Abstract
The blue light receptors termed cryptochromes mediate photomorphological responses in seed plants. However, the mechanisms by which cryptochrome signals regulate plant development remain obscure. In this study, cryptochrome functions were analyzed using the moss Physcomitrella patens. This moss has recently become known as the only plant species in which gene replacement occurs at a high frequency by homologous recombination. Two cryptochrome genes were identified in Physcomitrella, and single and double disruptants of these genes were generated. Using these disruptants, it was revealed that cryptochrome signals regulate many steps in moss development, including induction of side branching on protonema and gametophore induction and development. In addition, the disruption of cryptochromes altered auxin responses, including the expression of auxin-inducible genes. Cryptochrome disruptants were more sensitive to external auxin than wild type in a blue light–specific manner, suggesting that cryptochrome light signals repress auxin signals to control plant development.
INTRODUCTION
Because of their sessile nature, various mechanisms have evolved in plants that enable them to adapt their growth and morphology to environmental changes. Light is one of the most important environmental stimuli in this regard. Several sets of photoreceptors have developed to enable plants to respond to light. For instance, Arabidopsis has at least five red/far-red light receptor phytochromes, two blue light receptor cryptochromes, and two of another class of blue light receptors termed phototropins (Briggs and Olney, 2001; Briggs et al., 2001). Phytochrome is the best characterized photoreceptor in plants because of its ability to switch between an active form and an inactive form by absorbing red light or far-red light, respectively. Phytochrome light signals control developmental processes from germination to flowering (Neff et al., 2000; Smith, 2000). A cryptochrome was the first identified blue light receptor in plants (Ahmad and Cashmore, 1993), and blue light signals via cryptochromes control various responses that accompany developmental changes (Briggs and Huala, 1999; Cashmore et al., 1999; Lin, 2000). In contrast, phototropins control responses that change the direction of growth or organelle movements, such as phototropism (Briggs and Huala, 1999) and the chloroplast avoidance response (Jarillo et al., 2001; Kagawa et al., 2001).
Cryptochromes show a high degree of homology in their N-terminal regions with the photoreactivating enzymes termed photolyases. They also possess C-terminal amino acid extensions of various lengths that photolyases lack. The recombinant Arabidopsis CRY1 protein contains two cofactors, flavin adenine dinucleotide and methenyltetrahydrofolate, the same as in photolyases (Lin et al., 1995; Malhotra et al., 1995). In Arabidopsis, two cryptochromes, cry1 and cry2, are involved in many photomorphological processes, such as the inhibition of hypocotyl elongation (Ahmad and Cashmore, 1993; Lin et al., 1998), the induction of anthocyanin synthesis (Ahmad et al., 1995), and the control of flowering time (Guo et al., 1998; Mockler et al., 1999). Both cryptochromes also work as photoreceptors that input light signals to the circadian clock (Somers et al., 1998; Devlin and Kay, 2000). It was shown recently that the C-terminal domains are necessary to mediate photomorphogenic responses (Yang et al., 2000) and that the C-terminal domains interact directly with COP1, which is a negative regulator of photomorphogenesis and may repress COP1 activity (Wang et al., 2001).
Cryptochromes also have been found in other organisms, such as insects and mammals (Cashmore et al., 1999; Hall, 2000). Drosophila cryptochrome may function as a photoreceptor and as part of the circadian clock (Stanewsky et al., 1998; Ceriani et al., 1999). Two mouse cryptochromes are central clock components (Kume et al., 1999; van der Horst et al., 1999) and also may be photoreceptors (Selby et al., 2000). Although all cryptochromes are presumed to have evolved from photolyases, according to sequence comparisons, it is likely that these animal cryptochromes evolved differently from plant cryptochromes (Cashmore et al., 1999). To date, cryptochrome sequences have been reported in several plants (Cashmore et al., 1999). However, their functions have been characterized only in Arabidopsis and tomato (Ninu et al., 1999; Weller et al., 2001). It is of great interest to determine how and when plant cryptochromes obtain the functions that are seen in dicots. To answer this question, the functions of cryptochromes in many plant species from various taxa must be elucidated.
The moss Physcomitrella patens was chosen for this study because of its gene-targeting capability (Schaefer and Zryd, 1997; Schaefer, 2001) and its simple organization (Cove and Knight, 1993; Reski, 1998). Because cryptochromes regulate light-dependent developmental changes in Arabidopsis, the simple organization of the moss is a great advantage for the investigation of developmental changes. At least two similar cryptochrome genes exist in the genome of Physcomitrella. This study, using cryptochrome disruptants, revealed that both cryptochromes have redundant functions and that light signals via cryptochromes regulate some developmental steps throughout the moss life cycle. Moreover, we found that the cryptochrome signals suppress auxin signals, leading to the regulation of developmental steps.
RESULTS
Two Moss Cryptochrome Paralogs, Physcomitrella PpCRY1a and PpCRY1b, Are Nucleus-Localized Proteins
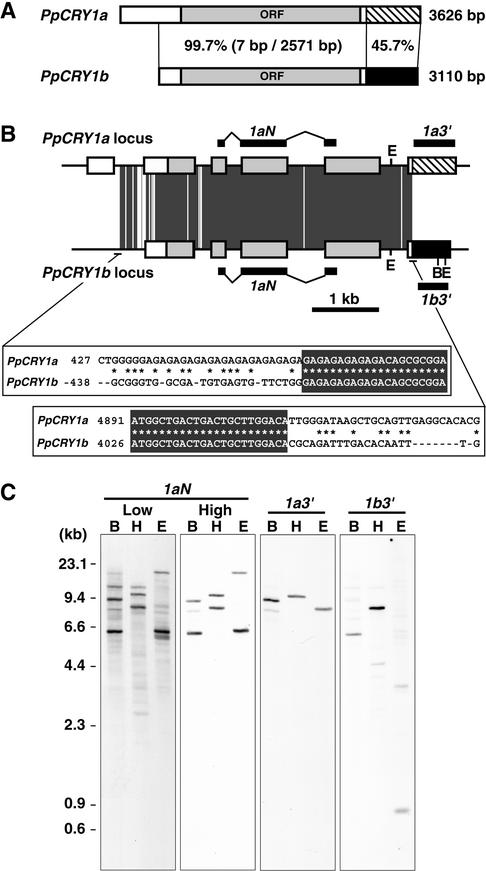
To analyze the function of the blue light receptor, cryptochrome, in the moss Physcomitrella, we determined two CRY cDNA sequences. They have almost identical nucleotide sequences except for the 3′ regions of the predicted 3′ untranslated region; therefore, they were designated PpCRY1a and PpCRY1b (Figure 1A). The genomic DNA sequences corresponding to each cDNA revealed that these cDNAs are derived from two different genes. Both genomic DNA sequences shared almost identical nucleotide sequences from the middle of the first intron to the beginning of the sixth exon of the PpCRY1a gene (Figure 1B). Except for that region, they showed little identity with each other. The existence of two CRY genes was confirmed by DNA gel blot analysis (Figure 1C). When the probe specific to both cryptochrome genes was used, two different bands appeared under high-stringency conditions. Each specific 3′ untranslated region probe hybridized to one of each band. Except for the bands corresponding to the two CRY genes, one or two bands appeared in each lane under low-stringency conditions.
Figure 1.
Gene Structures of Moss Cryptochromes.
(A) Scheme of the PpCRY1a and PpCRY1b cDNAs. The striped and black regions show the nucleotide sequences that are different between the PpCRY1a and PpCRY1b cDNAs. The percentages of identical nucleotides in the regions are shown. In the PpCRY1b transcript, the 5′ untranslated region of the PpCRY1b cDNA might be truncated, based on its size (see Figure 3C). ORF, open reading frame.
(B) Scheme of the PpCRY1a and PpCRY1b genes. The boxes indicate exons, and each box pattern is the same as in (A). The positions of the probes used in (C) are indicated. The dark gray regions indicate sequences that are identical between PpCRY1a and PpCRY1b. The nucleotide sequence alignments at the beginning and the end of the identical regions are shown. The recognition sites of BglII (B) and EcoRI (E) are indicated.
(C) DNA gel blot analysis of the PpCRY1a and PpCRY1b genes. Genomic DNA was digested by BglII (B), HindIII (H), or EcoRI (E). Blots were hybridized with the probe (1aN) specific to both cryptochrome genes under either low- or high-stringency conditions. The data for each specific probe (1a3′ and 1b3′) were obtained under high-stringency conditions.
