Abstract
Tandem repeat arrays often are found in interstitial (i.e., normally gene-rich) regions on chromosomes. In maize, genes on abnormal chromosome 10 induce the tandem repeats that make up knobs to move poleward on the meiotic spindle. This so-called neocentromere activity results in the preferential recovery, or meiotic drive, of the knobs in progeny. Here we show that two classes of repeats differ in their capacity to form neocentromeres and that their motility is controlled in trans by at least two repeat-specific activators. Microtubule dynamics appear to contribute little to the movement of neocentromeres (they are active in the presence of taxol), suggesting that the mechanism of motility involves microtubule-based motors. These data suggest that maize knob repeats and their binding proteins have coevolved to ensure their preferential recovery in progeny. Neocentromere-mediated drive provides a plausible mechanism for the evolution and maintenance of repeat arrays that occur in interstitial positions.
INTRODUCTION
Many plants and animals have long arrays of tandem repeats in interstitial positions on chromosome arms (John and Miklos, 1979; Rodionov, 1999). Two such repeats in maize, one that is 180 bp and another that is 350 bp (TR-1), occupy condensed regions known as knobs (Peacock et al., 1981; Dennis and Peacock, 1984; Ananiev et al., 1998a). Knobs are found at 22 different positions in the karyotype and are strikingly polymorphic, making them excellent cytological markers (Longley, 1938; Kato, 1984). They also have the capacity to behave like centromeres, or “neocentromeres,” in the presence of an unusual form of chromosome 10 (Rhoades and Vilkomerson, 1942). In strains carrying normal chromosome 10 (N10), the knobs are quiescent, whereas in strains carrying abnormal chromosome 10 (Ab10), knobs at all positions in the genome move rapidly poleward on the meiotic spindle, dragging their chromosome arms with them (Rhoades and Vilkomerson, 1942). The mechanism of neocentromere activity remains a mystery, although it is known that neocentromeres lack two major kinetochore proteins, CENPC and MAD2 (Dawe et al., 1999; Yu, 2000), and interact with microtubules in a lateral manner instead of in the end-on manner typical of maize centromeres (Yu et al., 1997).
Neocentromere activity plays an integral role in an associated phenotype known as meiotic drive. Meiotic drive has been documented in a variety of organisms (Lyttle, 1993), in which it is usually associated with several linked loci that collectively confer a segregation advantage to the linkage group. Meiotic drive systems presumably have evolved to “beat Mendel's rules” and therefore maximize their representation in the population (Sandler and Novitski, 1957). In some organisms, meiotic drive is a result of unusual chromosome segregation in meiosis (Rhoades, 1952; Cazemajor et al., 2000), and in others, it is caused by events that follow meiosis (Raju, 1996; Merrill et al., 1999). In maize, meiotic neocentromere activity at the large knob on Ab10 is thought to preferentially pull Ab10 toward the only functional (basal) megaspore of the linear tetrad (Rhoades and Vilkomerson, 1942; Dawe and Cande, 1996). Other knobs also form neocentromeres and are subject to meiotic drive in the presence (but not the absence) of Ab10 (Longley, 1945), suggesting that knobs on chromosomes other than 10 evolved secondarily in response to the meiotic drive system in maize (Buckler et al., 1999). One trans-acting mutation that affects neocentromeres (smd1) has been recovered from a screen for mutants of meiotic drive. smd1 is a cytologically undetectable mutation on Ab10 that acts to reduce neocentromere activity throughout the genome (Dawe and Cande, 1996).
In addition to being found in interstitial positions on chromosome arms, tandem repeat arrays in the size range of the maize knob repeats (180 and 350 bp; i.e., multiples of ∼180 bp) are found at telomeric locations and within or surrounding most higher eukaryotic centromeres (Charlesworth et al., 1994; Choo, 1997; Csink and Henikoff, 1998). In maize, the major centromeric repeat appears to be a tandemly arrayed ∼156-bp sequence known as CentC (Ananiev et al., 1998b). Considerable speculation has centered on whether such centromeric repeats function in chromosome segregation. Evidence in favor of their involvement in chromosome segregation comes from work in mammals suggesting that tandem arrays known as satellites can organize a functional centromere/kinetochore complex in artificial chromosomes (Brown et al., 2000). On the other hand, the near absence of sequence homology among satellites from different species and the fact that not all centromeres have satellite DNA suggest that the role of tandem arrays in kinetochore function is epigenetic (Karpen and Allshire, 1997). Although the role of centromeric tandem arrays remains uncertain, the similarities between “true” centromeres and maize neocentromeres suggest that the meiotic drive system may have co-opted structures and functions that normally are associated with the centromere/kinetochore complex.
Here we investigate the sequence dependence, genetic control, and mechanism of neocentromere activity. We show that there are clear motility differences between the two major neocentromere repeats in maize and that at least two different trans-acting factors encoded on Ab10 differentiate between the repeats. Ab10 effectively mobilizes neocentromeres on taxol-stabilized spindles, suggesting that microtubule dynamics contribute little to the poleward movement of either class of repeat. These data support a model whereby tandem arrays and sequence-specific binding proteins are coadapted to confer their preferential segregation to progeny, most likely via the activity of microtubule-based motor proteins. These observations are generalizable to many other species with long interstitial tandem arrays, and in principle they provide an explanation for the amplification, homogenization, and maintenance of such repeats.
RESULTS
TR-1 Occurs in Clusters on Knobs and Is a Major Component of the Three Chromomeres on Ab10
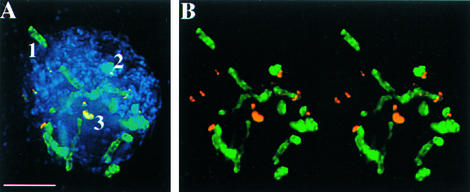
Maize lines vary with respect to knob number and size. Among the strains used here, the number of knobs varied from 2 (Knobless Wilbur's Flint [KWF]) to 13 (a strain from the Mescalero Apache Tribe). To determine how the 180-bp and TR-1 repeats are organized with respect to each other, we used three-dimensional light microscopy to analyze chromosomes at the prezygotene stage of meiotic prophase I. As shown in Figure 1, the knobs at prezygotene become long and extended such that they are amenable to substructural analysis (Dawe et al., 1994). Consistent with previous studies (Ananiev et al., 1998a), our data suggest that the TR-1 repeat occurs in long uninterrupted arrays. Many knobs are composed primarily of the 180-bp repeat (14 of 28 knobs in the strains used here), many are composed of a mixture of both repeats (10 of 28), and a minority are composed primarily of the TR-1 repeat (4 of 28). In mixed knobs, the 180-bp and TR-1 repeats appear to be separated into different domains within the knob (Figure 1).
Figure 1.
TR-1 and the 180-bp Repeat Occupy Discrete Domains in Knobs.
Images are projections of all of the optical sections in a cell at the prezygotene stage of meiotic prophase. The strain contained Ab10 and 13 other knobs.
(A) Three-color projection showing chromatin (blue), TR-1 (orange/yellow), and the 180-bp repeat (green). The knobs can be composed entirely of one repeat or can be mixed (1 = 180 bp only; 2 = mixed; 3 = TR-1 only).
(B) Stereo pair of (A) with only the knob labeling shown. The three chromomeres of Ab10 are visible in the upper left.
Bar in (A) = 5 μm for (A) and (B).
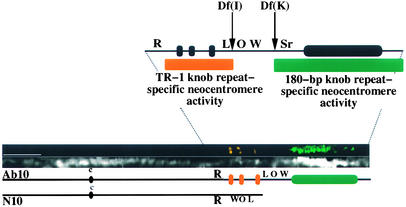
Based on knob distribution within the races of maize and teosinte (the presumed ancestor of maize), Buckler et al. (1999) argued that knobs evolved in response to the presence of Ab10 in the genome. Their hypothesis predicts that because both repeats are dispersed throughout the genome, Ab10 also will contain both repeats. Figure 2 illustrates the terminal portion of the long arm of Ab10 that is responsible for meiotic drive. It consists of a region with three prominent chromomeres: a central euchromatic region, a large knob, and a euchromatic tip (Rhoades and Dempsey, 1985). The central euchromatic region contains three known genes from N10 in an inverted orientation. The cytological features of Ab10 are best visualized in homozygous Ab10 strains at the pachytene substage of meiotic prophase I. When such pachytene chromosomes were hybridized with the TR-1 repeat, all three of the prominent chromomeres on Ab10 labeled brightly. In contrast, the large knob on Ab10 was composed almost entirely of the 180-bp repeat (as described by Peacock [1981]). With the sole exception that a small region containing the TR-1 repeat sometimes was visible near the tip of the large knob (data not shown), the chromomeres are essentially devoid of the 180-bp repeat and the large knob is essentially devoid of the TR-1 repeat (Figure 2). We conclude that Ab10 contains both knob repeats but that they are localized in separate regions of the chromosome.
