Abstract
Hemolytic uremic syndrome (HUS) is a disease characterized by microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure. Recent studies have identified a factor H–associated form of HUS, caused by gene mutations that cluster in the C-terminal region of the complement regulator factor H. Here we report how three mutations (E1172Stop, R1210C, and R1215G; each of the latter two identified in three independent cases from different, unrelated families) affect protein function. All three mutations cause reduced binding to the central complement component C3b/C3d to heparin, as well as to endothelial cells. These defective features of the mutant factor H proteins explain progression of endothelial cell and microvascular damage in factor H–associated genetic HUS and indicate a protective role of factor H for tissue integrity during thrombus formation.
Introduction
Hemolytic uremic syndrome (HUS) is a disease characterized by microangiopathic hemolytic anemia, thrombocytopenia, and acute renal failure, secondary to widespread microthrombi and reactive endothelial proliferation (1–3). The most prevalent form of HUS follows infection by some strains of Escherichia coli, which produce a powerful verocytotoxin (VTEC) or Shiga-like toxin. This form is self-limiting and nonrecurring, with complete recovery in about 90% of cases. This pattern contrasts with non-VTEC cases, which are rare, sometimes familial, often recurrent, and generally with a poor outcome. Emerging data indicate that the latter form may be caused by genetic deficiency of the multifunctional, multidomain complement regulator factor H (2, 4–9). Mutations in the factor H gene have been found in a subgroup of patients; consequently; the term factor H–associated HUS was created (10).
Factor H is a 150-kDa multifunctional single-chain plasma glycoprotein that plays a pivotal role in the regulation of the alternative pathway of complement. The protein acts as a cofactor for factor I in the degradation of newly formed C3b molecules and controls decay, formation and stability of newly formed C3 convertases C3bBb (11–13). The secreted plasma protein is organized in 20 homologous units, termed short consensus repeats (SCRs). In addition, factor H displays anti-inflammatory functions, acts as a ligand for plasma proteins, including C-reactive protein, adrenomedullin, osteopontin, or bone sialoprotein, and binds to integrins or ECM proteins (14, 15). Detailed structure-function studies have localized three distinct binding regions on factor H for the ligands C3b, glycosaminoglycans, or heparin. The presence of multiple interaction sites suggests a rather complex and probably highly controlled interaction of factor H with these molecules. Apparently, the individual sites operate in hierarchy: in the native factor H protein the C-terminal site is directly accessible and mediates initial contact with the ligand (16–18).
As a complement regulator, factor H maintains tissue integrity and possesses anti-inflammatory activities. A lack of functional factor H protein in plasma is associated with recurrent microbial infections, membranoproliferative glomerulonephritis type II, and HUS (19–21). An involvement of the alternative pathway of complement activation in HUS was initially reported in 1980 (22), and a role for the complement regulator factor H was identified in a familial association of two brothers, who both developed the disease and had low plasma levels of factor H (23). A case-controlled study in a large group of patients confirmed that mutations in the gene encoding factor H occur in HUS patients and their families (24), and HUS-associated point mutations in the factor H gene were reported in parallel (5, 25). Mutations within the gene-encoding factor H have been reported in a total of 21 cases of HUS, either sporadic or familial (7–9). These studies identify a factor H–associated form of HUS and indicate a central role of factor H in the pathophysiology of HUS. Interestingly, 78% of these mutations are clustered in one single SCR, i.e., SCR 20, the C-terminal domain of factor H. Mutations occurring at positions 1197, 1210, and 1215 of the protein have been found independently in four, three, and three unrelated cases, respectively (7, 8, 24, 25). This clustering within a single protein domain, and particularly the involvement of the same residues in unrelated cases, suggests that SCR 20, which includes binding sites for C3b/C3d and heparin, is important and central for factor H function. Consequently, exchanges of single amino acid residues may affect the structural integrity, the function of this domain, and that of the protein. Biochemical data show that the C terminus of factor H is central for target recognition and is involved in the differentiation of self and nonself structures (17, 26, 27).
