Abstract
Mutations in the AXR1 gene result in a reduction in auxin response and diverse defects in auxin-regulated growth and development. In a previous study, we showed that AXR1 forms a heterodimer with the ECR1 protein. This enzyme activates the ubiquitin-related protein RUB1 in vitro. Furthermore, we showed that the Skp1-Cul1/Cdc53-F-box (SCF) subunit AtCUL1 is modified by RUB1 in vivo. In this report, we demonstrate that the formation of RUB-AtCUL1 is dependent on AXR1 and ECR1 in vivo. The expression of AXR1 and ECR1 is restricted to zones of active cell division and cell elongation, consistent with their role in growth regulation. These results provide strong support for a model in which RUB conjugation of AtCUL1 affects the function of SCF E3s that are required for auxin response.
INTRODUCTION
Indole-3-acetic acid (IAA or auxin) is an important regulator of plant growth and development. Auxin is implicated in many growth processes, ranging from embryogenesis to floral development (Davies, 1995). In addition, auxin plays a principal role in the control of cell division and elongation (Evans, 1984; Gray et al., 1998, 1999). Despite the importance of this hormone, the molecular mechanisms of auxin action are poorly defined. In an attempt to elucidate these mechanisms, several groups have taken a genetic approach by screening for Arabidopsis mutants with defects in auxin response (Hobbie et al., 1994). One of the mutants recovered in these screens, called axr1, has a pleiotropic phenotype related to decreased auxin response. Mutant plants have reduced apical dominance, fewer lateral roots and root hairs, low fertility, and reduced gravitropic response (Lincoln et al., 1990). In addition, members of the Aux/IAA and SAUR families of auxin-regulated genes are not expressed normally in the mutant, suggesting that AXR1 functions in auxin signal transduction (Abel et al., 1995; Timpte et al., 1995).
AXR1 encodes a subunit of a heterodimeric RUB-activating enzyme (del Pozo et al., 1998). Biochemical studies suggest that a protein called ECR1 is the second subunit in this enzyme (del Pozo et al., 1998). RUB (or NEDD8 in humans) is a small ubiquitin-related protein that is conjugated to cellular proteins via a pathway similar to the ubiquitin conjugation pathway. At present, the only known targets of RUB modification are members of the cullin family (del Pozo et al., 1998; Lammer et al., 1998; Osaka et al., 1998; del Pozo and Estelle, 1999; Liakopoulos et al., 1999; Wada et al., 1999). Cullins are subunits of an E3-ubiquitin ligase complex called the Skp1-Cul1/Cdc53-F-box (SCF). The other subunits of the complex are SKP1, RBX1, and an F-box protein (Patton et al., 1998; Kamura et al., 1999). The SCF promotes the transfer of ubiquitin from a ubiquitin-conjugating enzyme (E2) to a target protein. The function of RUB modification of cullin is unknown. Unlike ubiquitination, RUB modification of the cullin does not affect its metabolic stability. Instead, genetic and biochemical evidence suggests that RUB modification regulates SCF function or subcellular localization (Lammer et al., 1998; Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000).
Genetic studies in Arabidopsis indicate that a ubiquitin-protein ligase called SCFTIR1 is required for auxin response (Gray et al., 1999). This E3 is composed of the cullin AtCUL1, the SKP1-related protein ASK1 or ASK2, and an F-box protein called TIR1. Mutations in either TIR1 or ASK1 result in a defect in auxin response, suggesting that the response depends on the ubiquitination and degradation of specific targets. On the basis of these results, we have proposed a model in which the normal activity of SCFTIR1 depends on RUB1 modification of AtCUL1 (del Pozo and Estelle, 1999; Gray and Estelle, 2000).
In this article, we show that both AXR1 and ECR1 are expressed specifically in growing cells throughout the plant. Expression of a mutant version of ECR1 in Arabidopsis plants produces a phenotype similar to the axr1 mutant. Furthermore, both AXR1 and ECR1 are required for the RUB1 modification of AtCUL1. These results strongly suggest that AXR1-ECR1–dependent RUB modification of AtCUL1 is required for normal auxin response.
RESULTS
AXR1 Is Expressed in Meristems and Organ Primordia
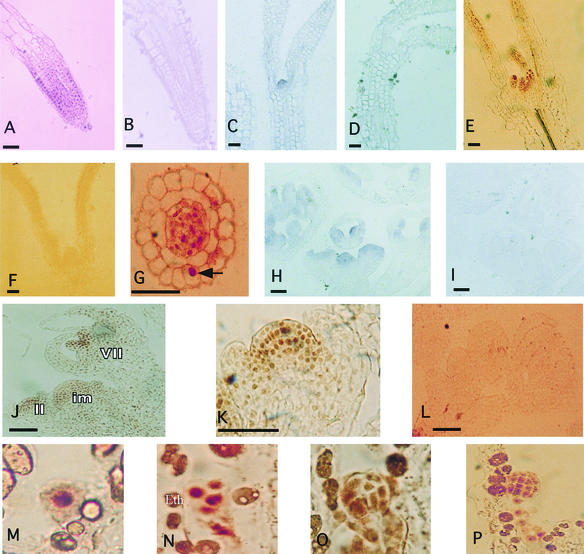
Mutations in the AXR1 gene affect the growth and development of most tissues in the plant (Lincoln et al., 1990; Cernac et al., 1997; Pitts et al., 1998). axr1 mutant seedlings display a number of defects, including an increase in the rate of primary root elongation, decreased hypocotyl elongation, and misshapen leaves (Lincoln et al., 1990). To learn more about the role of AXR1, we determined the pattern of expression by RNA in situ hybridization and/or immunolocalization of the AXR1 protein. In every case, the patterns of protein and mRNA accumulation were identical, suggesting the absence of post-transcriptional regulation of AXR1 expression. AXR1 RNA accumulated in the root and shoot meristems and in young leaves (Figures 1A and 1C). Similarly, the AXR1 protein was found in the shoot meristem and leaf primordium (Figure 1E) and in the root meristem (Figure 1G and data not shown). As reported previously, AXR1 was found primarily in the nuclei of these cells (Figures 1E and 1G) (del Pozo et al., 1998). This was seen most clearly in a section near the root tip (Figure 1G), where staining was observed in a single nucleus in the epidermal layer and in a number of nuclei in the stele. Figures 1B and 1D show the sense RNA control. Figure 1F shows AXR1 antibody staining of an axr1-12 seedling.
Figure 1.
The AXR1 Gene Is Expressed in Dividing and Elongating Cells throughout the Plant.
Expression was determined by in situ localization of RNA ([A] to [D], [H], and [I]) and immunolocalization of the AXR1 protein ([E] to [G] and [J] to [P]).
(A) Root meristem hybridized with antisense AXR1 RNA.
(B) Root meristem hybridized with sense AXR1 RNA.
(C) Seven-day-old seedling hybridized with antisense AXR1 RNA. Staining is visible on the leaf primordium.
(D) Seven-day-old seedling hybridized with sense AXR1 RNA.
(E) Section of a shoot meristem stained with antibodies against AXR1.
(F) Section of an axr1-12 shoot meristem stained with antibodies against AXR1.