Figure 2.
Structure and Genetics of the Ab10 Chromosome, Including the Repeat Composition of the Three Chromomeres and the Large Knob.
A computationally straightened Ab10 chromosome is shown in the middle of the figure, with the chromatin (4′,6-diamidino-2-phenylindole)-only image (bottom) separated from the in situ hybridization signals (top). In situ hybridization revealed that TR-1 (orange) occupies the three chromomeres and the 180-bp repeat (green) occupies the large knob. The scheme below shows a comparison of Ab10 and N10. The closely linked R locus is shown along with the relative positions of the genes L (luteus 13), O (opaque 7), and W (white 2). On Ab10, the region encompassed by the L, O, and W genes are inverted relative to N10. On the enlarged diagram of Ab10 (top), the breakpoints for the Df(I) and Df(K) deficiencies (Rhoades and Dempsey, 1985) and the locations of the repeat-specific neocentromere activators identified in this study are shown. The central euchromatin discussed in the text includes all of the chromatin between the chromomeres and the large knob. Bar = 5 μm.
TR-1 Repeats Form “Leaders” on Neocentromeres
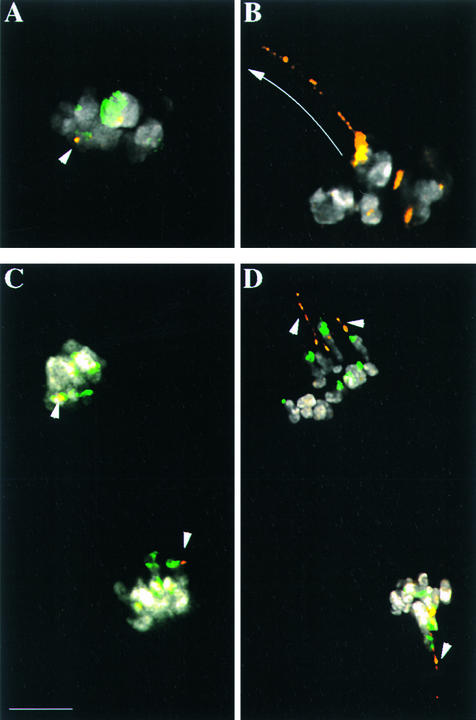
We used colocalization of the two knob repeats on neocentromeres to characterize their roles in neocentromere activity. As shown in Figures 3A and 3C, neither repeat array showed evidence of poleward motility in wild-type cells. In cells homozygous for Ab10, both classes of tandem repeats demonstrated poleward movement (Figures 3B and 3D). Neocentromeres were observed primarily in prometaphase (when spindles are formed) and anaphase of meiosis II, but they also were observed to a lesser degree in the same stages of meiosis I. Consistent with previous data, we found that arrays of the 180-bp repeat almost always were organized into compact globular domains on neocentromeres (Figure 3D). In contrast, arrays of the TR-1 repeat varied in shape from globular (rare), to visibly elongated with the spindle axis (common), to highly extended in long, thin threads stretching poleward (rare). In the most extreme cases, thread-like arrays of the TR-1 repeat followed the general outline of the spindle for more than 10 μm (Figures 3B and 3D). We were able to quantify the relative positions of the two different repeats by analyzing neocentromeres containing both repeats. An analysis of 95 mixed repeat neocentromeres indicated that in 77% of the cases the TR-1 repeat preceded the 180-bp repeat to the spindle pole. These data suggest that TR-1 repeats have a higher affinity for microtubules in the Ab10 background.
Figure 3.
The TR-1 Repeat Forms Neocentromere Leaders in the Presence of the Ab10 Chromosome.
Chromatin is shown in white, the 180-bp repeat is shown in green, and the TR-1 repeat is shown in orange. Images in (A) to (C) are single optical sections.
(A) Prometaphase II in an N10 background. No neocentromere activity is observed (arrowhead).
(B) Prometaphase II in a homozygous Ab10 background. A long TR-1 leader (>10 μm in length) is visible stretching toward the pole (arrow). In this cell, the 180-bp repeat was not labeled.
(C) Anaphase II in an N10 background. The knobs, which are located in interstitial positions on chromosome arms, lag behind the main mass of the chromosomes as they move poleward (arrowheads).
(D) Anaphase II in a heterozygous Ab10 background (image created by the projection of several optical sections). Neocentromeres led by long TR-1 leaders are visible (arrowheads).
Bar in (C) = 5 μm for (A) to (D).
The relative motility of a neocentromere is not necessarily related to the quantity of TR-1 repeats in the knob (Figure 3D). Rather, the position of a neocentromere in the spindle probably is a reflection of the overall size of the knob (Yu et al., 1997) and the relative proportion of the TR-1 repeat. We assume that each knob has its own characteristic motility pattern. This is consistent with the meiotic drive hypothesis of knob evolution (Buckler et al., 1999), which predicts that any particular knob is in competition only with other knobs at the same locus and not with the knobs at the ∼21 other sites in the genome.
Neocentromere Activity of the 180-bp and TR-1 Repeats Is Controlled Independently by Separate Loci on Ab10
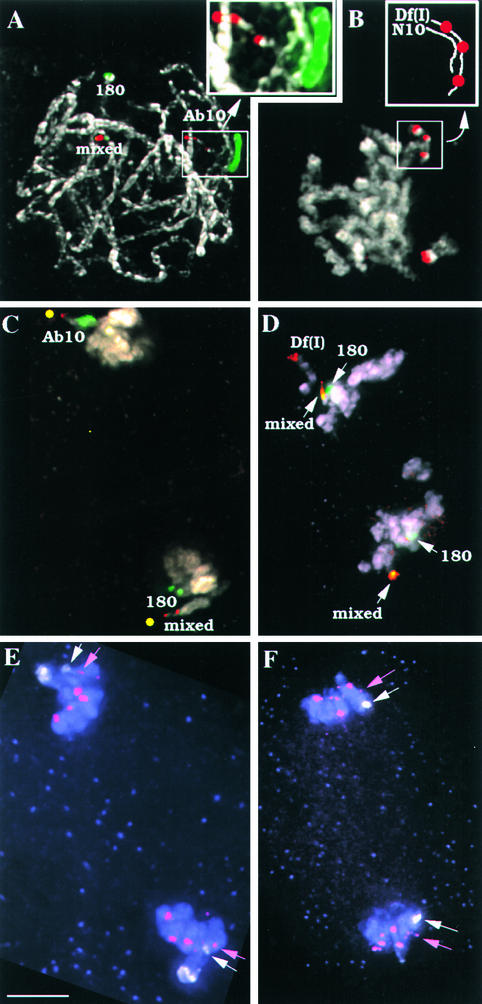
The deletion derivatives Ab10-Df(I) and Ab10-Df(K), which separate the chromomeres from the large knob on Ab10 (Rhoades and Dempsey, 1985), were used to determine whether the neocentromere activities of the knob repeats are under separate genetic control. As shown in Figures 2 and 4B, Df(I) has a breakpoint within the proximal portion of the central euchromatin and Df(K) has a breakpoint within the distal portion of the central euchromatin. Both deficiencies lack the large knob but contain the three chromomeres, portions of the central euchromatin, and any trans-acting functions that may be encoded there. To simplify the analysis, Ab10 and the two deletion derivatives were backcrossed to KWF, which contains two small knobs that are not linked to each other or to Ab10. As shown in Figure 4A, one of the knobs consists primarily of 180-bp repeats (referred to as the KWF-180 knob) and the second contains both repeats (referred to as the KWF-mixed knob). These small knobs can be distinguished readily from each other and from Ab10 (and its derivatives) when the knob repeats are labeled. Neocentromeres were scored during chromosome segregation at anaphase (I and II), when they could be detected in both halves of the spindle.
Figure 4.
Different Regions of the Ab10 Chromosome Regulate the 180-bp and TR-1 Repeats.
All images are from KWF backcrossed material. In (A) to (D), the chromatin is shown in white, the 180-bp repeat is shown in green, and the TR-1 repeat is shown in red. In (E) and (F), the chromatin is shown in lavender, the centromere repeat CentC is shown in magenta, and the 180-bp knob repeat is shown in white. Unless noted, all images are partial projections of several optical sections.