We analyzed the functional consequences of three separate point mutations associated with factor H–associated HUS. The E1172Stop is a novel mutation, and the R1210C and R1215G/Q mutations have each been reported in three independent cases. The mutated factor H protein with the R1210C mutation is identified in serum of two heterozygous patients due to its unusual mobility. All three mutations are heterozygous, and consequently the mutant and the native protein are present in serum and can be directly compared. By heparin chromatography we demonstrate that the mutant forms elute with a different profile. In addition, proteins with the R1210C and the R1215G mutations were generated on a background of SCRs 8–20 of factor H and recombinantly expressed in the baculovirus system. Both mutants show reduced binding to the central complement component C3b/C3d and to heparin, and they bind with lower affinity to the surface of endothelial cells. In addition, the mutant protein with the E1172Stop mutation was purified from patient plasma and shown to have reduced binding to heparin, C3b/C3d, and endothelial cells. This study provides, to our knowledge, the first functional characterization of three mutant factor H proteins that occur in HUS patients.
Methods
Patients.
Patient F106 from family 24 with familial factor H–associated HUS, who is heterozygous for the C3701T/R1210C exchange, and patient F34 from family 01, who has a heterozygous G3717A transition and R1215Q mutation, have been introduced previously (7). Patient R043 has a heterozygous G3587T mutation that results in E1172Stop within SCR 20.
Generation of mutant factor H fragments.
Generation of the wild-type factor H fragment FH 8-20 was described previously (17, 28). After restriction with PstI/SmaI, the FH 8-20 fragment was subcloned into Topo-TA cloning vector (Invitrogen Corp., San Diego, California, USA) and single nucleotide exchanges in SCR 20, according to the sequence reported (7, 25), representing R1210C and R1215G exchange (position of the intact full-length protein) were introduced by the Quik Change site-directed mutagenesis technique (Stratagene, La Jolla, California, USA) according to the manufacturer’s instructions. The indicated primers were used, and the modified nucleotides in the sequence are underlined: forward primers For-R1210C, 5′ CGTCTTTCATCA TGT TCTCACACATTGCGAACAAC 3′, For-R1215G, 5′ CGTTCTCACACATTG GGA ACAACATGTTGGGATGGG 3′; and reverse primers Rev-R1210C 5′, CGCAATGTGTGAGA ACA TGATGAAAGACGATATCC 3′, Rev-R1215G, 5′ CAGTTTCCCATCCCAACATGTTGT TCC CAATGTGTG 3′. The generated fragments were excised by digestion with PstI and SmaI and were subsequently cloned into expression vector pBSV-8His (28).
Insect cell culture.
Spodoptera frugiperda Sf9 cells were grown in monolayer cultures at 27°C in 140-mm2 cell-culture flasks using Insect Xpress medium (BioWhittaker Inc., Walkersville, Maryland, USA) containing 4% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), and Fungizone (250 ng/ml). Cells were transfected with recombinant pBSV-8His carrying the coding sequence for either recombinant wild-type (FH 8-20), the R1210C mutation (FH 8-20/R1210C), or the R1215G mutation (FH 8-20/R1215G), together with baculogold DNA as described (28). After initial infection, single plaques of recombinant virus were isolated, expressed, and propagated as described (28). Following purification, recombinant proteins were desalted into 0.5× PBS using PD 10 columns (Amersham Biosciences, Freiburg, Germany). Proteins were concentrated using Ultrafree-Centrifugal devices (Millipore Corp., Bedford, Massachusetts, USA), protein concentration was measured by the method of Bradford, and purity of samples was determined by SDS-PAGE in combination with silver staining.
Cultivation of endothelial cells.