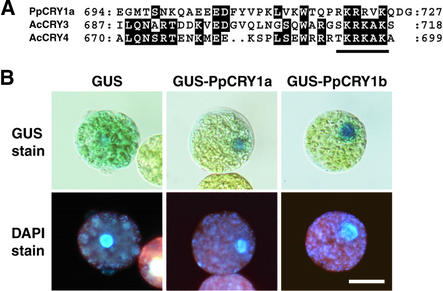
These cDNAs have just one nucleotide base difference in their predicted open reading frames. Both cDNAs encode proteins with 727 deduced amino acids. One single base difference led to PpCRY1a having threonine and PpCRY1b having methionine at amino acid 80. The amino acid at position 80 in other plant cryptochromes also varied. The N-terminal regions of the amino acid sequences of moss CRY genes showed the highest homology with cryptochromes from the fern Adiantum capillus-veneris (Kanegae and Wada, 1998; Imaizumi et al., 2000). These gene products possess the specific C-terminal extension. At the ends of the C termini of PpCRY proteins, we found the putative monopartite nuclear localization signals, which also are present at the same regions of the nucleus-localized fern cryptochromes AcCRY3 and AcCRY4 (Figure 2A). Therefore, we examined the intracellular localization of the β-glucuronidase (GUS)-PpCRY1a and GUS-PpCRY1b fusion proteins in a transient expression assay. Because the AcCRY3 protein showed differential localization depending on the light conditions (Imaizumi et al., 2000), we incubated the transfected protoplasts under white, blue, or red light or in the dark. Both GUS-PpCRY1a and GUS-PpCRY1b proteins localized to the nucleus under all light conditions examined (Figure 2B). Both CRY transcripts accumulated at low levels in protonemata (Figures 3B and 3C, lane WT) and in gametophores (data not shown). We also examined the light dependence of the accumulation levels of both CRY mRNAs and found that 2-day incubation under various light conditions did not alter the mRNA levels in protonemata (data not shown).
Figure 2.
Intracellular Distribution of GUS-PpCRY Fusion Proteins in Moss Protoplasts.
(A) Alignment of amino acid sequences of the C termini of PpCRY1a, AcCRY3, and AcCRY4. The conserved amino acid residues are highlighted. The putative nuclear localization signal is underlined.
(B) Protoplasts incubated under white light. The top row shows GUS activity, and the bottom row shows the positions of the nuclei in the same cells via 4′,6-diamidino-2-phenylindole (DAPI) fluorescence. Bar = 20 μm for all panels.
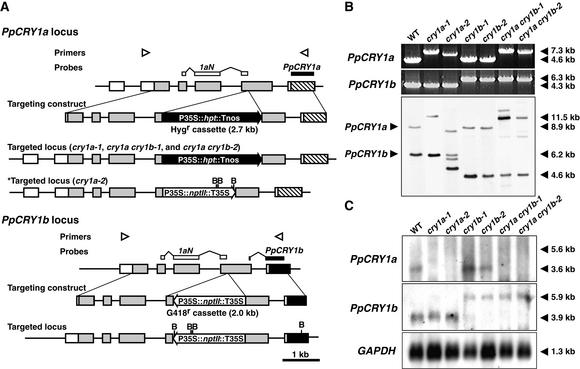
Figure 3.
Cryptochrome Targeting Constructs and Confirmation of Disruption.
(A) Scheme of the disruption of the PpCRY1a and PpCRY1b loci. The locations of primers used in (B) are shown by arrowheads. The probe that is supposed to hybridize both PpCRY1a and PpCRY1b (1aN) and that is used in (B) is indicated by white boxes, and the probes specific to each cryptochrome transcript used in (C) are indicated by closed boxes. The recognition sites of BglII (B) are indicated as well. Tnos, nopaline synthase terminator; Hygr, hygromycin-resistant; nptII, neomycin phosphotransferase II; hpt, hygromycin B phosphotransferase.
(B) PCR genotyping analysis and genomic DNA gel blot analysis of cryptochrome disruptants. Genomic DNA was digested with BglII. The bands corresponding to either native PpCRY1a or PpCRY1b loci are indicated at left. The lengths of major bands are shown at right. WT, wild type.
(C) RNA gel blot analysis of CRY transcripts in the cryptochrome disruptants. Poly(A)+ RNA derived from white light–grown protonemata were analyzed. As an internal control, the 1.1-kb fragment of Physcomitrella glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA was used (Leech et al., 1993).
Generation of PpCRY1a and PpCRY1b Single and Double Disruptants
To determine which responses these cryptochromes mediate, cryptochrome disruptants were generated by homologous recombination using targeting constructs (Figure 3A). We generated eight strains of PpCRY1a gene disruptants (designated cry1a strains), five strains of PpCRY1b gene disruptants (cry1b), and four strains of PpCRY1a and PpCRY1b gene double disruptants (cry1a cry1b). Every strain of each disruptant showed very similar phenotypes with each other, so we used two strains of each disruptant for further analyses. We confirmed by polymerase chain reaction (PCR) analyses that the expected CRY loci of these lines were replaced with targeting constructs (Figure 3B). We also performed DNA gel blot analysis (Figure 3B). The 8.9-kb band derived from the PpCRY1a locus shifted to approximately the 11.5-kb band in cry1a-1, cry1a cry1b-1, and cry1a cry1b-2. This finding indicated that the targeting construct that contains 2.7 kb of hygromycin cassette is integrated into the original PpCRY1a locus. The 8.9-kb band also disappeared in cry1a-2, indicating that the PpCRY1a locus was disrupted. There are three shifted bands (those except for the 6.2-kb band corresponding to the PpCRY1b locus), indicating that at least three copies of targeting constructs might be integrated in the genome. When we used both primers directed outward from the PpCRY1 locus, PCR fragments were amplified in cry1a-2, indicating that some targeting constructs were integrated in tandem in cry1a-2 (data not shown). Tandem insertion of targeting constructs in the moss genome have been reported previously (Schaefer and Zryd, 1997; Hofmann et al., 1999). The 6.2-kb band corresponding to the PpCRY1b locus shifted down to the 4.6-kb band in cry1b-1, cry1b-2, cry1a cry1b-1, and cry1a cry1b-2. This is reasonable because the G418 cassette contains BglII sites.
The expression of CRY transcripts in these strains was analyzed (Figure 3C). The PpCRY1a transcript was detected only in wild-type and cry1b strains. We hypothesize that transcription of the PpCRY1a transcript from the native promoter stopped at the 35S or nopaline synthase terminators present in the selection marker of the PpCRY1a-disrupted strains. The PpCRY1b transcript was detected in all strains, and larger bands (5.9 kb) were detected in cry1b and cry1a cry1b strains. The selection marker was inserted in the antisense direction in the PpCRY1b-targeted locus. On the basis of the size of the transcript, transcription must proceed through the selection marker (2.0 kb) and stop at the native PpCRY1b terminator. These data suggest that cry1a and cry1a cry1b strains do not contain intact PpCRY1a proteins, and they also show that cry1b and cry1a cry1b strains do not contain intact PpCRY1b proteins.
Blue Light Signals via Cryptochromes Induce Side Branches in Protonemata
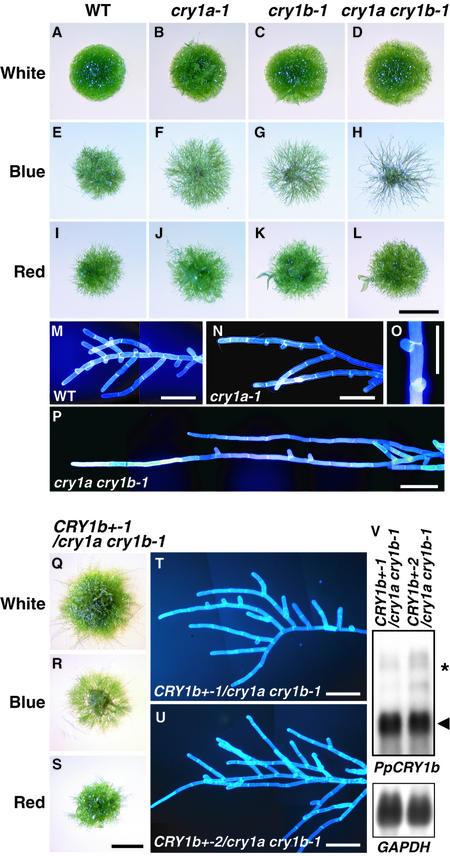
The growth of protonemal colonies of wild type and cryptochrome disruptants was observed under various light conditions. Colonies of similar diameters were inoculated onto new agar plates. These plates were incubated under white, blue, or red light or in darkness. Both wild-type and disruptant protonemata showed almost no growth in the dark under our culture conditions, so we present the results obtained under white, blue, or red light. The appearances of cry1a, cry1b, and cry1a cry1b colonies were not distinguishable from those of wild-type colonies under white and red light, whereas a clear difference was observed under blue light (Figures 4A to 4L). The colony diameter of each disruptant was larger than that of the wild type, and the densities of the protonemata in each colony decreased in order from wild type, to cry1a strains, to cry1b strains, to cry1a cry1b strains.
Figure 4.