(G) Cross-section of the root elongation zone stained with antibodies against AXR1. The arrow indicates a stained nucleus in the epidermis.
(H) Inflorescence and floral meristems hybridized with antisense AXR1 RNA.
(I) Inflorescence and floral meristems hybridized with sense AXR1 RNA.
(J) Inflorescence meristem (im) and floral meristems stained with AXR1 antibodies. II and VII indicate stages of flower development.
(K) Higher magnification of a stage II flower stained with AXR1 antibodies.
(L) Inflorescence meristem and flower section of axr1-12 stained with AXR1 antibodies.
(M) Wild-type zygote cell stained with AXR1 antiserum.
(N) Proembryo stage stained with AXR1 antibodies. The brown color in the endothecium (Eth) is not the result of AXR1 staining.
(O) Forty-hour-old embryo stained with AXR1 antibodies.
(P) Early globular embryo stained with AXR1 antibodies. Stained nuclei in the suspensor cell are detected.
Bars in (A) to (L) = 50 μm.
The inflorescence of axr1 plants is shorter and bushier than that of the wild type, indicating a reduction in apical dominance (Lincoln et al., 1990; Stirnberg et al., 1999). In addition, axr1 flowers are smaller than wild-type flowers. In accordance with these defects, AXR1 RNA (Figure 1H) and protein (Figures 1J and 1K) levels were high in the inflorescence meristem, the floral meristem, and developing flowers. In flowers at stage 2, AXR1 staining was highest in the floral apex and lower in developing sepals (Figures 1J and 1K). In stage 7 flowers, the strongest staining was observed in the developing gynoecium and medial stamen (Figure 1J). The specificity of AXR1 immunolocalization was confirmed by labeling axr1-12 floral sections with the AXR1 antiserum (Figure 1L). Figure 1I shows the mRNA sense control.
The overall structure of the plant, including the establishment of the apical–basal axis and the formation of the shoot, root, and cotyledons, is determined during embryogenesis. Because auxin has been implicated in these processes, we examined the pattern of AXR1 accumulation during embryogenesis. High levels of AXR1 protein were found in the zygote and throughout embryogenesis (Figures 1M to 1P and data not shown). A similar distribution of AXR1 RNA was observed (data not shown).
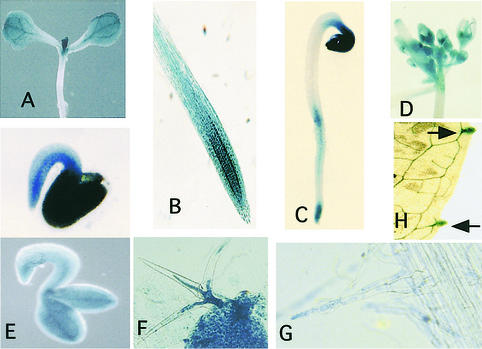
To further study AXR1 gene expression, we placed the β-glucuronidase reporter gene (GUS) adjacent to the promoter region of the AXR1 gene and introduced this construct into Arabidopsis plants (Jefferson et al., 1987). In general, the results of these experiments were similar to those obtained by RNA hybridization and immunolocalization indicating that GUS staining accurately reflects the activity of the AXR1 promoter. For example, intense GUS staining was observed in the shoot, root, and floral meristems (Figures 2A, 2B, and 2D; cf. with Figures 1C, 1A, and 1H, respectively). Several features of the staining pattern were particularly noteworthy. In 3-day-old etiolated seedlings, GUS staining was more intense on the underside of the apical hook region, suggesting that AXR1 is expressed differentially in this region of the hypocotyl (Figure 2C). This is consistent with the absence of an apical hook in the axr1-12 mutant (Cernac et al., 1997). Differential staining also was observed in the emerging root during germination, with more intense staining on the underside of the root (Figure 2E). Staining also was observed in the trichomes, root hairs, and hydathodes, suggesting a role for AXR1 in these cells (Figures 2F to 2H). Previous studies have demonstrated a root hair defect in axr1 plants (Pitts et al., 1998). Defects in hydathode or trichome development have not been documented.
Figure 2.
Pattern of AXR1 Expression as Revealed by Staining AXR1::GUS Seedlings.
(A) GUS staining of a 7-day-old light-grown seedling.
(B) GUS staining of a 7-day-old light-grown root.
(C) Three-day-old dark-grown seedling. Note the higher expression of AXR1 on the lower side of the apical hook than on the upper zone.
(D) Developing flowers.
(E) A 0.5-day-old seedling. The bottom panel shows the same seedling after mechanically removing the seed coat.
(F) Primary leaf of a 7-day-old light-grown seedling stained for GUS activity. Note that AXR1 is highly expressed in the young developing trichomes.
(G) Root hairs at the beginning of the differentiation zone of a 7-day-old seedling.
(H) Mature leaf (20 days old) of a light-grown plant stained for GUS activity. The arrows indicate the hydathodes.
To determine if transcription of the AXR1 gene responds to auxin treatment, transgenic seedlings were treated with 1 mM 2,4-D for 14 hr before staining. No differences were observed between these seedlings and control seedlings (data not shown). Similar results were obtained in RNA gel blot and GeneChip experiments using RNA from control and auxin-treated seedlings (data not shown; V. Godoy and M. Estelle, unpublished results).
Physiological studies have demonstrated a role for auxin in vascular development, and defects in vascular morphology have been reported in the axr1 mutants (Lincoln et al., 1990). Consistent with this finding, we observed GUS staining in vascular tissues in cotyledons and mature leaves (Figures 2A and 2H). In addition, the AXR1 protein was detected in the stele of the root (Figure 1G).
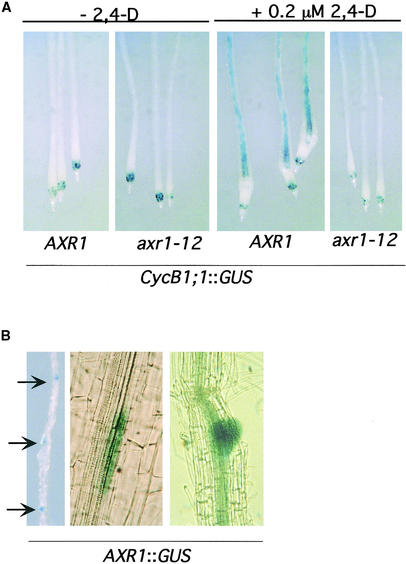
It is well known that auxin has an important role in lateral root development. Exogenous auxin stimulates the development of adventitious roots and lateral roots. In addition, the auxin response mutants axr1, axr4, and tir1 have a reduced number of lateral roots compared with those of the wild type (Lincoln et al., 1990; Hobbie and Estelle, 1995; Ruegger et al., 1998). In a previous study, we used a CycB1;1::GUS translational fusion to study the function of TIR1 in lateral root formation. Expression of this mitotic cyclin occurs in the lateral root founder cell before the first division; therefore, it is a convenient marker for the formation of lateral root primordia (Colon-Carmona et al., 1999). To assess the role of AXR1 in lateral root formation, we crossed the CycB1;1::GUS transgene into axr1-12 plants. In untreated wild-type and mutant seedlings, the fusion was expressed in the root tip (Figure 3A). In a wild-type seedling transferred to 0.2 μM 2,4-D for 24 hr, extensive GUS staining was observed throughout the pericycle (Figure 3A). This additional staining was completely absent in auxin-treated axr1-12 seedlings, indicating that AXR1 is required for the first cell divisions of the lateral root founder cells (Figure 3A).