(A) Pachytene chromosomes from a heterozygous Ab10 plant. Shown is a full projection of all of the optical sections in the data set. The KWF-180 knob (180), the KWF-mixed knob (mixed), and Ab10 are indicated. The inset shows a ×2 enlargement of the Ab10 chromosome. The distal segment of Ab10 is unpaired and appears as a single thread of chromatin (the rest of the chromosomes are paired, as is characteristic of pachytene).
(B) Pachytene chromosomes from a heterozygous Df(I) plant. The Df(I) breakpoint is just distal to the three chromomeres. The inset shows a scheme of the distal tips of N10 and Df(I), which are paired (nonhomologously).
(C) Anaphase I from a heterozygous Ab10 plant. The spindle was curved in this cell; presumed spindle poles are indicated with yellow circles. A single Ab10 chromosome, two KWF-180 knobs, and two KWF-mixed knobs are shown moving poleward as neocentromeres.
(D) Anaphase II from a heterozygous Df(I) plant. The TR-1 chromomeres of the Df(I) chromosome can be seen moving poleward in one of the half spindles. The KWF-mixed knobs, but not the KWF-180 knobs, are moving poleward.
(E) Anaphase II in an Ab10 heterozygote. TR-1 is not labeled here; instead, the centromere repeat CentC is shown (magenta). The large 180-bp knob of Ab10 (white) is shown moving poleward. The KWF-180 knobs (white arrows) precede their linked centromeres to the poles (magenta arrows), as expected if the knobs are showing neocentromere activity.
(F) Anaphase II in a Df(K) heterozygote. The KWF-180 knobs (white arrows) lag behind their linked centromeres (magenta arrows), indicating the absence of neocentromere activity.
Bar in (E) = 5 μm for (A) to (F).
We observed high levels of neocentromere activity in heterozygous Ab10 plants, with Ab10 or the KWF-mixed knob showing a poleward orientation in all of 74 half-spindles and the KWF-180 knob showing neocentromere activity in 71 of the 74 (96%) half-spindles (Figure 4C). Knobs containing TR-1 repeats showed strong neocentromere activity in heterozygous Df(I) and Df(K) strains as well. The chromomeres of Ab10 and/or the KWF-mixed knobs showed poleward movement in all of 42 Df(I) and all of 35 Df(K) half-spindles. In contrast, neocentromere activity of the KWF-180 knob was reduced significantly in the deletion strains. A poleward orientation of the KWF-180 knob was observed in only 9 of 42 (21%) Df(I) half-spindles and 3 of 35 (9%) Df(K) half-spindles. These data suggest that a gene (or genes) involved in generating TR-1–mediated neocentromere activity lies proximal to the Df(I) breakpoint and that a second gene (or genes) that activates the 180-bp repeat is localized distal to the Df(K) breakpoint. An image illustrating the phenotype of the Df(I) derivative is shown in Figure 4D.
A subset of the Ab10 and Df(K) cells were double labeled with the 180-bp knob repeat and the centromere repeat CentC (Ananiev et al., 1998b); TR-1 was not stained. This labeling scheme and the use of three-dimensional light microscopy allowed us to locate (in many cases) the centromere of the chromosome carrying the KWF-180 knob. If the knob was located closer to the pole than the centromere at anaphase (I or II), the knob was considered to be showing neocentromere activity; if it lagged behind the centromere, it was scored as lacking neocentromere activity. In Ab10 strains, the KWF-180 knob was located in a poleward position relative to the centromere in 15 of 18 (83%) half-spindles. On the other hand, in the Df(K) strain, the same knob was located in a poleward position in only 1 of 27 (4%) half-spindles. These data confirm that the KWF-180 knob is relatively inactive in the presence of Df(K). Images illustrating the relative positions of the centromeres and KWF-180 knobs in the Ab10 and Df(K) strains are shown in Figures 4E and 4F.
Our observations indicate that there are at least two different genes that regulate neocentromere activity on Ab10 (Figure 2). The neocentromere activity of TR-1 in a Df(I) deletion strain suggests that a gene(s) sufficient for TR-1 motility lies proximal to the Df(I) breakpoint. The fact that the KWF-180 knob did not show neocentromere activity in a Df(K) deletion strain (but did in Ab10 controls) indicates that a gene(s) necessary for 180-bp repeat motility lies distal to the Df(K) breakpoint. However, we make these statements with some caution. Because the Df(I) strain used here was backcrossed only four times to KWF, the TR-1 neocentromere-promoting gene(s) cannot be mapped with certainty. We favor the location distal to the R locus (Figure 2) on Ab10 because neocentromeres have never been observed in strains lacking this region (Rhoades, 1952; Miles, 1970; Dawe and Cande, 1996). We also note that although the 180-bp gene(s) is not required for the motility of the TR-1 repeat, our data leave open the possibility that both the 180-bp repeat gene(s) and the TR-1 gene(s) are required to mobilize the 180-bp repeats.
Neocentromeres Are Insensitive to Perturbations in Microtubule Dynamics
On the basis of the asynchronous poleward movement of neocentromeres and their tangential interaction with spindles, we proposed previously that neocentromeres move poleward via the activity of microtubule-based motors (Yu et al., 1997). An alternate mechanism invoked to explain an example of poleward chromosome arm movement in crane flies (Adames and Forer, 1996) is that the chromosomes do not actively move poleward but instead are caught up in the poleward flux of tubulin subunits in the spindle (LaFountain et al., 2000). Flux occurs because tubulin is added primarily to the plus ends of microtubules and removed from the minus ends (Inoué and Salmon, 1995). Because the minus ends are directed toward the poles, flux generates a poleward force in the spindle. Under a flux model for neocentromere movement, Ab10 would recruit microtubule binding proteins but not necessarily motor proteins that pull the neocentromeres poleward. In an effort to differentiate between the motor-based and flux-based models for neocentromere motility, we treated intact anthers with the microtubule-stabilizing drug taxol (paclitaxel). Spindle dynamics can be slowed or stopped with taxol (Wilson et al., 1985; Bokros et al., 1993; Waters et al., 1996; LaFountain et al., 2000). We anticipated that if the differential poleward motility were a function of flux, taxol would cause a measurable reduction in neocentromere activity. Taxol is not expected to have any effect on motor activity per se, but it could generally slow the anaphase movement of the chromosomes (which may be driven in part by flux [Bajer et al., 1982; Waters et al., 1996]). The neocentromere-mediated stretching of chromosome arms could be increased by taxol if kinetochore-mediated movement is slowed but neocentromere activity is not.
In a previous study of mitosis in the higher plant Haemanthus, concentrations of taxol ranging from 10 nM to 50 μM were shown to disrupt the distribution of microtubules (Molè-Bajer and Bajer, 1983). Our experiments with maize anthers established that an ∼2-hr treatment with 5 to 30 μM taxol also induced reproducible and dramatic effects on meiotic spindles. The primary effects included aberrant spindle formation and extra microtubule arrays in prometaphase, a heavier packing of microtubules (visualized as a more uniform staining of tubulin), and a widening of the spindle midzone and premature clearing of microtubules from the future cell plate at anaphase (as if a phragmoplast were beginning to form at anaphase). These also were the primary effects of taxol noted by Bajer and colleagues in Haemanthus (Bajer et al., 1982; Molè-Bajer and Bajer, 1983). A noteworthy difference is that we observed more pronounced effects at prometaphase than did the previous authors. This discrepancy may be attributable to differences in spindle morphogenesis between the two cell types (Yu et al., 1999).
To test the effects of taxol on neocentromere activity, Ab10 was made homozygous in a race of maize from Arizona that has 13 knobs of various sizes (the strain is from the Mescalero Apache Tribe and was chosen for its high knob content) (Figure 1; Longley, 1938). Anthers in various stages of meiosis I and II were incubated in a culture medium with or without taxol for 2 hr. Then they were fixed, stained with anti-tubulin antibodies, and observed using three-dimensional light microscopy. For each experiment, we also immediately fixed anthers to determine if there were any effects of culturing the meiocytes on neocentromere activity (see Methods for details). The results are illustrated in Figure 5. Because neocentromere activity is inherently variable during prometaphase, we did not attempt to quantify the data at this stage. Nonetheless, it is clear from the images (Figures 5A and 5B) that the profound effects of taxol on spindle structure were not paralleled by a reduction in neocentromere activity. At anaphase (Figures 5D and 5E), we were able to quantify neocentromere number as well as the degree of chromosome stretching conferred by the neocentromeres, as shown in Table 1. There were no significant differences in neocentromere number among the three treatments, indicating that there were no effects of taxol at this level. Neocentromere-mediated stretching in anaphase II was significantly higher in taxol-treated cells compared with cultured (no taxol) cells but not compared with cells that were fixed immediately. Double labeling of the neocentromeres for both knob repeats after taxol treatment (Figure 5G) demonstrated that the knobs were stretched poleward at anaphase and that the relative orientation of the two classes of tandem repeats was not affected by taxol (i.e., the TR-1 leaders still were present).