Protonemal Colonies of Cryptochrome Disruptants under Different Light Conditions and Complementation of Cryptochrome Disruptants by Overexpression of PpCRY1b cDNA.
Colonies of wild type, cry1a, cry1b, cry1a cry1b, and PpCRY1b cDNA overexpressors in the cry1a cry1b-1 background were inoculated on agar plates. These plates were incubated under continuous white, blue, or red light for 10 days.
(A), (E), (I), and (M) Wild type (WT).
(B), (F), (J), (N), and (O) cry1a-1.
(C), (G), and (K) cry1b-1.
(D), (H), (L), and (P) cry1a cry1b-1.
(Q) to (T) PpCRY1b cDNA overexpressor in cry1a cry1b-1 (CRY1b+-1/ cry1a cry1b-1).
(U) Different PpCRY1b cDNA overexpressor in cry1a cry1b-1 (CRY1b+-2/cry1a cry1b-1).
The light conditions are as given: white light ([A] to [D] and [Q]); blue light ([E] to [H] and [R]); and red light ([I] to [J] and [S]). The blue light–grown protonemata are shown in (M) to (P) and (T) and (U). The protonema cell walls were stained with 0.1% (v/v) Calcofluor white to help distinguish each cell. The appearance of protonemata of cry1b strains was not distinguished from that of cry1a strain protonemata. Bars in (L) and (S) = 2 mm for (A) to (L) and (Q) to (S); bars in (M) to (P) and (T) and (U) = 200 μm.
(V) Overexpression of PpCRY1b transcripts (arrowhead) in CRY1b+-1/ cry1a cry1b-1 strains was confirmed by RNA gel blot analysis. The asterisk shows the putative transcript derived from the disrupted PpCRY1b locus.
Close observation showed that the difference in density was a result of the inhibition of side branch formation on the protonemata (Figures 4M to 4P). In addition, even where side branches had been initiated, these branches often stopped growing under blue light (Figure 4O). This tendency is more obvious in the cry1a cry1b strains. We further analyzed the induction rate of the side branches using nonbranching protonemata (Table 1). When protonemata were cultured under weak unilateral red light, they grew toward the light with almost no branching (Kadota et al., 2000). When irradiated with strong blue light after preculturing in weak red light, approximately two branches emerged on the wild-type protonemata, whereas the average number of branches decreased to less than one in the cry1a strains. These numbers were even lower in the cry1b strains, and almost no branches were induced in the cry1a cry1b strains by blue light. The effects of strong red light were similar in both wild type and cry disruptants.
Table 1.
Induction of Side Branches on No-Branched Protonema of Cryptochrome Disruptants under Different Light Conditions
| Number of Induced Branches (n)
|
||
|---|---|---|
| Strain | Blue Light (28 μmol m−2·sec−1) |
Red Light (26 μmol m−2·sec−1) |
| Wild type | 2.15 ± 0.16 (106) | 0.88 ± 0.08 (104) |
| cry1a-1 | 0.65 ± 0.14 (40) | 0.68 ± 0.15 (37) |
| cry1a-2 | 0.63 ± 0.10 (81) | 0.72 ± 0.17 (32) |
| cry1b-1 | 0.41 ± 0.14 (29) | 0.69 ± 0.15 (26) |
| cry1b-2 | 0.29 ± 0.16 (21) | 0.75 ± 0.14 (40) |
| cry1a cry1b-1 | 0.08 ± 0.03 (74) | 0.50 ± 0.07 (103) |
| cry1a cry1b-2 | 0.02 ± 0.02 (62) | 0.80 ± 0.13 (74) |
Protonemata were precultured between cellophane and a cover slip on agar medium under unilateral red light (2.8 μmol m−2·sec−1) for 9 days. Protonemata of more than 1.5 mm in length, having no branch, were used for this experiment. Stimulus light was given continuously for 2 additional days, and the number of branches induced was scored. Two-day dark incubation did not produce any side branches on wild-type protonemata (n = 97).
Values shown are means ±se; n indicates number of protonemata counted.
Overexpressing PpCRY1b cDNA complemented the cry1a cry1b-1 strain (Figures 4Q to 4V). Two independent overexpressors in the cry1a cry1b-1 background (CRY1b+/cry1a cry1b-1) showed branching under blue light. These data further confirm that blue light signals via cryptochromes induce side branch formation.
Blue Light Signals via Cryptochromes Also Control Differentiation and Growth of Gametophores
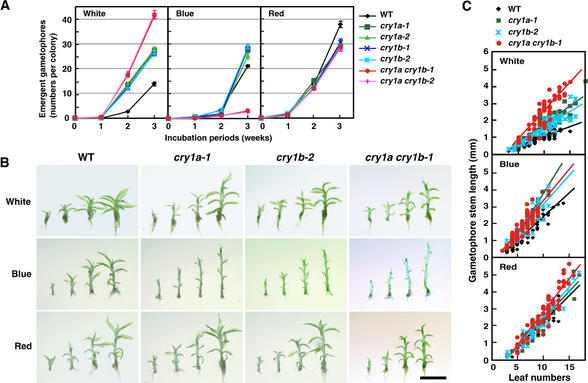
Less than 5% of the side branch initials on protonemata developed into gametophore buds (Cove and Knight, 1993). We found that cryptochromes also controlled the transition and growth of gametophores. Figure 5A shows the average number of gametophores that emerged in each wild-type and cryptochrome disruptant colony within 3 weeks under different light conditions. Under white light, a few gametophores were seen in wild type at 2 weeks, whereas more than 10 gametophores were counted in all cryptochrome disruptants. The number of gametophores emerging on the cry1a cry1b colonies was even higher than that on either the cry1a or cry1b colonies. Under blue light, in all strains a few gametophores were observed in the first 2 weeks. The number of gametophores increased to ∼25 per colony by 3 weeks in all strains except for the cry1a cry1b strains. The number in the cry1a cry1b strains was still less than five per colony in 3 weeks. Under red light, gametophore induction occurred to a similar extent in all of the strains examined. The increase in gametophore number for all strains under red light was similar to the increase for the cryptochrome disruptants under white light. When considering these data, it is important to remember that gametophore induction involves two light-controlled steps: (1) side branch formation, and (2) differentiation from the side branch initial to the bud. Under white and red light, it seemed that side branch formation occurred similarly in all strains (Figure 4, Table 1). The difference seen in Figure 5A between white and red light conditions may be ascribed to the difference that occurred at the second step. Therefore, these data indicate that cryptochrome light signals inhibit the transition to bud, whereas red light induces bud formation. Under blue light, the double disruptants were found to produce few side branches (Table 1). The limiting step on the emergence of gametophores in cry1a cry1b strains under blue light might be the first branching step. Although the inhibition of gametophore formation was observed in all strains examined by 2 weeks, it was overcome at 3 weeks in the strains that branched under blue light (wild type, cry1a, and cry1b).
Figure 5.
Gametophore Induction and Development Are Regulated by Cryptochromes.
(A) Number of gametophores that emerged on the colonies of the cryptochrome disruptants. The colonies were incubated under white, blue, or red light for up to 3 weeks. Gametophores that emerged were counted every week. Each point represents the mean ±se derived from the data of 30 independent colonies.
(B) Representative images of gametophores of one of each cryptochrome disruptant. Gametophores were collected from 3-week-old colonies after inoculation. The gametophores, which have 5, 7, 9, and 11 recognizable leaves, are placed from left to right in each image. Each image shows the gametophores of wild type, cry1a-1, cry1b-2, or cry1a cry1b-1 incubated under white, blue, or red light. The strains that are not shown here had the same phenotypes as each representative image of the cryptochrome disruptants. Bar = 2 mm for all panels.
(C) Relationship between leaf number and stem length. Tissue was considered as a leaf if it clearly looked separate from the stem or if the conical premature leaf bundle appeared at the gametophore apex. The stem length is the length from the position of the shoot apical meristem (presented in the apical leaf bundle) to the end of the stem, not including rhizoids. Slopes calculated by fitting a linear regression line for each group of data are shown in the graph.
Figure 5B shows gametophores grown under different light conditions. The cry1a, cry1b, and cry1a cry1b gametophores had short and narrow leaves compared with the wild type under blue light. Cell lengths and cell numbers of these leaves were less than those of the wild type (data not shown). In contrast, the size of the leaves of the wild-type, cry1a, and cry1b strains were almost the same under white and red light. Even under white or red light, however, the leaf sizes of cry1a cry1b strains were smaller than those of wild type. Stem length under the light conditions described for Figure 5B is shown in Figure 5C. Under white light, the stem lengths of both cry1a-1 and cry1b-2 were ∼1.5 times longer than those of wild type. The stems of cry1a cry1b-1 were approximately twice as long as those of wild type. Under blue light, the stem lengths of the cryptochrome single and double disruptants were similar and also were longer than those of wild type. However, the stem lengths of wild type and cryptochrome disruptants were almost identical under red light. PpCRY1b cDNA overexpressor (CRY1b+/cry1a cry1b-1) strains showed shorter stems than wild type not only under blue light but also under white and red light (data not shown). These findings indicate that blue light, acting through cryptochromes, induces leaf growth but inhibits stem growth.