Figure 3.
AXR1 Plays an Important Role in Lateral Root Development.
(A) AXR1 is required for the expression of CycB1;1 during lateral root formation. Five-day-old AXR1 CycB1;1::GUS, and axr1-12 CycB1;1::GUS seedlings were grown in the absence (−) or presence (+) of 0.2 μM 2,4-D for 24 hr and stained for GUS activity.
(B) Left, GUS expression in lateral root primordia formed after treatment with 0.01 μM 2.4-D for 24 hr. Arrows indicate the positions of lateral root primordia. Center, GUS staining in pericycle cells before cell division in a 2,4-D–treated root. Right, GUS staining in dividing cells in a later stage of lateral root development and in the vascular tissue close to the primordium.
Consistent with this result, analysis of the AXR1::GUS lines showed that AXR1 was expressed at the earliest stages of lateral root formation and throughout their development (Figure 3B). Lateral roots were induced by treating seedlings with 0.01 μM 2,4-D. As shown in Figure 3B, lateral root primordia were stained along the primary roots of these seedlings. Higher magnification revealed that low levels of GUS staining were observed in cells within the pericycle layer before any cell divisions were evident (Figure 3B, middle). At a later stage of development, when the pericycle cells start to divide and form a lateral root primordium, GUS staining increased in dividing cells as well as in the vascular tissue surrounding the primordium (Figure 3B, right).
ECR1 Protein Functions Together with AXR1 during Plant Development
In a previous study, we showed that AXR1 functions in vitro with the ECR1 protein to activate the ubiquitin-related protein RUB1 (del Pozo et al., 1998). To study the function of ECR1 in the plant, we first analyzed the pattern of ECR1 expression by in situ hybridization. As expected, ECR1 expression was observed in tissues that expressed AXR1. For example, in the inflorescence, the gene was highly expressed in the apical and floral meristems as well as the early stages of floral organ development (Figures 4A and 4B). We also observed expression in the root and shoot meristems, developing ovules, and embryo, all tissues in which AXR1 expression was observed (data not shown). ECR1 RNA was not detected in mature (nongrowing) tissues of the plant, such as in the stem distal to the apex or in fully expanded leaves (Figure 4 and data not shown). RNA gel blot studies indicated that ECR1 RNA did not accumulate in response to auxin (data not shown).
Figure 4.
ECR1 Functions with AXR1 during Plant Development.
(A) Antisense ECR1 probe hybridized to a section of inflorescence and developing flowers.
(B) Sense probe hybridized to a similar section.
(C) RNA gel blot showing ECR1 expression in 7-day-old wild-type (wt) seedlings and transgenic lines expressing ECR1C215A.
(D) Rosettes of ECR1-C4 and wild-type plants at 21 days.
(E) Phenotypes of ECR1C215A transgenic plants at 42 days.
(F) Phenotype of a wild-type plant at 42 days.
(G) Inflorescence with mature siliques of ECR1C215A transgenic plants.
(H) Flowers of wild type (i), axr1-12 (ii), ECR1-C1 (iii), and the axr1-12 ECR1-C1 double mutant (iv).
(I) Rosettes of axr1-12 ECR1-C1 double mutant plants at 42 days. Bars in (D) to (F), and (I) = 1 cm.
RUB activation involves the formation of a thiol ester bond between the C terminus of RUB and a cysteine residue within the activating enzyme. On the basis of sequence similarity, Cys-215 of ECR1 likely functions as the active site cysteine in a RUB-activating enzyme involving ECR1 (del Pozo et al., 1998). Replacement of this cysteine with alanine abolished ECR1 activity in vitro (del Pozo et al., 1998). To explore the function of ECR1 in the plant, we generated transgenic lines expressing the ECR1C215A mutant gene under the control of the 35S promoter of Cauliflower mosaic virus. A total of 15 lines were recovered that displayed a dwarf phenotype in the T1 and T2 generations. Two lines were selected for further study: a less severely affected line called ECR1-C1 and an extreme dwarf called ECR1-C4. RNA gel blot analysis revealed ECR1 transgene expression in both lines, with a higher level of expression in ECR1-C4 (Figure 4C).
The rosettes of both transgenic lines were smaller than those of the wild type, with small curled leaves (Figure 4D). The inflorescences were much shorter than those of the wild-type control, particularly in the case of ECR1-C4 (Figures 4E and 4F). Stem elongation between successive flowers was reduced severely, leading to the formation of a flower cluster at the end of the inflorescence (Figure 4G). The lateral branches of the transgenic lines behaved in a similar fashion, so that the mature plants looked like a small bush. In contrast to the aerial phenotype, the growth of the root system in transgenic seedlings grown on agar medium was similar to that of the wild type.
In many respects, the appearance of the transgenic lines resembles that of the axr1 mutants (Lincoln et al., 1990). One of the most distinctive aspects of the axr1 phenotype is floral morphology. The organs of axr1 flowers are smaller than those of the wild type and appear to be poorly developed (Lincoln et al., 1990). Because mutant stamens do not elongate normally, pollen is not deposited on the stigma, leading to a decrease in fertility. The flowers of the severe ECR1-C4 line have a similar appearance and produce fewer seed than do wild-type flowers (Figure 4H).
These data indicate that ECR1C215A has a dominant negative effect on plant growth and development. Because ECR1 and AXR1 function together in vitro, this effect probably is attributable to the formation of inactive AXR1-ECR1 heterodimers and a reduction in RUB activation.
The Arabidopsis genome contains a gene that is closely related to AXR1 that we have called AXL1. The existence of this gene leaves open the possibility that the axr1-12 mutation does not eliminate RUB E1 activity completely. To begin to address this possibility, we crossed the ECR1C215A transgene into the axr1-12 mutant. Homozygous axr1-12 plants were identified among the F2 plants by virtue of their auxin resistance. These plants then were scored for the presence of the transgene by polymerase chain reaction. The axr1-12 plants carrying the transgene were extremely small with tightly curled dark green/purplish rosette leaves and little inflorescence (Figure 4I). The flowers of these plants were severely deformed and infertile, with very short stamens and purplish petals and sepals (Figure 4H). These results suggest that axr1-12 plants possess some RUB E1 activity that can be reduced further by expression of the mutant ECR1 protein.
ECR1C215A Lines Are Deficient in Auxin Response
To determine if the ECR1C215A lines have a defect in auxin response, we first examined the effects of auxin on seedling root elongation. Unlike the axr1 mutant, the roots of the transgenic lines have a normal response to auxin by this assay (data not shown). This result is consistent with the absence of any root growth defect in the lines.