Human umbilical vein endothelial cells (HUVECs; American Type Culture Collection, Rockville, Maryland, USA) were grown at 37°C and 5% CO2 in DMEM medium (Invitrogen Corp.), supplemented with 10% FCS, penicillin (100 U/ml), streptomycin (100 μg/ml), Fungizone (250 ng/ml), and L-glutamine. For binding experiments, cells grown to confluence in either 140-mm2 cell-culture flasks or on Lab-Tek chamber slides (LabTek II; Nalge-Nunc International, Naperville, Illinois, USA) were washed and cultivated in serum-free DMEM medium for 20–30 h. For FACS analyses HUVECs were washed with sterile PBS and detached from the surface with 0.01% EDTA/PBS.
SDS-PAGE and Western blot analysis.
Human serum, cell culture supernatant from infected insect cells, or purified recombinant protein were separated by 7–10% SDS-PAGE under nonreducing conditions. Proteins were visualized by silver staining or transferred to a nitrocellulose membrane using a semi-dry system (28).
Membranes were treated with blocking buffer (3% BSA in PBS) for 30 min at room temperature. Primary Ab’s, polyclonal goat anti human factor H antiserum (Calbiochem-Novabiochem Corp., San Diego, California, USA), or mAb’s were diluted in blocking buffer. After incubation at 4°C overnight, the membrane was washed three times in PBS followed by incubation with HRP-conjugated secondary Ab for 2 h at room temperature. The membrane was washed thoroughly with PBS before 0.3% (wt/vol) 4-chloro-1-naphtol and 1% (vol/vol) hydrogen peroxide were added as substrates for HRP.
Surface plasmon resonance binding assays.
Protein-protein interactions were analyzed by the surface plasmon resonance technique using a Biacore 3000 instrument (Biacore AB, Uppsala, Sweden), essentially as described earlier (26). Briefly, C3b or C3d were coupled by a standard amine-coupling procedure to the flow cells of a sensor chip (carboxylated dextran chip CM5; Biacore AB). Two flow cells were activated, and C3d (50 μg/ml, dialyzed against 10 mM acetate buffer, pH 5.0) was first injected into one flow cell until a level of coupling corresponding to 4,000 resonance units was reached. Unreacted groups were inactivated by ethanolamine-HCl injection. A reference flow cell was prepared using identical conditions by injecting coupling buffer without protein. Before the binding experiments, the flow cells were washed thoroughly with sequential injections of 2 M NaCl in 10 mM acetate buffer, pH 4.6, and running buffer (PBS, pH 7.4). Recombinant FH 8-20 and mutated proteins FH 8-20/R1210C and FH 8-20/R1215G were dialyzed against running buffer. Each ligand was injected separately into the flow cell coupled with C3d or into a control cell using a flow rate of 5 μl/min at 25°C. The final concentrations of the fluid-phase ligands in the C3d-binding assay ranged from 20 to 30 ng. Each binding interaction was assayed at least twice using independently prepared sensor chips. In addition, mutant factor H protein isolated from plasma of patient R043, which shows the E1172Stop mutation, and factor H (Calbiochem-Novabiochem Corp.) were tested for binding to C3b and C3d immobilized on the chip surface.
Heparin chromatography and binding of native and mutant plasma factor H.
Serum (400 μl) from either patient F106 or a healthy individual was analyzed by heparin affinity chromatography using an ÄKTAPrime system (Amersham Biosciences). Samples were diluted in starting buffer (PBS: 137 mM NaCL, 2.7 mM KCl, 10 mM Na2HPO4, and 2 mM KH2PO4, pH7.4) to a volume of 4 ml and applied to the heparin column (HiTrap; Amersham Biosciences) at a flow rate of 0.5 ml/min. The flow-through was collected and reloaded onto the column four times (17). Columns were extensively washed with starting buffer (50 ml), and bound proteins were eluted using a salt gradient, ranging from 100 to 500 mM NaCl, in a total volume of 15 ml and at a flow rate of 1 ml/min. Fractions of 300 μl were collected and analyzed by SDS-PAGE in combination with silver staining or immunoblotting.