Cryptochrome Disruptants Show Altered Auxin Responses
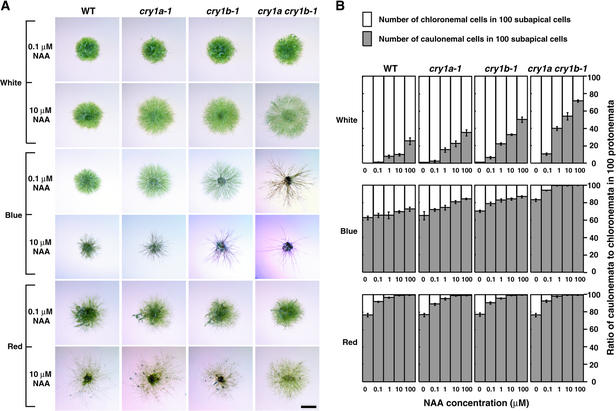
It has been reported that the developmental stages shown to be regulated by cryptochromes in this study also are controlled by plant hormones such as auxin and/or cytokinin (Cove and Knight, 1993; Reski, 1998). Although several articles have reported the light dependence of auxin and cytokinin responses in the mosses (Simon and Neaf, 1981; Lehnert and Bopp, 1983; Bhatla and Bopp, 1985), the molecular mechanisms of the interaction between light and hormones have not been established. To determine whether cryptochrome signals interact with the auxin signals, we examined the light-dependent phenotypes of cryptochrome disruptants incubated on agar containing synthetic auxin, 1-naphthalene acetic acid (NAA). We examined the colonies grown on agar plates containing NAA under white, blue, or red light. Although the colonies of wild type and cryptochrome disruptants looked identical under white light without auxin (Figures 4A to 4D), the addition of a high concentration (>1 μM) of NAA to the medium led cryptochrome disruptants to be distinguishable from wild type (Figure 6A). The diameters of both wild-type and cryptochrome disruptant colonies incubated under red light increased at almost the same rates as those of cry1a cry1b strains incubated under white light (Figure 6A). Because the difference in appearance between colonies grown in white light and red light might be partly the result of the different branching numbers induced, these data led us to infer that cryptochrome light signals inhibit auxin responses, whereas red light either induces or does not affect auxin responses. Under blue light, cry1a and cry1b strains grown on agar containing 10 μM NAA looked similar to the cry1a cry1b strains grown on agar without auxin (Figure 4H) or with 0.1 μM NAA (Figure 6A). These data indicate that adding exogenous auxin to the cry1a and cry1b strains mimics the phenotype of the cry1a cry1b strain at lower doses of NAA.
Figure 6.
Cryptochrome Light Signals Inhibit Auxin Responses.
Protonemal inocula were cultured for 10 days under different light conditions in the presence of 0.1 to 100 μM NAA in the medium.
(A) Representative colony images of cryptochrome disruptants grown on medium containing different concentrations of NAA. Data obtained from the addition of 0.1 and 10 μM NAA are shown. The colonies incubated under the same light conditions without NAA are shown in Figure 4. Bar = 2 mm for all panels.
(B) The ratio of caulonemal cells to chloronemal cells in 100 protonemal subapical cells incubated under different light conditions in the presence of NAA. Subapical cells of the protonemata were classified into chloronemal cells and caulonemal cells. The mean ±se of the number of caulonemal cells obtained from eight independent colonies, and 100 protonemal cells in each colony, were classified.
WT, wild type.
Moss protonemal cells are classified into two cell types (chloronema and caulonema) according to their morphology (Cove and Knight, 1993; Reski, 1998). Chloronema has cell walls perpendicular to the filament axis, whereas caulonema has cell walls oblique to the filament axis. After spore germination, chloronema emerge first and a transition to caulonema occurs. The transition from chloronema to caulonema is induced by exogenous auxin in mosses (Schumaker and Dietrich, 1997). We calculated the ratio of caulonemal cells to chloronemal cells in 100 protonemal subapical cells of a single colony as another index of auxin cellular effects under different light conditions (Figure 6B). When NAA was not added, all subapical cells were classified into chloronemal cells in wild type and cryptochrome disruptants under white light. Under either blue or red light, the number of caulonemal cells was more than the number of chloronemal cells in wild type and disruptants even without NAA. In proportion to the concentration of exogenous NAA in the medium, the ratio of caulonemal cells was increased in wild type and disruptants under all light conditions. It is noteworthy that the increasing ratio of caulonemal cells was lowest in wild type, higher in cry1a, higher still in cry1b, and highest in cry1a cry1b strains under white and blue light. Such a difference was not observed under red light. These results are in accord with the hypothesis that the inhibition of the auxin responses is specific to the cryptochrome signal.
Cryptochrome Light Signals Suppress Auxin-Induced Gene Expression
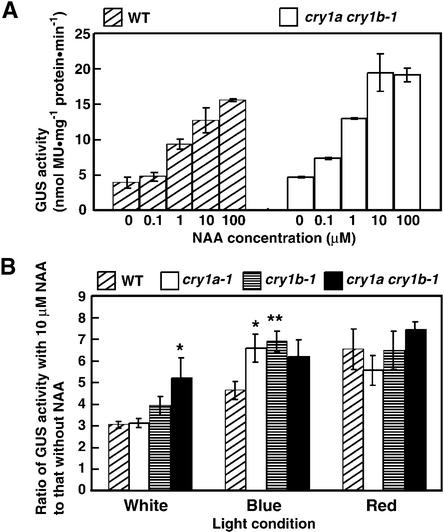
To further investigate the relationship between cryptochrome and auxin communication, an auxin-inducible soybean GH3 promoter–GUS fusion gene construct (Li et al., 1999) was introduced to protoplasts. Protoplasts transfected with GH3::GUS genes were treated with different concentrations of NAA under white light. GUS activity increased depending on the NAA dose in wild type and cry1a cry1b-1 (Figure 7A), indicating that auxin signals were transmitted to the GH3 promoter in Physcomitrella protoplasts. To analyze different light effects on auxin signal transduction, GUS activities of transfected protoplasts with or without NAA were assayed under white, blue, and red light. Adding NAA caused a three times greater induction of GUS activity in wild type under white light (Figure 7B). The GUS activity was induced at a five times greater ratio in cry1a cry1b-1. The induction of the GH3 promoter in wild type under blue light was higher than that under white light, whereas the cryptochrome disruptants showed an even greater induction ratio. The amount of induction in cry1a cry1b-1 was similar to that seen in cry1a-1 and cry1b-1, possibly because GUS activity was saturated in this condition. Adding NAA under red light led both wild type and cryptochrome disruptants to show similar GUS activity. These data suggest that blue light signals via cryptochrome inhibit auxin signals.
Figure 7.
Measurements of Auxin Signals in Cryptochrome Disruptants.
(A) Soybean GH3 promoter activity in moss. The GH3::GUS vector was transformed into moss protoplasts and incubated under white light for 24 hr in the presence of different doses of NAA. The data show the mean ±se derived from one trial, which consisted of two sets of group data at each point. Similar data were obtained in different trials. MU, 4-methylumbelliferone.
(B) Induction rates when exogenous NAA was added to the cryptochrome disruptants. The data show the induction values (mean ±se), which were calculated by dividing the means obtained from the data without NAA into the means obtained from the data in the presence of 10 μM NAA. These data were obtained from three different trials, each trial of which constituted of three different sets of group data (total, nine groups of data). * P < 0.05, ** P < 0.01 (Student's t test).
WT, wild type.