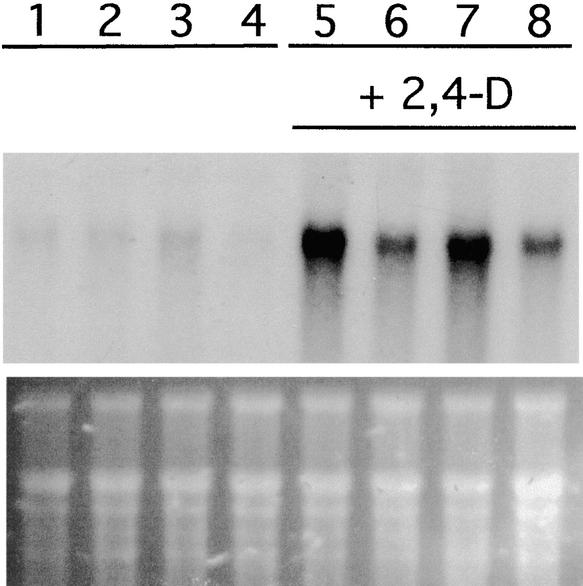
One of the earliest effects of auxin treatment is transcription of members of the Aux/IAA family of genes (Abel and Theologis, 1996). The axr1 mutants are deficient in this response (Abel et al., 1995; Timpte et al., 1995). To determine if expression of the ECR1C215A transgene affects the auxin-regulated expression of the IAA genes, we performed RNA gel blot analysis using RNA isolated from auxin-treated control and transgenic seedlings. Figure 5 shows that the ECR1C215A lines are deficient in auxin-induced expression of the IAA2 gene. This defect is more severe in the ECR1-C4 line, consistent with the increased expression of the transgene and the more severe phenotype. Thus, the affects on morphology observed in these lines are associated with a reduction in auxin response.
Figure 5.
Auxin-Induced Gene Expression Is Reduced in ECR1C215A Transgenic Plants.
RNA gel blot analysis of total RNA extracted from wild-type (lanes 1 and 5), ECR1-C4 (lanes 2 and 6), ECR1-C1 (lanes 3 and 7), and axr1-12 (lanes 4 and 8) plants treated with or without 20 mM 2,4-D. The blot was hybridized to IAA2 probe. An ethidium bromide–stained gel is shown at bottom.
AXR1-ECR1 Enzyme Is Required for the RUB Modification of AtCUL1
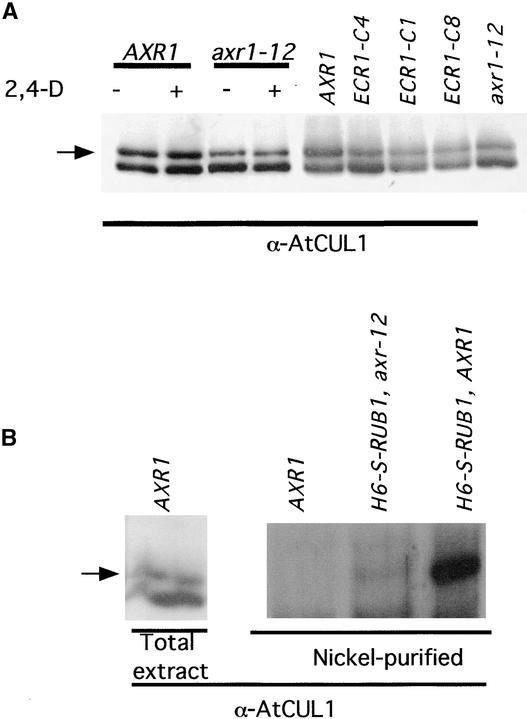
Previous studies have shown that the Arabidopsis cullin AtCUL1 is modified by RUB1 (del Pozo and Estelle, 1999). To determine if this modification is AXR1 and ECR1 dependent, we first characterized AtCUL1 by protein blot in the axr1-12 and ECR1C215A lines. The results are shown in Figure 6. In wild-type seedlings, AtCUL1 exists in two forms: the unmodified form with a molecular mass of 86 kD and the slightly larger modified form (Figure 6A). At least some, and perhaps all, of this larger species is RUB-AtCUL1 (del Pozo and Estelle, 1999). In axr1-12 seedlings, there is a decrease in the relative amount of the larger form of AtCUL1, indicating that AXR1 is required for the formation of modified AtCUL1. To determine if exogenous auxin affects the relative abundance of modified AtCUL1, we treated wild-type and axr1-12 seedlings with 20 μM 2,4-D. Extracts were prepared at time intervals between 10 and 120 min. No differences were observed during the course of the experiment (Figure 6A and data not shown), suggesting that auxin does not act by regulating the levels of RUB-AtCUL1. To confirm that ECR1 also is required for modification of AtCUL1, we performed a similar analysis with the ECR1C215A lines, including an additional line called ECR1-C8. As with axr1-12, the expression of the mutant ECR1 reduced the relative amount of modified AtCUL1 (Figure 6A).
Figure 6.
AXR1 and ECR1 Are Required for Modification of AtCUL1.
(A) Protein gel blot analysis of total proteins extracted from wild-type, axr1-12, and ECR1C215A transgenic plants. Extracts were prepared from 6-day-old seedlings, and either 10 mg (first four lanes) or 5 mg (next five lanes) was loaded onto the gel. α-AtCUL1, AtCUL1 antiserum.
(B) Protein gel blot analysis of total extract or nickel-purified proteins recovered from AXR1 and axr1-12 5-day-old seedlings carrying the H6-S–RUB1 transgene. The blots were probed with AtCUL1 antiserum (α-AtCUL1). The arrow indicates the position of RUB-AtCUL1.
The RUB family in Arabidopsis consists of three isoforms. RUB1 and RUB2 are almost identical, whereas RUB3 is diverged significantly (Rao-Naik et al., 1998). In addition, the Arabidopsis proteome includes a number of additional ubiquitin-related proteins, including a protein closely related to SUMO/Smt3 and a number of uncharacterized putative proteins. AtCUL1 may be modified by any of these proteins. Alternatively, AtCUL1 could be a target for other types of protein modification. Thus, it is important to demonstrate that AXR1-ECR1 is required in vivo for the RUB1 modification of AtCUL1. To do this, we used a transgenic line expressing a RUB1 derivative with an H6 S-peptide at its N terminus. The H6-S–RUB1 transgenic line was crossed into the axr1-12 mutant, and H6-S–RUB1 was purified from mutant and wild-type plants using nickel-Sepharose beads. When the eluate was analyzed by protein blotting using the AtCUL1 antibody, a single protein species was detected in the wild-type lane, confirming that AtCUL1 is modified by RUB1 in vivo. However, the amount of AtCUL1 detected in the axr1-12 extract was reduced dramatically, indicating that AXR1 is required to form RUB1-AtCUL1 in vivo. As expected, no AtCUL1 was detected in extracts from control plants that do not express H6-S–RUB1.