Figure 5.
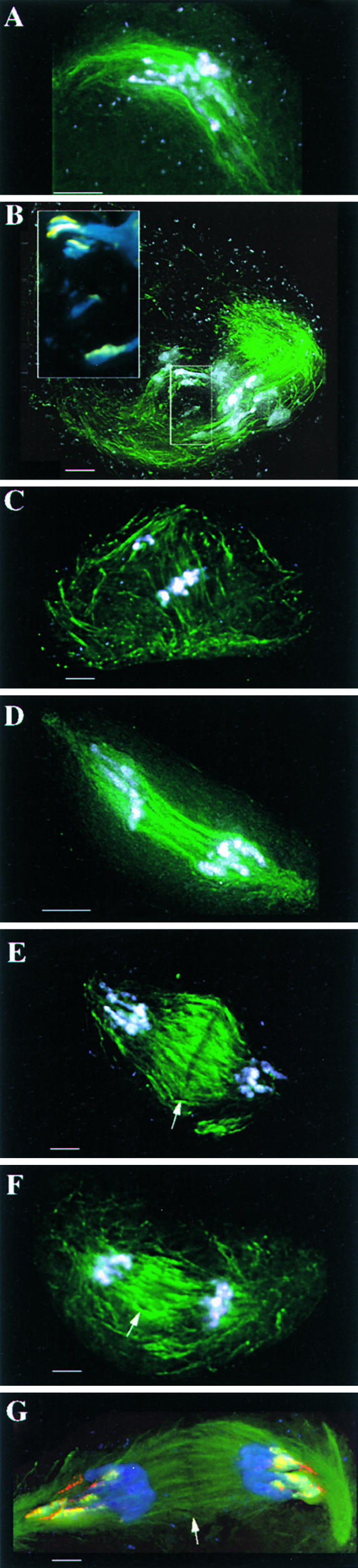
Taxol Does Not Impair Neocentromere Activity.
In (A) to (F), the chromatin is shown in white and the spindle (tubulin) is shown in green. All images are partial projections of several optical sections. In (E) to (G), arrows indicate the spindle midzones that have been cleared of microtubules prematurely (a characteristic effect of taxol treatment during anaphase). Bars = 5 μm.
(A) Prometaphase II cell from a homozygous Ab10 plant cultured without drug treatment. Note neocentromere activity.
(B) Prometaphase I cell from a homozygous Ab10 plant cultured with taxol. Note that the spindle morphology is aberrant but that there is no obvious difference in neocentromere activity. The inset (×2 magnification) shows the knob signal (yellow = the 180-bp repeat) at the tips of the chromosome arms (blue = chromosomes).
(C) Prometaphase II cell from a homozygous N10 plant cultured with taxol. Note the absence of neocentromere activity. In prometaphase, the chromosomes are not yet aligned, and in this case two chromosomes are separated from the others.
(D) Anaphase II cell from a homozygous Ab10 plant cultured without drug treatment. Note distinct neocentromere activity.
(E) Anaphase II cell from a homozygous Ab10 plant cultured with taxol, showing spindle morphology defects but no obvious effects on neocentromere activity.
(F) Anaphase II cell from a homozygous N10 control plant cultured with taxol. Note the absence of neocentromere activity.
(G) Anaphase II cell treated with taxol and stained for both knob repeats. Chromatin is shown in blue, spindle is shown in green, the 180-bp repeat is shown in yellow, and TR-1 is shown in red. Neocentromere leaders are still apparent after taxol treatment.
Table 1.
Effects of Taxol on Neocentromere Length and Number in a High-Knob Strain Homozygous for Ab10
| Fixeda
|
Cultured without Taxol
|
Cultured with Taxol
|
||||
|---|---|---|---|---|---|---|
| AIb | AII | AI | AII | AI | AII | |
| Neocentromere length μm ±sd |
7.3 ± 3.7 | 11.8 ± 2.6 | 10.2 ± 3.5 | 6.9 ± 2.14 | 9.1 ± 2.8 | 10.6 ± 2.3c |
| No. of cells analyzedd | 5 | 9 | 7 | 6 | 4 | 17 |
| Neocentromere number Mean no. of neocentromeres per cell ±sd |
9.7 ± 2.4 | 11.8 ± 4.9 | 11.1 ± 1.8 | 8.5 ± 2.3 | 9.0 ± 7.2 | 9.5 ± 2.6 |
| No. of cells analyzedd | 6 | 9 | 6 | 3 | 3 | 15 |
See Methods for a description of treatments.
AI, anaphase I; AII, anaphase II.
At anaphase II, neocentromere lengths in the taxol treatment were significantly different from those in the cultured treatment (P < 0.01) but not the fixed treatment. No significant differences were observed in anaphase I.
Data taken only from cells showing neocentromere activity.
The pronounced neocentromere activity observed in taxol treatments prompted us to consider whether taxol could induce neocentromeres in N10 strains. Therefore, in a second series of experiments, we investigated the effects of taxol in a family segregating for the presence or absence of Ab10 (the plants compared were full siblings). These data are shown in Figure 5 and Table 2. Fewer neocentromeres were observed in this study because there are fewer knobs in this strain (approximately eight) and because Ab10 induces fewer neocentromeres when heterozygous (Rhoades and Vilkomerson, 1942; Dawe and Cande, 1996). Consistent with the large body of evidence indicating that neocentromere activity is contingent on the presence of Ab10, taxol did not induce neocentromeres in N10 plants (Figures 5C and 5F). As for the effects in Ab10 plants (Table 2), taxol appeared to have a mild (but significant) enhancing effect on neocentromere number, although only when compared with cultured cells in anaphase II. There were no significant differences between the taxol-treated cells and those that were fixed immediately (Table 2).
Table 2.
Effects of Taxol on Neocentromere Number in Ab10 Heterozygotes and N10 Segregants
| Fixeda
|
Cultured without Taxol
|
Cultured with Taxol
|
||||
|---|---|---|---|---|---|---|
| Genotypeb | AIc | AII | AI | AII | AI | AII |
| N10/N10 Mean no. of neocentromeres per cell |
0 | 0d | 0 | 0 | N/Ae | 0 |
| No. of cells analyzed | 3 | 116 | 3 | 3 | N/A | 17 |
| Ab10/N10 Mean no. of neocentromeres per cell ±sd |
2.2 ± 1.2 | 2.0 ± 1.4 | 0.6 ± 0.5 | 1.4 ± 1.2 | N/A | 2.6 ± 1.6f |
| No. of cells analyzed | 6 | 23 | 15 | 53 | N/A | 64 |
| Percent of cells with neocentromeres | 100 | 83 | 40 | 72 | N/A | 88 |
a See Methods for a description of treatments.
b Plants were full siblings, varying by the presence of the Ab10 chromosome.
c AI, anaphase I; AII, anaphase II.
d A single neocentromere (or what appeared to be one) was detected in 116 cells.
e N/A, data not available.
At anaphase II, the number of neocentromeres in the taxol treatment was significantly different from the number in the cultured treatment (P < 0.01) but not the fixed treatment.
We conclude that taxol does not cause a measurable reduction in neocentromere activity, as would be predicted from the flux model. Taxol may cause a slight increase in neocentromere activity under some conditions (e.g., meiocyte culture), which is consistent with the motor-based model.
DISCUSSION
Here we present data indicating that two major knob repeats have different neocentromere activities and that the difference is controlled genetically by separate genes on Ab10. We also provide evidence from taxol-treated meiocytes that the microtubule dynamics of the spindle are not directly responsible for neocentromere motility. In this discussion, we review the evidence supporting our conclusions and discuss the implications with respect to the mechanism of neocentromere motility and the evolution of knob repeats.
Neocentromere Activities of Two Different Knob Repeats Are under Separate Genetic Control
The first information on the molecular composition of knobs was published over 20 years ago with the identification of the 180-bp satellite repeat (Peacock et al., 1981). More recently, Ananiev and co-workers (1998a) demonstrated the existence of a second repeat known as TR-1, which is found in many knobs in the maize genome. TR-1 is localized in large blocks apparently by itself or, more commonly, in close proximity to the 180-bp repeat. TR-1 also is found on the Ab10 chromosome, where it is a major component of three small knobs known previously as chromomeres (Rhoades, 1952). We show that, like the 180-bp repeat, the TR-1 repeat demonstrates distinct neocentromere activity. However, the neocentromere activity displayed by TR-1 is much more pronounced than that of the 180-bp repeat, often extending in long, thin threads toward the spindle poles (Figures 3B, 3D, and 5G). In knobs containing both repeats, the portion of the knob containing TR-1 is oriented toward the spindle pole in the majority of cases (77%). The 180-bp knob repeat appears to have a lower affinity or weaker interaction with fibers, but it is present in much greater abundance at knobs, which may serve to compensate for the slower/weaker neocentromere activity.