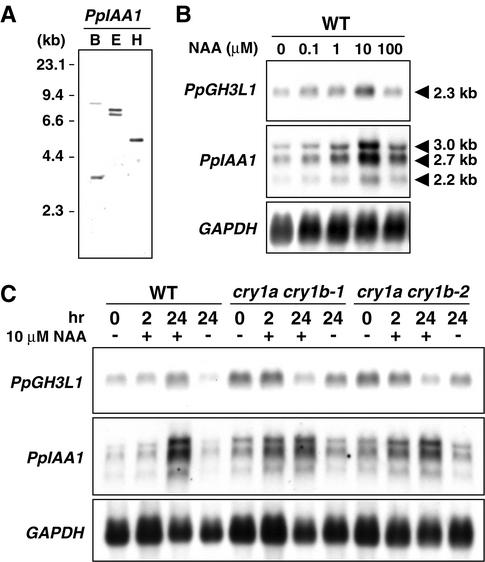
We further compared the amount of native auxin-inducible gene transcripts between wild type and cryptochrome disruptants. The GH3 and indole-3-acetic acid (IAA) gene homologs were isolated as candidates for the auxin-inducible genes in Physcomitrella. A cDNA (2304 bp) of the P. patens GH3-like protein 1 (PpGH3L1) gene encoded a deduced polypeptide of 599 amino acids with remarkable similarity (∼40% identity) to the entire sequences of flowering plant GH3 homologs. Two cDNAs encoding putative IAA genes were isolated. They contained identical sequences spanning ∼1.2 kb of the 3′ ends, whereas the 5′ ends were different. The longer cDNA, designated PpIAA1, is 2594 bp long and encodes a deduced polypeptide of 503 amino acids. The amino acid sequence of PpIAA1 contained the II, III, and IV domains typical of IAA proteins (Abel et al., 1995). When the 3′ identical sequence was used as a probe for DNA gel blot analysis, a few bands were detected (Figure 8A), whereas three different bands were detected in RNA gel blot analysis (Figures 8B and 8C). The 2.7-kb band may correspond to PpIAA1 transcripts. The amount of PpGH3L1 and PpIAA1 transcripts was induced by exogenous auxin within 24 hr (Figures 8B and 8C), and induction showed a biphasic response depending on the auxin concentration (Figure 8B), as reported in flowering plants (Hagen et al., 1991; Abel et al., 1995). Figure 8C shows that the amount of PpGH3L1 and PpIAA1 transcripts in cry1a cry1b strains was higher than that of the wild type in the medium without NAA. By the addition of 10 μM NAA, PpIAA1 transcripts in cry1a cry1b strains were accumulated faster than in the wild type. Adding 10 μM NAA suppressed the accumulation of PpGH3L1 transcripts in cry1a cry1b strains. On the basis of the results shown in Figure 8B, 10 μM NAA might be greater than the concentration required for maximal induction in cry1a cry1b strains. In summary, we confirmed that cryptochrome light signals interact with auxin signals to suppress the expression of auxin-inducible genes.
Figure 8.
Expression Levels of Native Auxin-Inducible Genes in Cryptochrome Disruptants.
(A) DNA gel blot analysis of the PpIAA1 gene. B, BglII; E, EcoRI; H, HindIII.
(B) Dose response of the accumulation of auxin-inducible genes in Physcomitrella. Five-day-old protonemata were treated with different concentrations of NAA for 24 hr.
(C) Accumulation patterns of auxin-inducible genes in cryptochrome disruptants. Five-day-old protonemata of wild-type and cry1a cry1b strains were treated with 10 μM NAA (+) or mock solution (−) for 2 and 24 hr.
WT, wild type.
DISCUSSION
Cryptochromes Regulate Developmental Processes throughout the Moss Life Cycle
Two Physcomitrella CRY genes are almost identical in both exon and intron sequences (Figure 1B), indicating that one of these CRY genes was generated by a recent gene duplication. The 5′ ends of the identical regions of both genes start from guanine-adenine (GA) repeat sequences (Figure 1B). Because homologous recombination occurs at high frequency in Physcomitrella (Schaefer, 2001), one of the CRY genes might have been generated by homologous recombination that occurred between the GA repeat sequence existing in the original CRY gene locus and another GA repeat sequence in the genome.
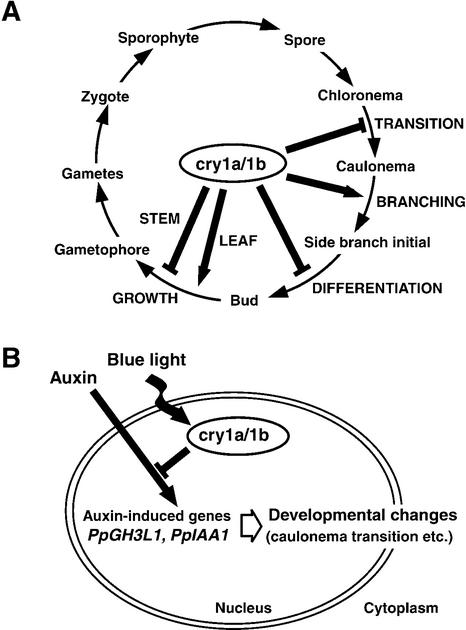
The regulation of moss development by blue light signals via cryptochrome was assessed using cryptochrome disruptants. The cry1a cry1b strains showed more severe phenotypes than did the single disruptants (Figures 4 to 8), indicating that PpCRY1a and PpCRY1b act additively to regulate moss development. A model of the effects of CRY genes on the moss life cycle is shown in Figure 9A. Cryptochrome light signals inhibited the transition from chloronema to caulonema (Figure 6B) but induced side branching on caulonemata (Table 1). In the fern A. capillus-veneris, physiological analyses indicated that blue light receptors localized in or close to the nucleus induce longitudinal cell division to the protonemal axis, which leads to the developmental change from protonema to prothallium (Wada and Furuya, 1971, 1978). Two cryptochromes were shown previously to localize in the nucleus in the fern (Imaizumi et al., 2000). Unlike moss protonema, the fern protonema does not have any side branches under the appropriate light conditions (Wada and Sugai, 1994). The orientation of the first cell plate of the side branch in the moss protonemata also is longitudinal to the growth axis. Therefore, blue light induced longitudinal cell division on both moss and fern protonemata, and both responses may be interpreted as the transition from first dimensional growth to second dimensional growth. These two phenomena display similar characteristics. If the fern nucleus-localized cryptochromes are involved in that phenomenon, similar cryptochrome signal transduction cascades, which are related to control switching of growth axes in the moss, may be retained in ferns.
Figure 9.
Summary of Cryptochrome Functions in the Moss Life Cycle and Possible Model of Cryptochrome Signal Transductions via Auxin.
(A) Cryptochrome functions revealed in this study are shown in this diagram of the moss life cycle.
(B) The model shows one predicted interaction between cryptochrome light signals and auxin signals.
Determination of the fate of the side branch initials is regulated by cryptochrome (Figure 5A). After the side branch initials differentiate into buds, the development of the gametophores also is controlled by cryptochrome (Figures 5B and 5C). Arabidopsis cryptochrome signals inhibit not only hypocotyl elongation (Ahmad and Cashmore, 1993) but also bolted stem elongation and control leaf and cotyledon expansion (Jackson and Jenkins, 1995). Cryptochromes seem to control the growth and development of analogous stems and leaves in both moss gametophores and seed plants. Hence, cryptochrome signal transduction involved in these phenomena in the moss might recruit diploid-phase analogous tissue in seed plants.
Cryptochrome Light Signals May Regulate Developmental Changes by Changing Auxin Sensitivities
It is of particular interest to determine how cryptochromes regulate the molecular events leading to changes in growth patterns. Here we have demonstrated one mechanism that may influence growth and developmental patterns controlled by cryptochromes via auxin signal transduction.
Previously, it was reported that the effect of external auxin is less obvious under high-intensity white light in the moss Funaria hygrometrica (Lehnert and Bopp, 1983). We showed that exogenous NAA works more effectively in cryptochrome disruptants than in wild type with respect to the transition from chloronema to caulonema (Figure 6B) and that blue light, but not red light, was involved in the suppression of auxin effects in Physcomitrella. These findings indicate that the effect of white light on the suppression of the auxin response in F. hygrometrica is the function of cryptochrome. Continuous blue light (100 μmol m−2·sec−1) also caused more than 50% inhibition of the auxin-induced elongation of hypocotyl segments of cucumber relative to red light controls. This inhibition was observed only when exogenous auxin (2 to 20 μM IAA) was added (Shinkle and Jones, 1988). These results suggest that a similar molecular mechanism for the suppression of auxin signals by cryptochrome signals might exist in seed plants as well.
There have been several articles reporting the interaction of light and auxin signal transduction pathways in Arabidopsis. Gain-of-function mutations in some IAA genes (SHY2/IAA3, AXR2/IAA7, and AXR3/IAA17) ectopically induce photomorphology in the dark (Tian and Reed, 1999; Nagpal et al., 2000), indicating that photomorphological responses are mediated via some IAA proteins. From the isolation of the suppressor of COP1, the GH3-like gene (FIN219) is believed to participate in phytochrome A signaling toward the COP1 repressor (Hsieh et al., 2000). We showed that the expression of those IAA and GH3 homologous genes is modulated by light via cryptochrome in the moss. Soybean GH3 promoter activity and the accumulation of moss IAA and GH3 homologous gene in cryptochrome disruptants are higher than those in wild type at the same dose of NAA (Figures 7 and 8). Because moss cryptochromes are nuclear proteins, we propose a model for the interaction between cryptochrome light signals and auxin signals occurring in the nucleus (Figure 9B). From the results shown in Figures 6, 7, and 8, it is likely that cryptochrome light signals control developmental changes by suppressing auxin signals at least at the point of transcription of auxin-regulated genes.