AtCUL1 Is Localized to the Nucleus in Wild-Type and axr1-12 Plants
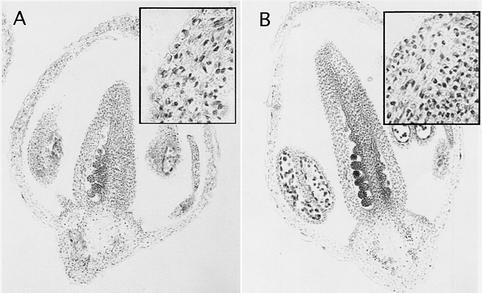
Farras et al. (2001) reported that AtCUL1 is localized predominantly to the nucleus of Arabidopsis cells. Because the RUB conjugation pathway also is localized to the nucleus (del Pozo et al., 1998; Yeh et al., 2000), it is possible that RUB modification of cullin is required for nuclear retention. To determine if AXR1 is required for this localization, we performed immunolocalization studies on wild-type and axr1-12 tissues. As reported above, we found that AtCUL1 is expressed in dividing cells throughout the plant (data not shown) and is localized primarily to the nucleus. We observed a similar distribution in the axr1-12 mutant. Longitudinal sections of wild-type and axr1-12 flowers are shown in Figure 7. AtCUL1 accumulates in most cells of the developing flower, with particularly high levels in the ovules. The insets show high magnification images from the carpel wall. AtCUL1 is found in the nuclei of both wild-type and axr1-12 cells. These results show that AXR1 is not required for the gross localization of AtCUL1. However, a significant fraction of AtCUL1 is still modified in the axr1-12 mutants, as indicated by the data shown in Figure 6A.
Figure 7.
The axr1 Mutation Does Not Affect the Nuclear Localization of AtCUL1.
Immunolocalization of AtCUL1 in wild-type (A) and axr1-12 (B) floral sections. The insets show high magnification images from the carpel walls, demonstrating normal nuclear localization in the mutant. The AtCUL1 antiserum was characterized previously (Gray et al., 1999; Farras et al., 2001).
DISCUSSION
During the last several years, the identification of genes that function in auxin response has begun to provide insight into the mechanisms of auxin action (Gray and Estelle, 2000). One of these genes, called AXR1, encodes a subunit of a heterodimeric enzyme responsible for the activation of the ubiquitin-related protein RUB (NEDD8 in animal systems). In a previous study, we presented in vitro data suggesting that the second subunit of the enzyme is encoded by the ECR1 gene (del Pozo et al., 1998). The results reported here confirm that AXR1 and ECR1 function together in the plant. The patterns of AXR1 and ECR1 gene expression are identical. Furthermore, overexpression of a mutant form of ECR1 disrupts the activity of the RUB pathway and causes a phenotype that is very similar to that of the axr1 mutant. Presumably, the ECR1C215A protein forms a nonfunctional dimer with AXR1, reducing the total amount of RUB-activating enzyme in the cell. The one major difference between the ECR1C215A plants and axr1 plants is the lack of an auxin defect in the roots. This may be because there are higher levels of AXR1 and endogenous ECR1 in the seedling root compared with those in the developing shoot system. Alternatively, the ECR1C215A transgene may be expressed poorly in the root system.
AXR1 and ECR1 Are Expressed in Growing Tissues throughout Plant Development
The axr1 mutants are affected in most aspects of postembryonic growth and development (Lincoln et al., 1990). This pleiotropy is consistent with a pattern of expression that includes all growing tissues in the plant, including the major meristems (del Pozo et al., 1998) and organ primordia originating from each of these meristems. In addition, expression is observed in elongating hypocotyls, trichomes, root hairs, and vascular tissues of all organs examined. In contrast, little or no RNA or protein is observed in nongrowing tissues.
Auxin is thought to play an important role in the differential growth of organs in a variety of contexts, including tropic growth of the root and shoot and the formation and maintenance of the apical hook in dark-grown seedlings. Indeed, the axr1 mutants are deficient in root and shoot tropism and lack an apical hook (Lincoln et al., 1990; Cernac et al., 1997; Watahiki et al., 1999). In addition, germinating axr1 seedlings are slow to orient the radicle downward in response to gravity. Consistent with these defects, AXR1 is expressed at higher levels on the lower side of an emerging root during germination and at higher levels on the underside of the apical hook. At present, it is not clear if differential AXR1 expression contributes directly to differential growth or if this expression pattern is a consequence of the growth pattern. AXR1 expression in root hairs also is consistent with a defect in root hair elongation in axr1 plants (Pitts et al., 1998). It will be interesting to determine if auxin has a similar affect on trichome development.
There is substantial evidence that auxin is involved in the differentiation of vascular bundles (Berleth et al., 2000). Stem cross-sections of the axr1-12 mutant revealed that the vascular bundles were poorly defined (Lincoln et al., 1990). In agreement with this, we found that AXR1 is highly expressed in vascular tissues and that the AXR1 protein accumulates in the nucleus of these specialized cells.
AXR1 Is a Key Factor in Root Development
Genetic and physiological studies have shown that auxin is essential for root development (Scheres, 2000; Casimiro et al., 2001). In Arabidopsis, auxin application inhibits the elongation of the main root but stimulates cell division in the root meristem and promotes the formation of adventitious and lateral roots. The auxin response mutants axr1, tir1, and axr4 all have a characteristic root phenotype that includes increased root elongation on unsupplemented medium and fewer lateral roots (Hobbie and Estelle, 1995; Ruegger et al., 1998). Lateral root primordia arise from G2-arrested pericycle cells. Upon stimulation by exogenous auxin, small groups of pericycle cells undergo a series of divisions to form the primordium. The mitotic cyclin gene CycB1;1 is expressed before the earliest cell divisions in primordium formation. Using the promoter activity of this gene as a marker, we have shown that TIR1 activity is required before any cell division can occur (Gray et al., 1999). A similar study in the axr1-12 background shows that AXR1 also is required for CycB1;1 expression during lateral root formation, consistent with the very early expression of AXR1 we observed during the formation of a lateral root. The dependence of CycB1;1 expression on AXR1 suggests that AXR1 expression occurs before CycB1;1 expression. However, it is possible that AXR1 acts at an earlier time to establish conditions whereby pericycle cells respond normally to auxin. Alternatively, AXR1 may be expressed in pericycle cells before the arrival of the auxin signal but at a level that is below our limits of detection.
Function of AXR1-ECR1 during Embryogenesis
In higher plants, the basic body plan is established during embryogenesis via specific patterns of cell division, elongation, and cell–cell interaction (Jurgens, 1995). Recently, several genetic and physiological studies strongly suggested that auxin transport and auxin response are required for normal embryogenesis (Przemeck et al., 1996; Hadfi et al., 1998; Hamann et al., 1999; Chen et al., 2001). Our studies show that AXR1 and ECR1 are expressed during embryo development, despite the fact that axr1 embryos do not have an obvious defect. However, the axr1 mutation does enhance the apical–basal defect observed in the embryonic mutant bodenlos (bdl) (Hamann et al., 1999). This mutation also confers auxin resistance and affects the formation of the root and hypocotyl during embryogenesis. Double mutant bdl axr1 embryos were wider and more compressed in the apical–basal axis than were bdl or wild-type embryos, suggesting a possible defect in cell elongation. In addition, bdl axr1 embryos resemble the embryonic mutant monopteros (mp) or wild-type embryos exposed to auxin transport inhibitors during embryogenesis (Hadfi et al., 1998). MP encodes a transcription factor (IAA24) that binds to the promoter elements of auxin-regulated genes (Ulmasov et al., 1997; Hardtke and Berleth, 1998). We have shown recently that overexpression of a mutant stabilized form of IAA7/AXR2 called axr2-1 in an axr1-3 background results in a mp phenotype (Gray et al., 2001). These results, together with the AXR1 expression observed in embryos, indicate that AXR1 functions during embryogenesis. The lack of a significant defect in the axr1 embryos may be caused by gene redundancy. In fact, a gene closely related to AXR1 (called AXL1) is present in the Arabidopsis genome (N. Dharmaisiri and M. Estelle, unpublished results).