To determine the genetic basis for the differential neocentromere activity, we took advantage of a set of terminal deficiencies of Ab10 (Rhoades and Dempsey, 1985). The Ab10-Df(I) chromosome retains the three chromomeres of Ab10 and a small amount of the central euchromatin (Figure 2). In strains containing the Df(I) chromosome and two other knobs, we were able to determine that the truncated version of Ab10 was sufficient to confer motility to TR-1–containing knobs (Figure 4). By expanding the analysis to include the Ab10-Df(K) deficiency, which includes more of the central euchromatin but still lacks the large knob, we identified a second neocentromere-activating region distal to the breakpoint. The second region is not required for neocentromere activity of the TR-1 repeat, but it is required for the motility of the 180-bp repeat.
Our data indicate that at least two sequence-specific, trans-acting factors are required for neocentromere activity in maize: one that is sufficient to mobilize TR-1–containing knobs poleward, and a second that is necessary to move the 180-bp repeat poleward. The cytological data cannot tell us how many genes within each region are required for neocentromere activity, but we can conclude that the genes are linked to their respective repeat targets (Figure 2). Theoretical models and genetic data from other meiotic drive systems (Haig and Grafen, 1991) indicate that meiotic drive genes evolve in close linkage with their targets to reduce the possibility that they will be separated by recombination (which would eliminate their fitness advantage). The Ab10 chromosome appears to have two neocentromere-activating cassettes that independently confer neocentromere activity and meiotic drive to the two major knob repeats. Experiments are under way to test a prediction of this model, which is that the smd1 mutation (Dawe and Cande, 1996) specifically diminishes neocentromere activity at one or the other class of knob repeats. Why two independent drive systems evolved on the Ab10 chromosome is not yet clear. One possibility is that the second cassette evolved as a result of competition with Ab10 type II (Rhoades and Dempsey, 1985), which differs cytologically and does not appear to encode the TR-1–mediated drive system (Hiatt, 2000).
The Drosophila protein PROD, which binds specifically to the 1.686 g/cm3 satellite repeat (Török et al., 2000), provides a useful model for the maize neocentromere binding proteins. PROD localizes primarily to the centromeric heterochromatin (where the satellite is abundant; Platero et al., 1998), and prod mutants show chromosome condensation and segregation defects. Because the 1.686 g/cm3 satellite is not detectable in close Drosophila relatives, it has been argued that the essential condensation function of PROD evolved only recently (Csink and Henikoff, 1998). Although PROD is not a conserved protein, qualitatively similar proteins may recruit motility proteins such as the C-terminal kinesins (discussed below) to the different classes of satellite repeats. The fact that neocentromere activity is sequence specific will facilitate the identification of these factors.
Mechanism of Neocentromere Activity
On the basis of their asynchronous movement and lateral interaction with spindle fibers, Yu and co-workers (1997) proposed that neocentromeres are mobilized by microtubule-based motors. However, a recent report on crane flies, which shows occasional chromosome arm motility at anaphase (Adames and Forer, 1996), seemed to cast doubt on this interpretation. The authors suggested that chromosome arm movement in crane flies is a passive outcome of microtubule flux (LaFountain et al., 2000), which is a poleward force caused by the depolymerization of tubulin at the poles (Sawin and Mitchison, 1994; Waters et al., 1996). Indeed, plants appear to display a poleward flux of the type found in other organisms (Bajer and Molè-Bajer, 1963; Hard and Allen, 1977). There also is evidence from maize suggesting that acentric fragments (produced by the maize mutant absence of first division 1) move poleward at metaphase and anaphase of meiosis II (Yu and Dawe, 2000), suggesting that a poleward force may operate during the period in which neocentromeres are active. To test the idea that neocentromeres are mobilized by microtubule dynamics, we conducted an experiment using the microtubule-stabilizing drug taxol. Taxol treatments sufficient to induce gross alterations in spindle architecture caused no apparent reduction in the extent or number of neocentromeres (Figures 5B and 5E, Table 1), suggesting that microtubule dynamics contribute little to neocentromere motility. A slight enhancing effect of taxol on neocentromere activity (Tables 1 and 2) can be explained if taxol impairs kinetochore-mediated movement (Bajer et al., 1982; Waters et al., 1996) but has no effect on neocentromere movement.
The fact that neocentromeres are pronounced on the most disturbed spindles supports the view espoused previously (Yu et al., 1997) that Ab10 provides or recruits proteins to neocentromeres that generate their movement de novo. We believe that the best candidates for proteins that generate neocentromere motility are likely to be found in the kinesin superfamily (dynein, the only other known microtubule-based motor, is absent in flowering plants [Lawrence et al., 2001]). The kinesins have varied and robust roles in chromosome movement, both at the centromere/kinetochore and along chromosome arms (Sharp et al., 2000). Because neocentromeres move poleward (toward the minus ends of microtubules), we expect that the motor will be minus end directed. The minus end–directed kinesins are limited to a class of kinesins in which the motor domain is located near the C terminus of the protein (Endow and Waligora, 1998). On the basis of sequence similarity, there are a large number of plant kinesins with the C-terminal arrangement, most of which are uncharacterized (Vos et al., 2000; Lawrence et al., 2002).
Evolution of Knob Repeats
Of all the known repetitive elements in higher eukaryotes, long tandem arrays in interstitial positions are perhaps the most difficult to explain from an evolutionary perspective (Charlesworth et al., 1994). Population genetic models that assume that tandem repeats have no function and impart a slight fitness cost predict the inevitable loss of repeats through the action of recombination and genetic drift (Charlesworth et al., 1986; Stephan, 1986). Even when tandem arrays are considered to be neutral, the repeats should be lost as a result of intrastrand recombination between repeat units (Walsh, 1987). The rate of loss is dependent on the rate of crossing over, so that repeats are expected to persist only long enough to be observed in chromosomal locations (i.e., around centromeres) in which there is reduced recombination. In contrast to the expectations from theory, long tandem arrays are surprisingly common in interstitial regions, where recombination is typically high (John and Miklos, 1979). Such interstitial arrays are found throughout the angiosperms and in some animals, in which they are usually visible as knob-like domains of heterochromatin (Vershinin et al., 1995; Schmidt and Heslop-Harrison, 1996; Chen et al., 1997; Niedermaier and Moritz, 2000; Cheng et al., 2001).
In maize, the evolution of knobs is tied intimately to the meiotic drive system encoded on Ab10 (Buckler et al., 1999). As described by Rhoades (1952), the mechanism of meiotic drive in maize can be summarized as follows. First, recombination occurs between the Ab10 drive system and the centromere to create heteromorphic dyads (containing one chromatid with and one chromatid without a knob). Second, neocentromere activity pulls knobs quickly to the spindle poles. Therefore, knobs are positioned so that as the meiosis II spindle is assembled, the knobs tend to segregate to the upper and lower (basal) megaspores and not to the two central spores. In female flowers of maize, only the basal megaspore of meiosis survives to become a megaspore. All of the ∼22 other knobs appear to have evolved through a similar mechanism by exploiting the factors encoded by Ab10 (Buckler et al., 1999). Once a knob is established at a site, there is selection for larger knobs because they are driven more effectively (Kikudome, 1959). Hence, knobs evolved at positions >50 centimorgan from the centromeres (to maximize recombination) and contain thousands of copies of the same knob repeats that are present on Ab10 (Buckler et al., 1999).
The proposed mechanism of meiotic drive in maize is very similar to (and in fact was based on) examples in Drosophila in which chromosomes or segments of chromosomes are distributed nonrandomly to progeny for structural reasons. Because in animals, as in plants, only one product of meiosis (the pronucleus) becomes a female gamete, chromosomes that tend to drag during meiosis I can be excluded from gametes (reviewed by Novitski, 1967). This concept of “chromosomal meiotic drive” has been expanded to incorporate the idea that centromeric DNA and associated proteins may compete to confer an orientation on the spindle that directs the chromosome to the pronucleus (Zwick et al., 1999; Malik and Henikoff, 2001). Specifically, it has been argued that the Drosophila centromeric histone CID/CENP-A and the microtubule-based motor NOD are evolving in response to rapid changes in centromeric satellite DNAs and that the mechanism of adaptive evolution may involve processes akin to chromosomal meiotic drive (Zwick et al., 1999; Malik and Henikoff, 2001). We see clear parallels to this situation in the evolution of maize knobs. In the case of Ab10, at least two different knob arrays have evolved, and the mechanism of amplification and spread is based to a large extent on meiotic drive. The two knob repeats have 40% similarity overall, with two small regions of 31 and 12 bp that show 64 and 76% homology, respectively (Ananiev et al., 1998a), suggesting that they had a common ancestor. We provide evidence that trans-acting factors have evolved in concert with tandem repeats to confer their preferential orientation on the spindle, consistent with the ideas presented by Zwick and co-workers (1999) and Malik and Henikoff (2001).