METHODS
Plant Materials and Light Sources
Protonemata of Physcomitrella patens subsp patens were cultured aseptically on BCDATG medium (which is BCD medium [1 mM MgSO4, 10 mM KNO3, 45 μM FeSO4, 1.8 mM KH2PO4, pH 6.5] supplemented with 1 mM CaCl2, 5 mM ammonium tartrate, and 0.5% [w/v] glucose [Nishiyama et al., 2000]) under different light conditions at 25° ± 1°C. For the analysis of exogenous auxin effects on growth, 1-naphthalene acetic acid (NAA) was added to the autoclaved medium. The light sources were described by Imaizumi et al. (2000). The light intensities used throughout this study were 45 μmol m−2·sec−1 for white light, 17 μmol m−2·sec−1 for blue light, and 28 μmol m−2·sec−1 for red light, unless noted otherwise. Harvesting of light-irradiated tissues was performed under a dim green safelight.
Isolation and Detection of DNA and RNA
Genomic DNA was isolated using the cetyltrimethylammonium bromide method (Murray and Thompson, 1980). Trizol reagent (Invitrogen, Carlsbad, CA) was used to isolate total RNA derived from protonemata. To purify poly(A)+ RNA, oligo(dT) latex beads (Oligotex-dT30[Super]; Takara, Kyoto, Japan) were used. The digested genomic DNA (2 μg per lane) was separated on a 0.8% agarose gel. Poly(A)+ RNA (4 μg per lane) was electrophoresed on a 1% formaldehyde-agarose gel. DNA and RNA were blotted on nylon membranes. A chemifluorescence-labeled gene detection kit (Gene Images; Amersham Pharmacia Biotech) was used for detection. High-stringency genomic DNA gel blot hybridization and RNA gel blot hybridization were performed at 65°C with 0.1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% (w/v) SDS, and low-stringency genomic DNA gel blot hybridization was performed at 60°C with 0.5 × SSC and 0.1% SDS.
Cloning of Physcomitrella Cryptochrome PpCRY1a and PpCRY1b Genes
Partial cryptochrome fragments were amplified using cryptochrome-specific degenerate primers, and the remaining 5′ and 3′ regions of the cDNA were obtained by 5′ and 3′ rapid amplification of cDNA ends (RACE) methods (Imaizumi et al., 1999). Two 3′ RACE products with different sequences were obtained, and primers specific to each 3′ RACE product were used for reverse transcription of 5′ RACE. The primers were 5′-CGCCTCTTCATTATCACTGCTC-3′ for PpCRY1a and 5′-AGCCGGATCCATCTCATACG-3′ for PpCRY1b. Genomic DNA fragments corresponding to each CRY cDNA were amplified and sequenced. Further 5′ and 3′ genomic regions were cloned by the inverse polymerase chain reaction (PCR) method (Triglia et al., 1988). Physcomitrella genomic DNA was digested by BglII, DraI, HindIII, or XbaI, ligated, and used as PCR templates. The primer sets used for the first round of inverse PCR were 5′-AAACTCCCCGATATGTAC-TGCC-3′ and 5′-ACGGAGTTCAAGCAGGCAAT-3′ for PpCRY1a and 5′-CTCATCATACACCTCCCAAG-3′ and 5′-GCGTATCTAGTTCCA-GTGCA-3′ for PpCRY1b. Nested PCR was performed using the following primers: 5′-GCAAAAACGAATCCAGCGACTT-3′ and 5′-CCA-CCACATTTTAACCTCAGACGGCATG-3′ for PpCRY1a and 5′-CCA-AATGTGTCAAGCTCTGC-3′ and 5′-CAAAGGTTCGACTGGATTGC-3′ for PpCRY1b.
Construction of PpCRY1a and PpCRY1b Gene Disruption Vectors and the Vector for the PpCRY1b cDNA Overexpressor
PpCRY1a and PpCRY1b genomic DNA fragments (4.3 and 4.2 kb, respectively) were amplified using the primers 5′-AGTAGCGAGAAG-AGGGACAAGT-3′ and 5′-CGCCTCTTCATTATCACTGCTC-3′ for PpCRY1a, and the same forward primer and 5′-GCCCACAATTCA-TACTGCACTG-3′ for PpCRY1b, and cloned into a vector, pCRII-TOPO (Invitrogen). The hygromycin cassette containing the 35S promoter of Cauliflower mosaic virus, the hygromycin B phosphotransferase (hpt) gene, and the nopaline synthase (nos) polyadenylation signal was amplified originally from the binary vector pBIH1-IG (Okamoto et al., 1993) using the primers 5′-AGGATAAATTAT-CGCGCGCGGTGTC-3′ and 5′-CCCAGTCACGACGTTGTAAAA-CGAC-3′, cloned into a vector (pGEM-T Easy; Promega), and named pGEM-T Hyg. The hygromycin cassette in pGEM-T Hyg was excised as a SalI fragment. The neomycin phosphotransferase II expression cassette (Nishiyama et al., 2000) was excised by XhoI sites from pMBL5. Each excised resistant gene cassette was blunted and inserted into blunted SacII sites located in the middle of the PpCRY1a and PpCRY1b genomic fragment of each plasmid. These plasmids with disruption constructs were named pPpCRY1a-KO and pPpCRY1b-KO.
For ectopic expression of PpCRY cDNAs, the pPpMADS2-7133 plasmid was used. This plasmid contains a genomic fragment of the PpMADS2 gene, in the middle of which the neomycin phosphotransferase II expression cassette (Nishiyama et al., 2000), the E7133 promoter (Mitsuhara et al., 1996), and the nos terminator are inserted. Disruptants of the PpMADS2 locus are not distinguishable from wild type, indicating that this locus is useful as a neutral site to integrate an overexpression construct. The PpCRY1b cDNA fragment (nucleotides 304 to 3040) containing the expected open reading frame was inserted into the SmaI site located between the E7133 promoter and the nos terminator and named pPpCRY1b-7133.
Transformation of Moss Protoplasts
Isolation of protoplasts and polyethylene glycol–mediated transformation were performed according to Nishiyama et al. (2000). To obtain stable transformants in which an introduced DNA could be integrated into the Physcomitrella genome, all plasmids were digested with appropriate restriction enzymes to separate its insert fragment with vector sequences, and the digested plasmids were introduced into protoplasts. Transformed protoplasts were incubated for 4 days on BCDAT medium (which is BCDATG medium without glucose) without antibiotics and then transferred to BCDAT medium containing either 50 mg/L G418 or 30 mg/L hygromycin B for 3 weeks. The selected plants were incubated for 1 additional week without antibiotics and transferred again onto the selection medium. Genomic DNA of plants which still retained resistance to antibiotics was extracted, and integration of foreign DNA was confirmed by PCR analyses.
To obtain stable disruptants of the PpCRY1a and PpCRY1b genes, pPpCRY1a-KO and pPpCRY1b-KO were digested with ApaI and HindIII and with NotI and HindIII, respectively, and used for the transformation. cry1a cry1b strains were generated to retransform cry1a-1 with the pPpCRY1b-KO construct and selected by PCR analysis from G418– and hygromycin B–resistant colonies. To obtain stable transformants containing pPpCRY1b-7133, this plasmid was digested with NotI and introduced into protoplasts of cry1a cry1b-1. Protoplasts of cry1a cry1b-1 were resistant to G418 less than 100 mg/L but not to higher concentrations. Even though these protoplasts were resistant to 100 mg/L G418, they grew slower than they did in 50 mg/L G418. Transformants with pPpCRY1b-7133 were selected by growth rate on medium containing 100 mg/L G418, and then PpCRY1b transcript overexpressors were selected by PCR analysis and RNA gel blot hybridization.
β-Glucuronidase–PpCRY Plasmid Construction and Histochemical Assay of GUS Activity
Each PpCRY1a and PpCRY1b cDNA was fused to the 3′ end of the β-glucuronidase (GUS) coding region in modified pBI221 vector as described by Imaizumi et al. (2000). PpCRY1a and PpCRY1b cDNAs were amplified using the following primers: 5′-AGTTACTCTTCACAAATGGCGGCGTGCACAATAGTGTGG-3′ and 5′-AGTTAC-TCTTCAAGCGGCAGCCAAGGACGAATGCTTGTGC-3′ for PpCRY1a, and the same forward primer and 5′-AGTTACTCTTCAAGCACT-GGAACTAGATACGCCCCAAACCT-3′ for PpCRY1b. The HindIII-SmaI 0.8-kb fragment containing the 35S promoter of Cauliflower mosaic virus in pBI221 was replaced by the HindIII-SmaI 2.2-kb fragment containing the rice actin 1 (Act1) promoter derived from pAct1-D (McElroy et al., 1990), which is more active in Physcomitrella (Zeidler et al., 1999), in both plasmids and named GUS-PpCRY1a and GUS-PpCRY1b. These plasmids were introduced, without digestion of restriction enzymes, into protoplasts, incubated for 12 hr in the dark, and treated for 24 hr under different light conditions. The protoplasts were fixed under the same light conditions for 30 min with 0.37% (v/v) formaldehyde in 100 mM sodium phosphate buffer, pH 7.0, washed with phosphate buffer, incubated in phosphate buffer with 0.03% (v/v) Triton X-100 for 1 hr, and washed with phosphate buffer without detergent. The protoplasts then were stained with 100 mM sodium phosphate, 1 mM EDTA, 0.5 mM potassium ferricyanide, 0.5 mM potassium ferrocyanide, 10 ng/mL 4′,6-diamidino-2-phenylindole, and 0.5 mM 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, pH 7.0, for 6 hr at 37°C.