RUB Conjugation Pathway and Auxin Response
In a previous study, we showed that Arabidopsis RUB1 is conjugated to the cullin AtCUL1, a component of the ubiquitin protein ligase SCFTIR1 (del Pozo and Estelle, 1999). In addition, we have demonstrated that SCFTIR1 is required for auxin response and that AXR1 and TIR1 function in the same or overlapping pathways (Ruegger et al., 1998; Gray et al., 1999). On the basis of these results, we have proposed that the axr1 mutants are deficient in auxin response because RUB modification of AtCUL1 is required for the activity of SCFTIR1 (Gray and Estelle, 2000). Our current results provide strong support for this model. Both the axr1 mutations and the expression of ECR1C215A result in a decrease in RUB1 modification of AtCUL1.
If AXR1-ECR1 is required for SCFTIR1 function, then mutations in the pathway should stabilize substrates of the SCF. Indeed, recent results indicate that this is the case. The products of the Aux/IAA genes are unstable nuclear proteins that function to regulate auxin-dependent transcription (Abel and Theologis, 1996). Dominant gain-of-function mutations have been isolated in five of these genes, and in every case the mutation causes an amino acid substitution within a small region of the protein called domain II (Reed, 2001). Each mutant gene acts to disrupt various auxin-regulated growth processes. Two of the mutations, axr2-1 and axr3-1, act to stabilize their respective proteins, and it is likely that the others have the same effect (Worley et al., 2000; Ouellet et al., 2001). Our recent results indicate that members of the Aux/IAA family of proteins interact directly with SCFTIR1 (Gray et al., 2001). Furthermore, two members of this family, AXR3/IAA17 and AXR2/IAA7, are stabilized by the tir1 and axr1 mutations (Gray et al., 2001). These results demonstrate that auxin response depends on SCFTIR1-mediated degradation of the Aux/IAA proteins, a process that is dependent on AXR1-ECR1. However, it is important to note that auxin does not appear to regulate the activity of the RUB conjugation pathway because levels of RUB-modified AtCUL1 do not change in response to auxin. In addition, expression of the AXR1 and ECR1 genes is not affected directly by auxin treatment. Rather, auxin appears to promote the interaction between the Aux/IAA proteins and SCFTIR1 (Gray et al., 2001).
Function of the RUB-Cullin Conjugation
Accumulating evidence indicates that all eukaryotic cullins can be modified by RUB/NEDD8 except for the cullin-related proteins present in the anaphase-promoting complex (Yeh et al., 2000). In the case of the Arabidopsis cullin family, RUB modification has been demonstrated directly only for AtCUL1. However, the other members of the family also have the conserved RUB modification motif on their C termini, suggesting that they also are substrates for modification (del Pozo and Estelle, 1999). The precise function of RUB modification is unknown (Yeh et al., 2000). Several studies have reported that the NEDD8 pathway enhances E3 activity in vitro, but the basis for this enhancement is not clear (Morimoto et al., 2000; Podust et al., 2000; Read et al., 2000; Wu et al., 2000; Tanaka et al., 2001). NEDD8 does not appear to be required for the assembly or stability of the complex (Yeh et al., 2000). There is some evidence that the modification is important for cellular localization of the SCF (Freed et al., 1999). Although we do not see a difference in the localization of AtCUL1 in the axr1 mutant, this experiment is not conclusive because a significant fraction of AtCUL1 still is modified in the mutant.
Whatever its function, the modification is particularly important for certain SCF ligases. In budding yeast, the RUB pathway is not essential for viability but is required for the normal function of SCFCDC4, an E3 that promotes ubiquitination of the cyclin-dependent kinase inhibitor Sic1p (Lammer et al., 1998). Other SCFs do not appear to be affected by loss of the pathway. In contrast, the pathway is essential for viability in fission yeast (Osaka et al., 2000). The basis for these differences is not clear. It is possible that RUB modification confers additional functionality to some SCFs that is required specifically for their activity. Alternatively, it is possible that the modification affects the activity of all SCFs equally but that some ligases have a more important role in cellular regulation than others.
In Arabidopsis, the pathway appears to be critical for the activity of SCF ligases that function in auxin response. Most aspects of the axr1 phenotype can be explained by defects in auxin response (Walker and Estelle, 1998). In addition, defects associated with mutations that affect other SCFs are not observed in axr1 plants. For example, the F-box protein UFO is required for floral development and probably is part of an SCF complex, yet axr1 flowers do not have a ufo phenotype. Similarly, loss of the ASK1 protein, an SCF subunit, results in male sterility. axr1 plants still make viable pollen, although pollen yield is reduced (Yang et al., 1999). The situation with the F-box protein COI1 may be particularly instructive. The coi1 mutant is completely insensitive to jasmonic acid, indicating that an SCF functions in jasmonic acid response (Xie et al., 1998). The mutant also is male sterile. Although axr1 plants do not display either of these extreme phenotypes, recent results indicate that axr1-12 is slightly resistant to jasmonic acid (P. Staswick, personal communication). Thus, defects in RUB modification also appear to affect the activity of SCFCoi1, although less severely than SCFTIR1.
Our results indicate that the axr1-12 mutant is not completely deficient in RUB-AtCUL1. We expect that complete ablation of the pathway will require mutating the AXR1-related gene AXL1. It will be interesting to determine if this situation will result in additional effects on SCF activity.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana (ecotype Columbia) plants were grown on MS medium (Murashige and Skoog [1962] salts [Gibco BRL], 1% sucrose, 1 × Gamborg's B5 vitamins, and 1% agar, pH 5.7) or in soil under continuous light. For experiments requiring sterile conditions, seed were sterilized and grown as described previously (Ruegger et al., 1998). The synthetic auxin 2,4-D was filter-sterilized and added to autoclaved medium.
Preparation of AXR1:GUS and ECR1 Constructs and Transformation of Arabidopsis Plants
Two different AXR1::β-glucuronidase (GUS) translational fusions were made by inserting DNA fragments containing the 5′ upstream region of the AXR1 gene (3 or 1 kb upstream from the ATG to nucleotide +10) fused in frame with the GUS coding region in the binary vector pBI101.1 (Jefferson et al., 1987). Agrobacterium tumefaciens harboring these constructs was used to transform Arabidopsis plants by the vacuum infiltration method (Bechtold et al., 1993). Kanamycin-resistant T1 plants were selected by plating seed on MS medium supplemented with 1% sucrose and 50 μg/mL kanamycin. These plants were transferred to soil, and five independent T2 homozygous lines with one AXR1::GUS insertion were identified. Before GUS staining, plant material was fixed in 50 mM NaPO4, pH 7.0, 1% formaldehyde, and 0.5% Triton X-100 for 30 min and washed three times with large amounts of the same buffer without formaldehyde. This material was incubated in 50 mM NaPO4, pH 7.0, 0.5% Triton X-100, and 0.5 mg/mL 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (Rose Scientific, Edmonton, Canada) for 16 hr at 37°C in the dark. Afterward, this plant material was cleared with several changes in 70% ethanol. The expression pattern was identical in the transgenic lines harboring the 3.0-kb promoter or the 1.0-kb promoter. In this study, we use transgenic lines containing the 3.0-kb AXR1 promoter construction.
