A. (1985). Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82, 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101, 533–542. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: Transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., and Fire, A. (2000). Functional anatomy of a dsRNA trigger: Differential requirement for the two trigger strands in RNA interference. Mol. Cell 6, 1077–1087. [DOI] [PubMed] [Google Scholar]
- Ratcliff, F., Harrison, B.D., and Baulcombe, D.C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560. [DOI] [PubMed] [Google Scholar]
- Read, A.F., and Taylor, L.H. (2001). The ecology of genetically diverse infections. Science 292, 1099–1102. [DOI] [PubMed] [Google Scholar]
- Roux, L., Simon, A.E., and Holland, J.J. (1991). Effects of defective interfering viruses on virus replication and pathogenesis in vitro and in vivo. Adv. Virus Res. 40, 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, M., Burgyán, J., and Martelli, P.G. (1994). The molecular biology of Tombusviridae. Adv. Virus Res. 44, 382–424. [DOI] [PubMed] [Google Scholar]
- Schweizer, P., Pokorny, J., Schulze-Lefert, P., and Dudler, R. (2000). Double-stranded RNA interferes with gene function at the single-cell level in cereals. Plant J. 24, 895–903. [DOI] [PubMed] [Google Scholar]
- Sharp, P.A., and Zamore, P.D. (2000). RNA interference. Science 287, 2431–2433. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. (2000). EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10, 169–178. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. [DOI] [PubMed] [Google Scholar]
- Svoboda, P., Stein, P., Hayashi, H., and Schultz, R.M. (2000). Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127, 4147–4156. [DOI] [PubMed] [Google Scholar]
- Szittya, G., and Burgyán, J. (2001). Cymbidium ringspot tombusvirus coat protein coding sequence acts as an avirulent RNA. J. Virol. 75, 2411–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- Thomas, C.L., Jones, L., Baulcombe, D.C., and Maule, A.J. (2001). Size constraints for targeting post-transcriptional gene silencing and for RNA-directed methylation in Nicotiana bentha-miana using a potato virus X vector. Plant J. 25, 417–425. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., and Baulcombe, D.C. (1997). Systemic signalling in gene silencing. Nature 389, 553. [DOI] [PubMed] [Google Scholar]
- Voinnet, O., Pinto, Y., and Baulcombe, D.C. (1999). Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses. Proc. Natl. Acad. Sci. USA 96, 14147–14152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Lederer, C., and Baulcombe, D.C. (2000). A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103, 157–167. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95, 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Smith, N.A., and Wang, M.B. (1999). Virus resistance and gene silencing: Killing the messenger. Trends Plant Sci. 4, 452–457. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- White, J.L., and Kaper, J.M. (1989). A simple method for detection of viral satellite RNAs in small tissue samples. J. Virol. Methods 23, 83–94. [DOI] [PubMed] [Google Scholar]
- White, K.A. (1996). Formation and evolution of Tombusvirus defective interfering RNAs. Semin. Virol. 7, 409–416. [Google Scholar]
- Wu-Scharf, D., Jeong, B., Zhang, C., and Cerutti, H. (2000). Transgene and transposon silencing in Chlamydomonas reinhardtii by a DEAH-box RNA helicase. Science 290, 1159–1162. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. [DOI] [PubMed] [Google Scholar]