Physiological Analyses of Phenotypes of Cryptochrome Disruptants
The protonemata used for the physiological analyses were precultured under white light. To prepare protonemata that were at similar developmental stages, they were fragmented with a homogenizer and transferred onto new medium every 5 days (Nishiyama et al., 2000). On this culture schedule, the protonemata usually did not develop any buds. As starting materials for all experiments, protonemal colonies (∼1 mm in diameter) were inoculated the same distance apart on the agar plates. The protonemal colonies and the gametophores were observed with a stereomicroscope, and fine observations were made using a microscope. The gametophores that emerged on the colony were counted one by one using fine forceps under the stereomicroscope lens. For the determination of the ratio of caulonemata to chloronemata, protonemata were picked randomly from colonies of the same age and all subapical cells of the protonema that were more than four cells long were classified into caulonemal cells or chloronemal cells under a microscope lens.
Detection of Soybean GH3 Promoter Activity in Cryptochrome Disruptants
The activity of the GH3 promoter (Hagen et al., 1991) in Physcomitrella protoplasts was assayed by the activity of the uidA gene product (GUS) regulated by this promoter. The protoplasts were transformed with the GH3-GUS plasmid and incubated for 12 hr in the dark. The protoplasts then were incubated with different concentrations of NAA for 24 hr under different light conditions. Proteins were extracted from the protoplasts by repeating freeze-thaw cycles in the extraction buffer (1 mM Na2EDTA, 50 mM NaH2PO4, 10 mM 2-mercaptoethanol, 0.1% [w/v] sarcosine, and 0.1% [v/v] Triton X-100, pH 7.4). GUS activities were estimated by the fluorescence of the fluorogenic product, 4-methylumbelliferone, released from the GUS substrate 4-methylumbelliferyl glucuronide (Jefferson, 1987).
Cloning of Physcomitrella Auxin-Inducible PpGH3L1 and PpIAA1 cDNAs
Vegetatively propagated protonemata were cultured for 1 week, and 10 μM NAA solution was poured onto the medium after 2 hr of incubation. These protonemata were used for RNA extraction. PpGH3L1 and PpIAA1 cDNAs were amplified by nested reverse transcriptase–mediated PCR using degenerate primers. Degenerate primers for PpGH3L1 were 5′-GGIGARMGIAARYTIATGCC-3′ and 5′-ACRCAI-CKIGGIACYTTRTAYTG-3′ for first round PCR and 5′-ATGTAYRCI-CARATGYTITG-3′ and 5′-TCCCARWAIADIACRTARTG-3′ for nested PCR. Degenerate primers for PpIAA1 were 5′-ACNGARYTNMGN-YTNGGNYTNCC-3′ and 5′-CCANGGNACRTCNCCNACNARCAT-CCA-3′ for first round PCR and 5′-CARRTNGTNGGNTGGCCN-CCNGTNMG-3′ and 5′-ARCATCCARTCNCCRTCYTTRTCYTCRTA-3′ for nested PCR. Candidate clones of PpGH3L1 and PpIAA1 were sequenced, and whole cDNAs were obtained by 5′ and 3′ RACE. For 5′ RACE, the following primers were used (a primer for first-strand cDNA synthesis, first-round PCR primer, and the nested PCR primer, respectively): 5′-CCGCTGATGTACTGCT-3′, 5′-GTAGCTGTCGTT-CGTCTTCA-3′, and 5′-CGGAAGGGCTCGACAATCAT-3′ for PpGH3L1 and 5′-TCAGGGTCACTTACACG-3′, 5′-ACTGTCCCTTCCCGTATG-TC-3′, and 5′-TTCACAAGGTTCCCGCTCGA-3′ for PpIAA1. For 3′ RACE, the following primers were used (first-round PCR primer and the nested PCR primer, respectively): 5′-TTCTGAGGCATGGAT-AGGCA-3′ and 5′-GACTGACGAAGAAGAGCTAC-3′ for PpGH3L1 and 5′-GTTTCAGCAGTACATCAGCG-3′ and 5′-AGAGTCATTGGGTAGCGTCG-3′ for PpIAA1. Fragments (1053 and 938 bp; nucleotide sequence positions 1132 to 2184 of PpGH3L1 cDNA and 1621 to 2558 of PpIAA1 cDNA) were used for DNA gel blot and RNA gel blot hybridization analyses.
Accession Numbers
The accession numbers for the cDNAs described in this article are AB027528 (PpCRY1a), AB060693 (PpCRY1b), AB061221 (PpGH3L1), and AB061223 (PpIAA1).
Acknowledgments
We thank Dr. David McElroy, Dr. Ray Wu, and Dr. Jon Hughes for pAct1-D plasmid, Dr. Yi Li for GH3-GUS plasmid, Dr. Takeshi Kanegae for pGEM-T Hyg plasmid, and Dr. Rumiko Kofuji for pPpMADS2-7133 plasmid. pMBL5 was kindly provided by Dr. David Cove, and we thank Dr. Jesse Machuka as part of The Physcomitrella Expressed Sequence Tag Program at the University of Leeds (UK) and Washington University in St. Louis. We thank Futamura Chemical Industries Co., Ltd., for cellophane and Kyowa Hakko Kogyo Co., Ltd., for Driserase. We are grateful to Dr. Jane Silverthorne for critical reading of the manuscript. We thank Masae Umeda for protoplast transformation and technical assistance. This work was supported in part by a Grant-in-Aid for Scientific Research (B) (Grant No. 09440270) and the Program for the Promotion of Basic Research Activities for Innovative Biosciences to M.W., the Ministry of Education, Science, Culture, and Sports of Japan (M.W., M.H.), and research fellowships of the Japan Society for the Promotion of Science for Young Scientists (10639301) to T.I. This work was performed under the National Institute for Basic Biology Cooperative Research Program (00-133).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010388.