Human serum from patient R043 (500 μl) carrying the E1172Stop mutation was stepwise precipitated with 7 and 13% polyethylenglycol (PEG) 6000. The pellet obtained by precipitation with 13% PEG was dissolved in PBS and applied to the HiTrap column. The column was treated as described above. Individual fractions were assayed for the presence of the mutant and the wild-type protein by SDS-PAGE and Western blot analysis.
Heparin chromatography and binding of recombinant factor H (FH 8-20) and mutant proteins (FH 8-20/R1210C, FH 8-20/R1215Q).
Culture supernatants from insect cells, expressing either recombinant wild-type (FH 8-20), the R1210C mutation (FH 8-20/R1210C), or the R1215G mutation (FH 8-20/R1215G) was diluted in 0.5× PBS, and 5–10 ml of supernatant was applied to a heparin column (HiTrap; Amersham Biosciences) at a flow rate of 1 ml/min. After loading, columns were washed with 100 ml of buffer A (0.5× PBS, 75 mM NaCl) followed by a short wash with 1% of buffer B (500 mM NaCl in PBS). Subsequently, bound proteins were gradually eluted in a total volume of 10 ml using a linear salt gradient ranging from 100 to 500 mM NaCl. Fractions of 500 μl were collected, separated by SDS-PAGE, and used for silver staining or immunoblotting. To compare the elution of all three proteins an overlay of the individual profiles was performed using the Prime View software (Amersham-Biosciences Biotech). For each protein the separation was repeated at least four times, and in all cases identical elution profiles were obtained.
Immunofluorescence staining.
Endothelial cells were grown on eight-well chamber slides, as described above. All subsequent incubations were performed at 4°C. Serum-free medium was removed, and the cell monolayers were washed three times with PBS before fixation with 4% paraformaldehyde for 1 h. Cells were washed thoroughly with buffer, treated with 1% BSA/PBS for 30 min to prevent unspecific binding, and incubated for 3–5 h with purified recombinant protein diluted in 0.5× PBS. Identical amounts of recombinant proteins (wild-type FH 8-20, FH 8-20/R1210C, FH 8-20/R1215G), and FH 15-20 or FH 8-11 as controls, were used. Cells were washed with PBS and incubated overnight with 150 μl of primary Ab, i.e., polyclonal anti–factor H antiserum (Calbiochem-Novabiochem Corp.), or mAb T13, which binds an epitope located within SCRs 15–18 of factor H diluted 1:100 in blocking buffer (final concentration 1.5 μg). To remove excess Ab, cells were washed with PBS and further incubated at room temperature with Alexa fluor-488–conjugated donkey anti-goat or goat anti-mouse antiserum (Molecular Probes, Eugene, Oregon, USA). After 2 h, cells were washed with PBS, stained with propidium iodide (0.5 μg/ml), washed further with PBS, and distilled water, and mounted in fluorescence-preserving medium. Fluorescence staining was visualized with appropriate filter settings using an Olympus BX51 microscope (Olympus Optical Co., Tokyo, Japan). An Olympus ColorView digital camera and analysis software (Soft Imaging System GmbH, Münster, Germany) were used for photography.
FACS analyses.
HUVECs were kept in serum-free medium for 20–30 h. Cells were removed from the growth surface, washed twice with PBS, and 5 × 105 cells were transferred into plastic tubes (Eppendorf, Hamburg, Germany). Unspecific binding sites were blocked with 1% BSA/0.5× PBS for 15 min prior to incubation with recombinant proteins. Duplicate samples were incubated either with 100 μl of recombinant wild-type FH 8-20, FH 8-20/R1210C, or FH 8-20/R1215G for 1 h at 37°C with gentle rocking. Proteins were diluted in 0.5× PBS to final concentrations of 2.5, 5, 10, or 20 μg of wild-type FH 8-20 or to 5 μg for both mutant proteins. In addition, cells were incubated with 5 μg purified mutant factor H isolated from patient R043 or native factor H (Calbiochem-Novabiochem Corp.). Control experiments were performed in the absence of recombinant protein. Cells were thoroughly washed in 0.5× PBS, then 1 μg of mAb T13 (diluted in blocking buffer) was added and the cells were incubated on ice for 30 min. After washing, the secondary antiserum (Alexa-fluor 488–conjugated goat anti-mouse antiserum, diluted 1:100 in blocking buffer) was added. Cells were examined by fluorescence-activated cell sorter (FACScan, Becton-Dickinson Immunocytometry Systems, Mountain View, California, USA). Forward and sidewise scatters were used to define the fluorescent cell population, and 10, 000 events were routinely counted.