Is neocentromere-mediated drive a general mechanism for the evolution of interstitial tandem arrays? Evidence from a variety of sources suggests that the answer may be yes. Knobs in interstitial positions appear to have evolved many times in nature (Rodionov, 1999), and in general they tend to be highly polymorphic (Döbel et al., 1973; Vosa, 1973; Árnason, 1974; Marks and Schweizer, 1974; Lelley et al., 1978; Belyayev et al., 1995). Neocentromeres have been observed in at least eight plant species, including the “enormous stretching of the chromatin” in Pennisetum orientale and the “fantastically elongated” chromosomes in Elymus wiegandii (Vilkomerson, 1950; Walters, 1952; Hayman, 1955; Zohary, 1955; Bosemark, 1956; Vardhan and Lakshmi, 1983; Viinikka, 1985). In maize, rye, Pennisetum (millet), and Festuca (meadow fescue), the sites of neocentromere activity appear to be knobs. Dramatic neocentromeres also are found during meiosis in the parasitic nematode Parascaris univalens (Goday and Pimpinelli, 1989), and the sites of neocentromere activity are large knobs composed primarily of two classes of tandem repeat arrays (Niedermaier and Moritz, 2000). There also is an intriguing meiotic and mitotic drive associated with expansions of large trinucleotide repeat arrays at the myotonic dystrophy type I locus in humans (Chakraborty et al., 1996; Khajavi et al., 2001) that may have a similar mechanistic basis.
It is likely that many other examples of neocentromere activity exist in nature and that subtle cases have not been noticed. Even a slight transmission advantage would be sufficient to drive a chromosome segment to fixation (in the absence of other forces). In addition, the neocentromere activity that led initially to the amplification and spread of a tandem array may now be present only in subpopulations or may be suppressed by host modifiers that have evolved to counteract the negative fitness consequences associated with meiotic drive (Ardlie, 1998). The idea that neocentromere-mediated meiotic drive systems may be genetically isolated or suppressed is supported by the observation that in some species neocentromeres appear to be activated only in hybrids (Walters, 1952; Vardhan and Lakshmi, 1983).
Modeling experiments suggest that unequal recombination is not sufficient to account for long repeat arrays and that some bias toward gene amplification is required (Walsh, 1987). Neocentromere-mediated meiotic drive provides a mechanical basis for the predicted bias in regions of the chromosome with typically high recombination (reduced recombination may impart a bias in pericentromeric regions) (Walsh, 1987; Charlesworth et al., 1994). Selection for meiotic drive can oppose the action of recombination and genetic drift and can lead to the expansion of the arrays. The high turnover of repeats between closely related species, which has been cited as evidence for their evolution via recombination and drift (Walsh, 1987; Charlesworth et al., 1994), also is compatible with a meiotic drive model in which the repeats and their binding proteins are evolving rapidly in concert (Malik and Henikoff, 2001). Meiotic drive can not only provide the selective force for the persistence and rapid expansion of tandem arrays (Buckler et al., 1999), but the sequence specificity of the drive systems can serve as a homogenizing force that reduces variation among the individual repeats in an array.
METHODS
Maize Strains
Abnormal chromosome 10 (Ab10) was made homozygous in either the W23 inbred line (with 5 knobs on other chromosomes) or a race of maize (Zea mays) from the Mescalero Apache Tribe in the American Southwest that segregates at least 13 knobs (Plant Introduction number 213736; North Central Regional Plant Introduction Station, Ames, IA). Another line of mixed origin, containing eight knobs, was used to produce homozygous normal chromosome 10 (N10) and heterozygous Ab10 siblings for the comparative study (Table 2).
Ab10 and the deletion derivatives Df(I) and Df(K) (Rhoades and Dempsey, 1985) were also backcrossed to Knobless Wilbur's Flint (KWF; obtained from the Maize Genetics Cooperation Stock Center, University of Illinois, Urbana), which has only two small knobs. Df(I) was backcrossed three times (i.e., introgressed four times), Df(K) was backcrossed five times, and Ab10 was backcrossed six times to KWF. The resulting strains carried only the two small KWF knobs and one copy of Ab10, Df(I), or Df(K).
All plants processed for the analysis of neocentromere activity were grown in the Department of Botany greenhouses of the University of Georgia during the months of December through March. We have found that winter months produce the most consistent neocentromere activity. Even at this time of year, however, many cells do not show cytologically visible neocentromere activity. Only those cells that showed at least one neocentromere were included in the analysis (except when wild-type cells were included in the analysis, as shown in Table 2; in this case, all of the data are shown).
In Situ Hybridization and Immunolocalization
Anthers were dissected out of tassels and fixed in modified 1 × buffer A plus 0.32 M sorbitol (80 mM KCl, 20 mM NaCl, 0.5 mM EGTA, 2 mM EDTA, and 15 mM Pipes buffer, pH 7.0) with 4% paraformaldehyde and 0.1% Triton X-100 for a minimum of 2 hr under ambient conditions (Dawe et al., 1994). In situ hybridization was performed as described previously (Dawe and Cande, 1996; Yu et al., 1997; Dawe et al., 1999). Immunolocalization of spindles was performed using a primary antibody to α-tubulin (Asai et al., 1982) as described previously (Yu et al., 1997; Dawe et al., 1999).
Hybridization Probes
The 180-bp knob repeat probe consisted of a mixture of 10 fluorescein isothiocyanate–labeled oligonucleotides, each 18 nucleotides in length (Yu et al., 1997). The oligonucleotide sequences are nonoverlapping and together cover the entire repeat (Peacock et al., 1981; Dennis and Peacock, 1984). For each cover slip, the probe mixture contained 0.8 ng/μL of each oligonucleotide, giving a total DNA concentration of 8.0 ng/μL. For Figure 5G, the 180-bp repeat was labeled with a rhodamine-labeled oligonucleotide as well as with the fluorescein isothiocyanate–labeled oligonucleotides (making the knob appear yellow on the merged image).
Two different probes were used for the TR-1 repeat. For the first probe, polymerase chain reaction primers (5′-CAGTTCACTCAC-ACAATTTGGC-3′ and 5′-GATGTTTCCCTGATGTCAAGGG-3′) were used to amplify the TR-1 knob repeat from the maize inbred line W23. The polymerase chain reaction product was labeled using a random priming kit (Yu et al., 1997). The second probe for the TR-1 repeats consisted of five rhodamine-labeled oligonucleotides, each 23 nucleotides in length. The TR-1 repeat probes covered approximately one-third of the total sequence. The individual oligonucleotide sequences were as follows: (1) 5′-TTAGAGTACAACTAGTGGATG-AA-3′; (2) 5′-GTTTCCTATAATCCCCTCTATTC-3′; (3) 5′-TCCACTCAAGTAAAACACCACAC-3′; (4) 5′-GAACTGTCCAAACATAGGTTA-AG-3′; and (5) 5′-AAGTTGGAATATAAAGAATTCAA-3′. For each cover slip, the probe mixture contained 0.2 ng/μL of each oligonucleotide, giving a total DNA concentration of 1.0 ng/μL.
A probe for the maize centromere repeat CentC (Ananiev et al., 1998b) was used to localize the centromere. The CentC probe was composed of eight fluorescein isothiocyanate–labeled oligonucleotides, each 18 nucleotides in length. The nonoverlapping oligonucleotides essentially cover the entire repeat. For each cover slip, the probe mixture contained 1.3 ng/μL of each oligonucleotide, giving a total DNA concentration of 10.4 ng/μL.