Generation of the ECR1C215A mutant was described previously (del Pozo et al., 1998). The mutant gene was inserted into the pBI121 vector, replacing the GUS coding region and putting the gene under the control of the 35S promoter of Cauliflower mosaic virus. The construct was introduced into Arabidopsis plants as described above. A total of 15 severely affected lines were recovered. These lines exhibited variable stability into the T2 and T3 generations, with a gradual diminishing of the severity of the phenotype. Analysis was performed on the T2 and T3 generations.
Construction of axr1-12 Lines Carrying the ECR1C215A Transgene
The ECR1C215A transgene was introduced into axr1-12 by crossing with the ECR1-C1 line. Homozygous axr1 plants were identified in the F2 generation by screening for auxin resistance. Lines carrying the transgene were identified by polymerase chain reaction amplification of the NPTII gene.
Immunolocalization and RNA in Situ Studies
Both the AXR1 and AtCUL1 antiserum were described previously (del Pozo et al., 1998; Gray et al., 1999). For this work, the immunoglobulins were immunoaffinity purified against AXR1 recombinant protein as described by Gray et al. (1999). Immunolocalization analyses were performed as described by Perry et al. (1996) using the ABC kit (Vectastain, Vectorlabs, Burlingame, CA). The AXR1 and AtCUL1 antisera were used at dilutions of 1:500 and 1:1000, respectively. The full-length ECR1 cDNA was used to prepare the sense and antisense probes for RNA in situ hybridizations as described by Gray et al. (1999).
In Vivo Conjugation of H6-S–RUB1 to AtCUL1
The construction of the H6-S–RUB1 transgene was described by del Pozo and Estelle (1999). Purification of H6-S–RUB1 was performed as follows. Soluble protein from 5-day-old wild-type and H6-S–RUB1 seedlings (0.4 g of tissue) was extracted in 1 mL of buffer E [100 mM Tris-HCl, pH 7.5, 400 mM (NH4)2SO4, 10 mM MgCl2, 1 mM EDTA, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 × proteases inhibitor cocktail (Boehringer Mannheim)] for 30 min at 4°C. The extracts were clarified by centrifugation for 30 min at 4°C. The soluble fraction was precipitated with (NH4)2SO4 at 50% saturation. Proteins in the pellet were dissolved in 0.4 mL of binding buffer (50 mM NaPO4, pH 8, 400 mM NaCl, 0.3% Triton X-100, 1 mM PMSF, and 1 × proteases inhibitor cocktail), and an aliquot (crude extract) was taken and mixed with SDS loading buffer with 5% β-mercaptoethanol. Then the extracts were incubated with prewashed nickel beads (Qiagen, Valencia, CA) in binding buffer for 3 hr at 4°C. Afterward, the beads were washed five times for 10 min each with 1.5 mL of washing buffer (50 mM NaPO4, pH 6, 400 mM NaCl, 40 mM imidazole, 0.3% Triton X-100, 1 mM PMSF, and 1 × proteases inhibitor cocktail). Proteins were liberated from the beads in binding buffer/1 × SDS loading buffer with β-mercaptoethanol and boiled for 10 min.
Standard Molecular Biology Procedures
All standard molecular biology techniques were performed as described by Ausubel et al. (1990). For protein gel blot analysis, proteins were resolved on a 9% acrylamide gel and transferred to a nitrocellulose membrane (Ausubel et al., 1990). The AtCUL1 protein was detected using immunoaffinity-purified antibodies at a 1:1000 dilution as described by Gray et al. (1999).
For RNA gel blot hybridization, RNA was extracted from seedlings treated with or without 20 μM 2,4-D for 60 min using Tri Reagent (Sigma). Ten micrograms of total RNA was loaded in each lane, and the blot was hybridized with IAA2 cDNA carrying the entire coding region labeled with α-32P-dCTP using the RadPrime labeling kit (Gibco BRL).
Acknowledgments
J.C.d.P was supported by a long-term fellowship from the Spanish government. H.H. was supported by a grant from the Deutsche Forschungsgemeinshcaft (No. HE 3224/1-1). This work was supported by National Institutes of Health Grant No. RO1-GM43644 to M.E.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010282.
References
- Abel, S., and Theologis, A. (1996). Early genes and auxin action. Plant Physiol. 111, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel, S., Nguyen, M.D., and Theologis, A. (1995). The PS-IAA4/ 5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J. Mol. Biol. 251, 533–549. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1990). Current Protocols in Molecular Biology. (New York: Wiley).
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 15–18. [Google Scholar]
- Berleth, T., Mattsson, J., and Hardtke, C.S. (2000). Vascular continuity and auxin signals. Trends Plant Sci. 5, 387–393. [DOI] [PubMed] [Google Scholar]
- Casimiro, I., Marchant, A., Bhalerao, R.P., Beeckman, T., Dhooge, S., Swarup, R., Graham, N., Inze, D., Sandberg, G., Casero, P.J., and Bennett, M. (2001). Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac, A., Lincoln, C., Lammer, D., and Estelle, M. (1997). The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development 124, 1583–1591. [DOI] [PubMed] [Google Scholar]
- Chen, J.G., Ullah, H., Young, J.C., Sussman, M.R., and Jones, A.M. (2001). ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 15, 902–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Carmona, A., You, R., Haimovitch-Gal, T., and Doerner, P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cycline-GUS fusion protein. Plant J. 20, 503–508. [DOI] [PubMed] [Google Scholar]
- Davies, P.J. (1995). The plant hormones: Their nature, occurrence and functions. In Plant Hormones: Physiology, Biochemistry and Molecular Biology, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–12.
- del Pozo, J.C., and Estelle, M. (1999). The Arabidopsis cullin AtCUL1 is modified by the ubiquitin-related protein RUB1. Proc. Natl. Acad. Sci. USA 96, 15342–15347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo, J.C., Timpte, C., Tan, S., Callis, J., and Estelle, M. (1998). The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- Evans, M.L. (1984). Functions of hormones at the cellular level of organization. In Hormonal Regulation of Development II, Vol. 10, T.K. Scott, ed (Berlin: Springer-Verlag), pp. 23–79.