Results
Identification of a mutated factor H protein in patient serum.
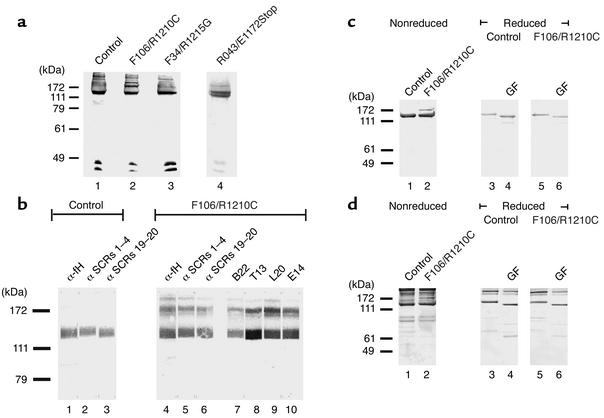
Sera derived from patients F106, F34, and R043, which display distinct, single amino acid exchanges (F106: R1210C; F34: R1215Q and R043 E1172Stop) in one allele of the factor H gene were separated by SDS-PAGE and subjected to Western blot analysis. This assay identified the normal 150-kDa factor H protein in the patients’ samples, as well as in control serum (Figure 1a, lanes 1–4). A band with a mobility of 175 kDa reacting with the factor H antiserum was identified in serum from patient F106, who has the R1210C exchange (Figure 1a, lane 2). This additional band was absent in serum derived from patient F34, the healthy control, and patient R043 (Figure 1a, lane 1, 3, and 4), but was also detected in plasma of a second patient (R16) who has the same R1210C mutation (7, 24). Due to heterozygosity, both the mutated 175-kDa and wild-type 150-kDa form of factor H are detected in sera of patient F106 (Figure 1a, lane 2). Similarly, serum obtained from patient R043, who has a heterozygous G3587T mutation that results in an E1172Stop within SCR 20, was separated by SDS-PAGE. An additional band of higher mobility is identified. This band represents the mutant protein, which lacks most of SCR20 and consequently has a lower molecular weight (Figure 1a, lane 4).
Figure 1.
Identification of mutant factor H protein in serum of HUS patients. (a) Sera from normal individual (control) (lane 1) and HUS patients with the R1210C (F106, lane 2), the R1215G (F34, lane 3), and the E1172Stop mutation (R043, lane 4) were separated by SDS-PAGE and assayed by Western blotting using anti–factor H antiserum. Note the band with 175 kDa depicted by the arrow in serum of patient F106 (lane 2). Serum derived from patient F34 with the R1215G mutation showed normal factor H, and no additional band was detected. In serum from patient with the E1172Stop mutation (R043), a band of higher mobility with a reduced molecular weight is identified. (b) Reactivity of the normal plasma factor H and the 175 band with antisera and mAb’s specific for factor H. Sera from a healthy individual (lanes 1–3) and patient F106 with the R1210C mutation (lanes 4–10) were separated by SDS-PAGE, and after Western blotting, reacted with the indicated polyclonal and monoclonal Ab’s (lanes 1, 4). (c) Western blotting and (d) silver staining of reduced and deglycosylated (N-glycosidase F; GF) control and patient samples.
Characterization of the mutated 175-kDa factor H isoform of patient F106.