References
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366, 162–166. [DOI] [PubMed] [Google Scholar]
- Ahmad, M., Lin, C., and Cashmore, A.R. (1995). Mutations throughout an Arabidopsis blue-light photoreceptor impair blue-light-responsive anthocyanin accumulation and inhibition of hypocotyl elongation. Plant J. 8, 653–658. [DOI] [PubMed] [Google Scholar]
- Bhatla, S.C., and Bopp, M. (1985). The hormone regulation of protonema development in mosses. III. Auxin-resistant mutants of the moss Funaria hygrometrica Hedw. J. Plant Physiol. 120, 233–243. [Google Scholar]
- Briggs, W.R., and Huala, E. (1999). Blue-light photoreceptors in higher plants. Annu. Rev. Cell Dev. Biol. 15, 33–62. [DOI] [PubMed] [Google Scholar]
- Briggs, W.R., and Olney, M.A. (2001). Photoreceptors in plant photomorphogenesis to date: Five phytochromes, two cryptochromes, one phototropin, and one superchrome. Plant Physiol. 125, 85–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs, W.R., et al. (2001). The phototropin family of photoreceptors. Plant Cell 13, 993–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Ceriani, M.F., Darligton, T.K., Staknis, D., Más, P., Petti, A.A., Weitz, C.J., and Kay, S.A. (1999). Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285, 553–556. [DOI] [PubMed] [Google Scholar]
- Cove, D.J., and Knight, C.D. (1993). The moss Physcomitrella patens, a model system with potential for study of plant reproduction. Plant Cell 5, 1483–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin, P.F., and Kay, S.A. (2000). Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hagen, G., Martin, G., Li, Y., and Guilfoyle, T.J. (1991). Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 17, 567–579. [DOI] [PubMed] [Google Scholar]
- Hall, J.C. (2000). Cryptochromes: Sensory reception, transduction, and clock functions subserving circadian systems. Curr. Opin. Neurobiol. 10, 456–466. [DOI] [PubMed] [Google Scholar]
- Hofmann, A.H., Codon, A.C., Ivascu, C., Russo, V.E.A., Knight, C., Cove, D., Schaefer, D.G., Chakhparonian, M., and Zryd, J.-P. (1999). A specific member of the Cab multigene family can be efficiently targeted and disrupted in the moss Physcomitrella patens. Mol. Gen. Genet. 261, 92–99. [DOI] [PubMed] [Google Scholar]
- Hsieh, H.-L., Okamoto, H., Wang, M., Ang, L.-H., Matsui, M., Goodman, H., and Deng, X.-W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Imaizumi, T., Kiyosue, T., Kanegae, T., and Wada, M. (1999). Cloning of the cDNA encoding the blue-light photoreceptor cryptochrome from the moss Physcomitrella patens (accession No. AB 027528). Plant Physiol. 120, 1205. [Google Scholar]
- Imaizumi, T., Kanegae, T., and Wada, M. (2000). Cryptochrome nucleocytoplasmic distribution and gene expression are regulated by light quality in the fern Adiantum capillus-veneris. Plant Cell 12, 81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, J.A., and Jenkins, G.I. (1995). Extension-growth re-sponses and expression of flavonoid biosynthesis genes in the Arabidopsis hy4 mutant. Planta 197, 233–239. [DOI] [PubMed] [Google Scholar]
- Jarillo, J.A., Gabrys, H., Capel, J., Alonso, J.M., Ecker, J.R., and Cashmore, A.R. (2001). Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A. (1987). Assaying chimeric genes in plants: The GUS gene fusion system. Plant Mol. Biol. Rep. 5, 387–405. [Google Scholar]
- Kadota, A., Sato, Y., and Wada, M. (2000). Intracellular chloroplast photolocation in the moss Physcomitrella patens is mediated by phytochrome as well as by a blue-light receptor. Planta 210, 932–937. [DOI] [PubMed] [Google Scholar]
- Kagawa, T., Sakai, T., Suetsugu, N., Oikawa, K., Ishiguro, S., Kato, T., Tabata, S., Okada, K., and Wada, M. (2001). Arabidopsis NPL1: A phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141. [DOI] [PubMed] [Google Scholar]
- Kanegae, T., and Wada, M. (1998). Isolation and characterization of homologues of plant blue-light photoreceptor cryptochrome genes from the fern Adiantum capillus-veneris. Mol. Gen. Genet. 259, 345–353. [DOI] [PubMed] [Google Scholar]
- Kume, K., Zylka, M.J., Sriram, S., Shearman, L.P., Weaver, D.R., Jin, X., Maywood, E.S., Hastings, M.H., and Reppert, S.M. (1999). mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205. [DOI] [PubMed] [Google Scholar]
- Leech, M.J., Kammerer, W., Cove, D.J., Martin, C., and Wang, T.L. (1993). Expression of myb-related genes in the moss, Physcomitrella patens. Plant J. 3, 51–61. [DOI] [PubMed] [Google Scholar]
- Lehnert, B., and Bopp, M. (1983). The hormonal regulation of protonema development in mosses. I. Auxin-cytokinin interaction. Z. Pflanzenphysiol. 110, 379–391. [Google Scholar]
- Li, Y., Wu, Y.H., Hagen, G., and Guilfoyle, T. (1999). Expression of the auxin-inducible GH3 promoter/GUS fusion gene as a useful molecular marker for auxin physiology. Plant Cell Physiol. 40, 675–682. [Google Scholar]
- Lin, C. (2000). Plant blue-light receptors. Trends Plant Sci. 5, 337–342. [DOI] [PubMed] [Google Scholar]
- Lin, C., Robertson, D.E., Ahmad, M., Raibekas, A.A., Jorns, M.S., Dutton, P.L., and Cashmore, A.R. (1995). Association of flavin adenine dinucleotide with the Arabidopsis blue light receptor CRY1. Science 269, 968–970. [DOI] [PubMed] [Google Scholar]
- Lin, C., Yang, H., Guo, H., Mockler, T., Chen, J., and Cashmore, A.R. (1998). Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 95, 2686–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra, K., Kim, S.-T., Batschauer, A., Dawut, L., and Sancar, A. (1995). Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry 34, 6892–6899. [DOI] [PubMed] [Google Scholar]
- McElroy, D., Zhang, W., Cao, J., and Wu, R. (1990). Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuhara, I., et al. (1996). Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol. 37, 49–59. [DOI] [PubMed] [Google Scholar]
- Mockler, T.C., Guo, H., Yang, H., Duong, H., and Lin, C. (1999). Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082. [DOI] [PubMed] [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal, P., Walker, L.M., Young, J.C., Sonawala, A., Timpte, C., Estelle, M., and Reed, J.W. (2000). AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol. 123, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Ninu, L., Ahmad, M., Miarelli, C., Cashmore, A.R., and Giuliano, G. (1999). Cryptochrome 1 controls tomato development in response to blue light. Plant J. 18, 551–556. [DOI] [PubMed] [Google Scholar]
- Nishiyama, T., Hiwatashi, Y., Sakakibara, K., Kato, M., and Hasebe, M. (2000). Tagged mutagenesis and gene-trap in the moss Physcomitrella patens by shuttle mutagenesis. DNA Res. 7, 9–17. [DOI] [PubMed] [Google Scholar]
- Okamoto, H., Hirano, Y., Abe, H., Tomizawa, K., Furuya, M., and Wada, M. (1993). The deduced amino acid sequence of phytochrome from Adiantum includes consensus motifs present in phytochrome B from seed plants. Plant Cell Physiol. 34, 1329–1334. [Google Scholar]
- Reski, R. (1998). Development, genetics and molecular biology of mosses. Bot. Acta 111, 1–15. [Google Scholar]
- Schaefer, D. (2001). Gene targeting in Physcomitrella patens. Curr. Opin. Plant Biol. 4, 143–150. [DOI] [PubMed] [Google Scholar]
- Schaefer, D., and Zryd, J.-P. (1997). Efficient gene targeting in the moss Physcomitrella patens. Plant J. 11, 1195–1206. [DOI] [PubMed] [Google Scholar]
- Schumaker, K.S., and Dietrich, M.A. (1997). Programmed changes in form during moss development. Plant Cell 9, 1099–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby, C.P., Thompson, C., Schmitz, T.M., Van Gelder, T.M., and Sancar, A. (2000). Functional redundancy of cryptochrome and classical photoreceptors for nonvisual ocular photoreception in mice. Proc. Natl. Acad. Sci. USA 97, 14697–14702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkle, J.R., and Jones, R.L. (1988). Inhibition of stem elongation in Cucumis seedlings by blue light requires calcium. Plant Physiol. 86, 960–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, P.E., and Neaf, J.B. (1981). Light dependency of cytokinin-induced bud initiation in protonemata of the moss Funaria hygrometrica. Physiol. Plant. 53, 13–18. [Google Scholar]
- Smith, H. (2000). Phytochromes and light signal perception by plants? An emerging synthesis. Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Devlin, P.F., and Kay, S.A. (1998). Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science 282, 1488–1490. [DOI] [PubMed] [Google Scholar]
- Stanewsky, R., Kaneko, M., Emery, P., Beretta, B., Wager-Smith, K., Kay, S.A., Rosbash, M., and Hall, J.C. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. [DOI] [PubMed] [Google Scholar]
- Tian, Q., and Reed, J.W. (1999). Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126, 711–721. [DOI] [PubMed] [Google Scholar]
- Triglia, T., Peterson, M.G., and Kemp, D.J. (1988). A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 16, 8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst, G.T.J., et al. (1999). Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature 398, 627–630. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Furuya, M. (1971). Photocontrol of the orientation of cell division in Adiantum. II. Effect of the direction of white light on the apical cell of gametophytes. Planta 98, 177–185. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Furuya, M. (1978). Effects of narrow-beam irradiations with blue and far-red light on the timing of cell division in Adiantum gametophytes. Planta 138, 85–90. [DOI] [PubMed] [Google Scholar]
- Wada, M., and Sugai, M. (1994). Photobiology of ferns. In Photomorphogenesis in Plants, 2nd ed, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 783–802.
- Wang, H., Ma, L.-G., Li, J.-M., Zhao, H.-Y., and Deng, X.W. (2001). Direct interaction of Arabidopsis cryptochromes with COP1 in mediation of photomorphogenic development. Science 294, 154–158. [DOI] [PubMed] [Google Scholar]
- Weller, J.L., Perrotta, G., Schreuder, M.E.L., van Tuinen, A., Koornneef, M., Giuliano, G., and Kendrick, R.E. (2001). Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochrome A, B1 and B2. Plant J. 25, 427–440. [DOI] [PubMed] [Google Scholar]
- Yang, H.-Q., Wu, Y.-J., Tang, R.-U., Liu, D., Liu, Y., and Cashmore, A.R. (2000). The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell 103, 815–827. [DOI] [PubMed] [Google Scholar]
- Zeidler, M., Hartmann, E., and Hughes, J. (1999). Transgene expression in the moss Ceratodon purpureus. J. Plant Physiol. 154, 641–650. [Google Scholar]