Taxol Study
The three anthers of a floret were removed and placed into one of three different treatments. Treatments were as follows: (1) immediately fixed (in PHEMS buffer [60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, 0.32 M Sorbitol, pH 6.8] with 4% paraformaldehyde and 0.1% Triton X-100) for a minimum of 2 hr under ambient conditions (Dawe and Cande, 1996); (2) placed in meiocyte culture medium (Yu et al., 1997) with no taxol added; and (3) placed in culture medium plus 5, 10, or 30 μM taxol (from a 10-mM stock solution of paclitaxel [Sigma catalog number T-7402] dissolved in DMSO). For treatments 2 and 3, the anthers were shaken gently in 400 μL of medium for 90 to 150 min and then fixed in the same manner as the anthers in treatment 1. Anthers were maintained and tracked individually. By comparing 12 to 24 florets per experiment (eight experiments total), we were able to obtain good representation of all of the relevant stages of meiosis. For the data in Table 1, neocentromere length was measured as the total distance from the plateward side of the closest chromosome to the poleward end of the most advanced neocentromere. Length measurements and neocentromere counts were analyzed for statistical significance using one-way analysis of variance and t tests.
Microscopy and Image Analysis
All data except those in Table 2 were collected using a DeltaVision multidimensional light microscope system (Applied Precision, Inc., Issaguah, WA; for specific methods, see Yu et al., 1997). Software included with the DeltaVision system provided the three-dimensional constrained iterative deconvolution algorithms that were used to remove out-of-focus information from the raw data. The deconvolved images were scaled to improve contrast. DeltaVision software also was used to model and straighten pachytene chromosomes (Dawe et al., 1994).
Acknowledgments
We thank Edward Buckler for originally suggesting that we check for the localization of TR-1 on Ab10. This work was supported by a grant from the National Science Foundation (9513556) to R.K.D. Additional support was provided to E.N.H. and E.K.K. by a National Science Foundation interdisciplinary research training grant (BIR9220329).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010373.
References
- Adames, K.A., and Forer, A. (1996). Evidence for polewards forces on chromosome arms during anaphase. Cell Motil. Cytoskeleton 34, 13–25. [DOI] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998. a). A knob-associated tandem repeat in maize capable of forming fold-back DNA segments: Are chromosome knobs megatransposons? Proc. Natl. Acad. Sci. USA 95, 10785–10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananiev, E.V., Phillips, R.L., and Rines, H.W. (1998. b). Chromosome-specific molecular organization of maize (Zea mays L.) centromeric regions. Proc. Natl. Acad. Sci. USA 95, 13073–13078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardlie, K.G. (1998). Putting the brake on drive: Meiotic drive of t haplotypes in natural populations of mice. Trends Genet. 14, 189–193. [DOI] [PubMed] [Google Scholar]
- Árnason, U. (1974). Comparative chromosome studies in Cetacea. Hereditas 77, 1–36. [DOI] [PubMed] [Google Scholar]
- Asai, D.J., Brokaw, C.J., Thompson, W.C., and Wilson, L. (1982). Two different monoclonal antibodies to tubulin inhibit the bending of reactivated sea urchin spermatozoa. Cell Motil. 2, 599–614. [DOI] [PubMed] [Google Scholar]
- Bajer, A., and Molè-Bajer, J. (1963). Cine-analysis of some aspects of mitosis in endosperm. In Cinemicrography in Cell Biology, G.G. Rose, ed (New York: Academic Press), pp. 357–409.
- Bajer, A.S., Christopher, C., Molè-Bajer, J., and Howard, H.M. (1982). Taxol-induced anaphase reversal: Evidence that elongating microtubules can exert a pushing force in living cells. Proc. Natl. Acad. Sci. USA 79, 6569–6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyayev, A., Punina, E., and Grif, V. (1995). Intrapopulation and individual polymorphism of heterochromatin segments in Trillium camschatcense Ker.-Gawl. Caryologia 48, 157–164. [Google Scholar]
- Bokros, C.L., Hugdahl, J.D., Hanesworth, V.R., Murthy, J.V., and Morejohn, L.C. (1993). Characterization of the reversible taxol-induced polymerization of plant tubulin into microtubules. Biochemistry 32, 3437–3447. [DOI] [PubMed] [Google Scholar]
- Bosemark, N.O. (1956). On accessory chromosomes in Festuca pratensis. IV. Cytology and inheritance of small and large accessory chromosomes. Hereditas 42, 235–260. [Google Scholar]
- Brown, R.A., Mee, P.J., and Shen, M.H. (2000). Artificial chromosomes: Ideal vectors? Trends Biotechnol. 18, 218–223. [DOI] [PubMed] [Google Scholar]
- Buckler, E.S.I., Phelps-Durr, T.L., Buckler, C.S.K., Dawe, R.K., Doebley, J.F., and Holtsford, T.P. (1999). Meiotic drive of chromosomal knobs reshaped the maize genome. Genetics 153, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazemajor, M., Joly, D., and Montchamp-Moreau, C. (2000). Sex-ratio meiotic drive in Drosophila simulans is related to equational nondisjunction of the Y chromosome. Genetics 154, 229–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, R., Stivers, D.N., Deka, R., Yu, L.M., Shriver, M.D., and Ferrell, F.E. (1996). Segregation distortion of the CTG repeats at the myotonic dystrophy locus. Am. J. Hum. Genet. 59, 109–118. [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Langley, C., and Stephan, W. (1986). The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112, 947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, B., Sneglowski, P., and Stephan, W. (1994). The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371, 215–220. [DOI] [PubMed] [Google Scholar]
- Chen, C.-M., Wang, C.-T., Wang, C.-J., Ho, C.-H., Kao, Y.-Y., and Chen, C.-C. (1997). Two tandemly repeated telomere-associated sequences in Nicotiana plumbaginifolia. Chromosome Res. 5, 561–568. [DOI] [PubMed] [Google Scholar]
- Cheng, Z., Stupar, R.M., Gu, M., and Jiang, J. (2001). A tandemly repeated DNA sequence is associated with both knob-like heterochromatin and a highly decondensed structure in the meiotic pachytene chromosomes of rice. Chromosoma 110, 24–31. [DOI] [PubMed] [Google Scholar]
- Choo, K.H.A. (1997). The Centromere. (New York: Oxford University Press).
- Csink, A.K., and Henikoff, S. (1998). Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 15, 200–204. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K., and Cande, W.Z. (1996). Induction of centromeric activity in maize by suppressor of meiotic drive 1. Proc. Natl. Acad. Sci. USA 93, 8512–8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe, R.K., Sedat, J.W., Agard, D.A., and Cande, W.Z. (1994). Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell 76, 901–912. [DOI] [PubMed] [Google Scholar]
- Dawe, R.K., Reed, L., Yu, H.-G., Muszynski, M.G., and Hiatt, E.N. (1999). A maize homolog of mammalian CENPC is a constitutive component of the inner kinetochore. Plant Cell 11, 1227–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, E.S., and Peacock, W.J. (1984). Knob heterochromatin homology in maize and its relatives. J. Mol. Evol. 20, 341–350. [DOI] [PubMed] [Google Scholar]
- Döbel, P., Rieger, R., and Michaelis, A. (1973). The Giemsa banding patterns of the standard and four reconstructed karyotypes of Vicia faba. Chromosoma 43, 409–422. [DOI] [PubMed] [Google Scholar]
- Endow, S.A., and Waligora, K.W. (1998). Determinants of kinesin motor polarity. Science 291, 1200–1202. [DOI] [PubMed] [Google Scholar]
- Goday, C., and Pimpinelli, S. (1989). Centromere organization in meiotic chromosomes of Parascaris univalens. Chromosoma 98, 160–166. [DOI] [PubMed] [Google Scholar]
- Haig, D., and Grafen, A. (1991). Genetic scrambling as a defence against meiotic drive. J. Theor. Biol. 153, 531–558. [DOI] [PubMed] [Google Scholar]
- Hard, R., and Allen, R.D. (1977). Behaviour of kinetochore fibres in Heamanthus katherinae during anaphase movements of chromosomes. J. Cell Sci. 27, 47–56. [DOI] [PubMed] [Google Scholar]
- Hayman, D.L. (1955). Centromeric behaviour of the univalents in two Phalaris hybrids. Aust. J. Biol. Sci. 8, 241–253. [Google Scholar]
- Hiatt, E.H. (2000). Cytogenetic Analysis of Meiotic Drive in Maize. PhD Thesis. (Athens, GA: University of Georgia).