To clarify whether the 175-kDa band represents the mutated form of factor H, a set of antisera and mAb’s specific for factor H was employed. All polyclonal antisera and monoclonal anti–factor H Ab’s used reacted with the 175-kDa protein: i.e., polyclonal antisera specific for intact factor H (anti-fH), the N-terminal (anti–SCRs 1–4), and the C-terminal region (anti–SCRs 19–20), as well as mAb’s B22, which binds to the N-terminal region (i.e., SCR 5), T13, and L20 and E14, which bind to the C-terminal region (i.e., SCRs 15–18, SCR 19, and SCR 20) of factor H (Figure 1b). The reactivity with all antisera and Ab’s suggests that the 175-kDa band represents an isoform of factor H and that exchange of a single arginine to a cysteine residue at position 1210 causes the reduction in mobility.
Disulfide bonding is essential for the conformation of SCR-containing proteins, and factor H is also subject to posttranslational processing such as N-linked glycosylation (28). To study whether disulfide bonding and N-linked glycosylation are changed in the mutated factor H protein of patient F106, this isoform was analyzed under reducing and nonreducing conditions and following treatment with N-glycosidase F. Under nonreducing conditions the 175-kDa mutant factor H is identified by its unusual mobility, both by Western blot analysis (Figure 1c, lane 2) and silver staining (Figure 1d, lane 2). Normal factor H in the serum of patient 106 and in the control sample show identical mobility of 150 kDa. Upon reduction, the mobility of the wild-type factor H protein is decreased due to the disruption of disulfide bonds (Figure 1c, lane 3). Removal of N-linked sugar moieties by treatment with N-glycosidase F reduced the mass of the protein and consequently increased the mobility of factor H (Figure 1c, lane 4). As reduction decreases the number of epitopes recognized by the antiserum, which was raised against the intact native protein, the reduced native factor H showed lower intensity of staining (Figure 1c, lanes 3–6). Silver staining of the same samples confirmed that comparable amounts of proteins were used (Figure 1d). The factor H mutant was affected by this type of treatment. The aberrant band was detected under nonreducing conditions, but not under reducing conditions (Figure 1, c and d, lane 2 versus lanes 5 and 6). Thus, upon reduction, the mutated and the native wild-type factor H proteins have identical mobility. These results confirm further that the aberrant band represents the mutated factor H protein.
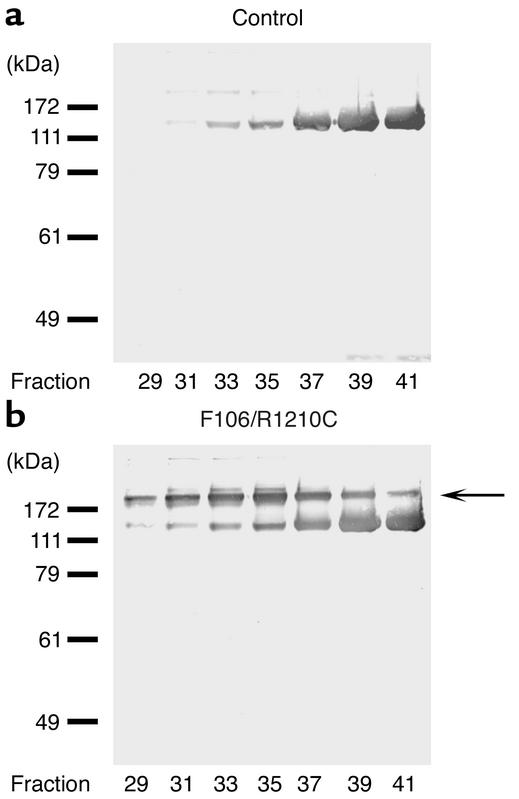
Heparin binding of mutant and native factor H: binding of mutant factor H (R1210C) to heparin is reduced.