- Farras, R., Ferrando, A., Jasik, J., Kleinow, T., Okresz, L., Tiburcio, A., Salchert, K., del Pozo, C., Schell, J., and Koncz, C. (2001). SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 20, 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed, E., Lacey, K.R., Huie, P., Lyapina, S.A., Deshaies, R.J., Stearns, T., and Jackson, P.K. (1999). Components of an SCF ubiquitin ligase localize to the centrosome and regulate the centrosome duplication cycle. Genes Dev. 13, 2242–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., and Estelle, M. (2000). Function of the ubiquitin-proteasome pathway in auxin response. Trends Biochem. Sci. 25, 133–138. [DOI] [PubMed] [Google Scholar]
- Gray, W.M., Ostin, A., Sandberg, G., Romano, C.P., and Estelle, M. (1998). High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 7197–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, W.M., Kepinski, S., Rouse, D., Leyser, O., and Estelle, M. (2001). Auxin regulates SCFTIR1-dependent degradation of the Aux/IAA proteins. Nature 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Hadfi, K., Speth, V., and Neuhaus, G. (1998). Auxin-induced developmental patterns in Brassica juncea embryos. Development 125, 879–887. [DOI] [PubMed] [Google Scholar]
- Hamann, T., Mayer, U., and Jurgens, G. (1999). The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbie, L., and Estelle, M. (1995). The axr4 auxin-resistant mutants of Arabidopsis thaliana define a gene important for root gravitropism and lateral root initiation. Plant J. 7, 211–220. [DOI] [PubMed] [Google Scholar]
- Hobbie, L., Timpte, C., and Estelle, M. (1994). Molecular genetics of auxin and cytokinin. Plant Mol. Biol. 26, 1499–1519. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens, G. (1995). Axis formation in plant embryogenesis: Cues and clues. Cell 81, 467–470. [DOI] [PubMed] [Google Scholar]
- Kamura, T., Conrad, M.N., Yan, Q., Conaway, R.C., and Conaway, J.W. (1999). The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 13, 2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer, D., Mathias, N., Laplaza, J.M., Jiang, W., Liu, Y., Callis, J., Goebl, M., and Estelle, M. (1998). Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 12, 914–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., Busgen, T., Brychzy, A., Jentsch, S., and Pause, A. (1999). Conjugation of the ubiquitin-like protein NEDD8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc. Natl. Acad. Sci. USA 96, 5510–5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, M., Nishida, T., Honda, R., and Yasuda, H. (2000). Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1). Biochem. Biophys. Res. Commun. 270, 1093–1096. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Osaka, F., Kawasaki, H., Aida, N., Saeki, M., Chiba, T., Kawashima, S., Tanaka, K., and Kato, S. (1998). A new NEDD8-ligating system for cullin-4A. Genes Dev. 12, 2263–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaka, F., et al. (2000). Covalent modifier NEDD8 is essential for SCF ubiquitin-ligase in fission yeast. EMBO J. 19, 3475–3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet, F., Overvoorde, P.J., and Theologis, A. (2001). IAA17/AXR3: Biochemical insight into an auxin mutant phenotype. Plant Cell 13, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton, E.E., Willems, A.R., and Tyers, M. (1998). Combinatorial control in ubiquitin-dependent proteolysis: Don't Skp the F-box hypothesis. Trends Genet. 14, 236–243. [DOI] [PubMed] [Google Scholar]
- Perry, S.E., Nichols, K.W., and Fernandez, D.E. (1996). The MADS domain protein AGL15 localizes to the nucleus during early stages of seed development. Plant Cell 8, 1977–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts, R.J., Cernac, A., and Estelle, M. (1998). Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 16, 553–560. [DOI] [PubMed] [Google Scholar]
- Podust, V.N., Brownell, J.E., Gladysheva, T.B., Luo, R.S., Wang, C., Coggins, M.B., Pierce, J.W., Lightcap, E.S., and Chau, V. (2000). A Nedd8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc. Natl. Acad. Sci. USA 97, 4579–4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przemeck, G.K., Mattsson, J., Hardtke, C.S., Sung, Z.R., and Berleth, T. (1996). Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Rao-Naik, C., delaCruz, W., Laplaza, J.M., Tan, S., Callis, J., and Fisher, A.J. (1998). The rub family of ubiquitin-like proteins: Crystal structure of Arabidopsis rub1 and expression of multiple rubs in Arabidopsis. J. Biol. Chem. 273, 34976–34982. [DOI] [PubMed] [Google Scholar]
- Read, M.A., et al. (2000). Nedd8 modification of cul-1 activates SCF(β(TrCP))-dependent ubiquitination of IκBα. Mol. Cell. Biol. 20, 2326–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, J.W. (2001). Roles and activities of Aux/IAA proteins in Arabidoposis. Trends Plant Sci. 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Ruegger, M., Dewey, E., Gray, W.M., Hobbie, L., Turner, J., and Estelle, M. (1998). The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev. 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres, B. (2000). Non-linear signaling for pattern formation? Curr. Opin. Plant Biol. 3, 412–417. [DOI] [PubMed] [Google Scholar]
- Stirnberg, P., Chatfield, S.P., and Leyser, H.M. (1999). AXR1 acts after lateral bud formation to inhibit lateral bud growth in Arabidopsis. Plant Physiol. 121, 839–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, K., Kawakami, T., Tateishi, K., Yashiroda, H., and Chiba, T. (2001). Control of IκBα proteolysis by the ubiquitin-proteasome pathway. Biochimie 83, 351–356. [DOI] [PubMed] [Google Scholar]
- Timpte, C.S., Lincoln, C., Pickett, F.B., Turner, J., and Estelle, M. (1995). The AXR1 and AUX1 function in separate auxin-response pathways. Plant J. 8, 561–569. [DOI] [PubMed] [Google Scholar]
- Ulmasov, T., Hagen, G., and Guilfoyle, T.J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. [DOI] [PubMed] [Google Scholar]
- Wada, H., Yeh, E.T., and Kamitani, T. (1999). Identification of NEDD8-conjugation site in human cullin-2. Biochem. Biophys. Res. Commun. 257, 100–105. [DOI] [PubMed] [Google Scholar]
- Walker, L., and Estelle, M. (1998). Molecular mechanisms of auxin action. Curr. Opin. Plant Biol. 1, 434–439. [DOI] [PubMed] [Google Scholar]
- Watahiki, M.K., Tatematsu, K., Fujihira, K., Yamamoto, M., and Yamamoto, K.T. (1999). The MSG1 and AXR1 genes of Arabidopsis are likely to act independently in growth-curvature re-sponses of hypocotyls. Planta 207, 362–369. [DOI] [PubMed] [Google Scholar]
- Worley, C.K., Zenser, N., Ramos, J., Rouse, D., Leyser, O., Theologis, A., and Callis, J. (2000). Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 21, 553–562. [DOI] [PubMed] [Google Scholar]
- Wu, K., Chen, A., and Pan, Z.Q. (2000). Conjugation of Nedd8 to CUL1 enhances the ability of the ROC1–CUL1 complex to promote ubiquitin polymerization. J. Biol. Chem. 275, 32317–32324. [DOI] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Yang, M., Hu, Y., Lodhi, M., McCombie, W.R., and Ma, H. (1999). The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc. Natl. Acad. Sci. USA 96, 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E.T., Gong, L., and Kamitani, T. (2000). Ubiquitin-like proteins: New wines in new bottles. Gene 248, 1–14. [DOI] [PubMed] [Google Scholar]