- Inoué, S., and Salmon, E.D. (1995). Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John, B., and Miklos, G.L.G. (1979). Functional aspects of satellite DNA and heterochromatin. Int. Rev. Cytol. 58, 1–114. [DOI] [PubMed] [Google Scholar]
- Karpen, G.H., and Allshire, R.C. (1997). The case of epigenetic effects on centromere identity and function. Trends Genet. 13, 489–496. [DOI] [PubMed] [Google Scholar]
- Kato, Y.T.A. (1984). Chromosome morphology and the origin of maize and its races. Evol. Biol. 17, 219–254. [Google Scholar]
- Khajavi, M., Tari, A.M., Patel, N.B., Tsuji, K., Siwak, D.R., Meistrich, M.L., Terry, N.H.A., and Ashizawa, T. (2001). ‘Mitotic drive’ of expanded CTG repeats in myotonic dystrophy type 1 (DM1). Hum. Mol. Genet. 10, 855–863. [DOI] [PubMed] [Google Scholar]
- Kikudome, G.Y. (1959). Studies on the phenomenon of preferential segregation in maize. Genetics 44, 815–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain, J.R., Cole, R.W., and Rieder, C.L. (2000). Taxol inhibits pole-directed transport of acentric fragments in crane-fly spindles. Mol. Biol. Cell 11 (suppl.), 371a.. [Google Scholar]
- Lawrence, C.J., Morris, N.R., Meagher, R.B., and Dawe, R.K. (2001). Dyneins have run their course in plant lineage. Traffic 2, 362–363. [DOI] [PubMed] [Google Scholar]
- Lawrence, C.J., Malmberg, R.L., Muszynski, M.G., and Dawe, R.K. (2002). Maximum likelihood methods reveal conservation of function among closely related kinesin families. J. Mol. Evol. 54, 42–43. [DOI] [PubMed] [Google Scholar]
- Lelley, T., Josifek, K., and Kaltsikes, P.J. (1978). Polymorphism in the Giemsa C-banding pattern of rye chromosomes. Can. J. Genet. Cytol. 20, 307–312. [Google Scholar]
- Longley, A.E. (1938). Chromosomes of maize from North American Indians. J. Agric. Res. 56, 177–195. [Google Scholar]
- Longley, A.E. (1945). Abnormal segregation during megasporogenesis in maize. Genetics 30, 100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyttle, T.W. (1993). Cheaters sometimes prosper: Distortion of mendelian segregation by meiotic drive. Trends Genet. 9, 205–210. [DOI] [PubMed] [Google Scholar]
- Malik, H.S., and Henikoff, S. (2001). Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics 157, 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, G.E., and Schweizer, D. (1974). Giemsa banding: Karyotype differences in some species of Anemone and in Hepatica nobilis. Chromosoma 44, 405–416. [Google Scholar]
- Merrill, C., Bayraktaroglu, L., Kusano, A., and Ganetsky, B. (1999). Truncated RanGAP encoded by the Segregation Distorter locus of Drosophila. Science 283, 1742–1745. [DOI] [PubMed] [Google Scholar]
- Miles, J.H. (1970). Influence of Modified K10 Chromosomes on Preferential Segregation and Crossing Over in Zea mays. PhD Thesis (Bloomington, IN: Indiana University).
- Molè-Bajer, J., and Bajer, A.S. (1983). Action of taxol on mitosis: Modifications of microtubule arrangements and function of the mitotic spindle in Haemanthus endosperm. J. Cell Biol. 96, 527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermaier, J., and Moritz, K.B. (2000). Organization and dynamics of satellite and telomere DNAs in Ascaris: Implications for formation and programmed breakdown of compound chromosomes. Chromosoma 109, 439–452. [DOI] [PubMed] [Google Scholar]
- Novitski, E. (1967). Nonrandom disjunction in Drosophila. Annu. Rev. Genet. 1, 71–86. [Google Scholar]
- Peacock, W.J., Dennis, E.S., Rhoades, M.M., and Pryor, A.J. (1981). Highly repeated DNA sequence limited to knob heterochromatin in maize. Proc. Natl. Acad. Sci. USA 78, 4490–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero, J.S., Csink, A.K., Quintanilla, A., and Henikoff, S. (1998). Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 140, 1297–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju, N.B. (1996). Meiotic drive in fungi: Chromosomal elements that cause fratricide and distort genetic ratios. J. Genet. 75, 287–296. [Google Scholar]
- Rhoades, M.M. (1952). Preferential segregation in maize. In Heterosis, J.W. Gowen, ed (Ames, IA: Iowa State College Press), pp. 66–80.
- Rhoades, M.M., and Dempsey, E. (1985). Structural heterogeneity of chromosome 10 in races of maize and teosinte. In Plant Genetics, M. Freeling, ed (New York: Alan R. Liss), pp. 1–18.
- Rhoades, M.M., and Vilkomerson, H. (1942). On the anaphase movement of chromosomes. Proc. Natl. Acad. Sci. USA 28, 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov, A.V. (1999). Evolution of the chromosomal banding pattern. Russian J. Genet. 35, 215–227. [Google Scholar]
- Sandler, L., and Novitski, E. (1957). Meiotic drive as an evolutionary force. Am. Nat. 91, 105–110. [Google Scholar]
- Sawin, K.E., and Mitchison, T.J. (1994). Microtubule flux in mitosis is independent of chromosomes, centrosomes, and antiparallel microtubules. Mol. Biol. Cell 5, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, T., and Heslop-Harrison, J.S. (1996). High-resolution mapping of repetitive DNA by in situ hybridization: Molecular and chromosomal features of prominent dispersed and discretely localized DNA families from the wild beet species Beta procumbens. Plant Mol. Biol. 30, 1099–1114. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000). Microtubule motors in mitosis. Nature 407, 41–47. [DOI] [PubMed] [Google Scholar]
- Stephan, W. (1986). Recombination and the evolution of satellite DNA. Genet. Res. 47, 167–174. [DOI] [PubMed] [Google Scholar]
- Török, T., Gorjánácz, M., Bryant, P.J., and Kiss, I. (2000). Prod is a novel DNA-binding protein that binds to the 1.686 g/cm3 10 bp satellite repeat of Drosophila melanogaster. Nucleic Acids Res. 28, 3551–3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardhan, Z.V., and Lakshmi, N. (1983). A new triploid race of Pennisetum orientale Rich exhibiting chromosomal numerical mosaicism and neo-centric activity. Proc. Indian Acad. Sci. 92, 259–264. [Google Scholar]
- Vershinin, A.V., Schwarzacher, T., and Heslop-Harrison, J. (1995). The large-scale genomic organization of repetitive DNA families at the telomeres of rye chromosomes. Plant Cell 7, 1823–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viinikka, Y. (1985). Identification of the chromosomes showing neocentric activity in rye. Theor. Appl. Genet. 70, 66–71. [DOI] [PubMed] [Google Scholar]
- Vilkomerson, H. (1950). The unusual meiotic behavior of Elymus wiegandii. Exp. Cell Res. 1, 534–542. [Google Scholar]
- Vos, J.W., Safadi, F., Reddy, A.S., and Hepler, P.K. (2000). The kinesin-like calmodulin binding protein is differentially involved in cell division. Plant Cell 12, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosa, C.G. (1973). Heterochromatin recognition and analysis of chromosome variation in Scilla sibirica. Chromosoma 43, 269–278. [Google Scholar]
- Walsh, J. (1987). Persistence of tandem arrays: Implications for satellite and simple-sequence DNAs. Genetics 115, 553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, M.S. (1952). Atypical chromosome movement in meiotic anaphase of Bromus pitensis × B. marginatus. Am. J. Bot. 39, 619–625. [Google Scholar]
- Waters, J.C., Mitchison, T.J., Rieder, C.L., and Salmon, E.D. (1996). The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, L., Miller, H.P., Farrell, K.W., Snyder, B., Thompson, W.C., and Purich, D.L. (1985). Taxol stabilization of microtubules in vitro: Dynamics of tubulin addition and loss at opposite microtubule ends. Biochemistry 24, 5254–5262. [DOI] [PubMed] [Google Scholar]
- Yu, H.-G. (2000). The Maize Kinetochore: Composition, Structure and Roles in Meiotic Chromosome Segregation. PhD Thesis (Athens, GA: University of Georgia).
- Yu, H.-G., and Dawe, R.K. (2000). Functional redundancy in the maize meiotic kinetochore. J. Cell Biol. 151, 131–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Hiatt, E.N., Chan, A., Sweeney, M., and Dawe, R.K. (1997). Neocentromere-mediated chromosome movement in maize. J. Cell Biol. 139, 831–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H.-G., Muszynski, M.G., and Dawe, R.K. (1999). The maize homologue of the cell cycle checkpoint protein MAD2 reveals kinetochore substructure and contrasting mitotic and meiotic localization patterns. J. Cell Biol. 145, 425–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohary, D. (1955). Secondary centric activity in Lilium formosanum. Am. Nat. 89, 50–52. [Google Scholar]
- Zwick, M.E., Salstrom, J.L., and Langley, C.H. (1999). Genetic variation in rates of nondisjunction: Association of two naturally occurring polymorphisms in the chromokinesin nod with in-creased rates of nondisjunction in Drosophila melanogaster. Genetics 152, 1605–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]