The fact that both the mutated and the wild-type form of factor H could be easily distinguished in serum of patient F106 allowed us to assay and compare the heparin-binding characteristics of the two proteins directly. To this end sera derived from patient F106 and from a healthy control were subjected to heparin affinity chromatography. Bound proteins were eluted with a linear salt gradient, and eluted fractions were collected and assayed by SDS-PAGE and Western blot analysis. Factor H from the healthy individual eluted at a salt concentration of approximately 200 mM NaCl and reached its peak in fraction 41 (Figure 2a). The two factor H forms in serum of patient F106 showed different profiles. Wild-type protein showed an elution profile very similar to factor H from control serum (Figure 2b, lower band); in contrast, the mutant protein eluted earlier, starting in fraction 29, and reached its peak in fraction 35 (Figure 2b, upper band, see arrow). This experiment shows that the single amino acid exchange (R1210C) within SCR 20 of factor H caused reduced binding to heparin.
Figure 2.
The mutant form of factor H binds less efficiently to heparin. Sera from a healthy control (a) and from HUS patient F106 with the R1210C mutation (b) were applied to heparin affinity chromatography, and after washing, bound proteins were eluted with the linear NaCl gradient. Individual fractions of 500 μl were collected starting at an NaCl concentration of 200 mM. The fractions were assayed by SDS-PAGE in combination with Western blotting. The 175-kDa mutant factor H eluted at a lower salt concentration (fraction 35) prior to the wild-type factor H protein (fraction 41). The arrow indicates the position of the mutant factor H protein.
Recombinant expression of mutant factor H in insect cells.
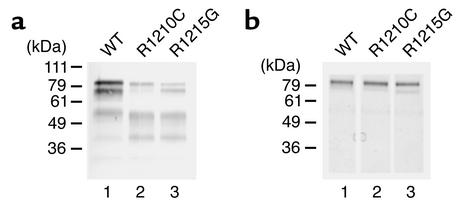
To further analyze whether the described amino acid exchanges affect protein function, proteins displaying the R1210C or the R1215G exchange and wild-type factor H were recombinantly expressed in the baculovirus system. The SCR 8–20 protein backbone was chosen in order to exclude the contribution of the complement regulatory domain located within SCRs 1–4, as well as the C3b- and heparin-binding domains, which are located within SCRs 1–7 (29–31). Mutant proteins with the R1210C (FH 8-20/R1210C) and the R1215G exchange (FH 8-20/R1215G) were generated by site-directed mutagenesis. Recombinant wild-type (FH 8-20), as well as mutant proteins, were expressed and secreted by insect cells, as detected by immunofluorescence staining of infected cells (data not shown) and by SDS-PAGE and Western blot analysis of the cell culture supernatant (Figure 3a). The His tag added to the C terminus of the recombinant proteins allowed purification of all three recombinant proteins by nickel chelate chromatography (Figure 3b). All three recombinant proteins have an apparent mass of approximately 80 kDa. The additional bands of lower mass are likely degradation products, because they are absent in the purified proteins (Figure 3b). The recombinant protein with the R1210C mutation has the same mobility as the wild-type protein.
Figure 3.
Recombinant expression of wild-type factor H SCRs 8–20 and HUS mutations of factor H. (a) Culture supernatants of insect cells infected with the recombinant viruses were harvested 5 days after infection, separated by SDS-PAGE, and used for Western blot analysis. (b) Purified recombinant proteins representing the wild-type factor H protein (SCRs 8–20) or the proteins with the indicated mutations were separated by SDS-PAGE and detected by silver staining. The mobility of the marker is indicated on the left.
Binding of wild-type and mutant proteins to C3d.
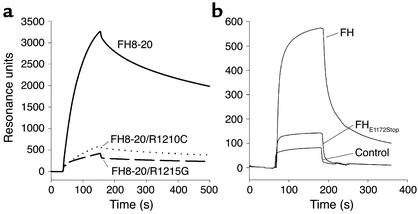
The C terminus of factor H is crucial for interaction with C3b/C3d. Therefore, binding of the recombinant proteins to C3d was measured by the surface plasmon-resonance technique. C3d was immobilized to the chip surface, and the wild-type and mutant FH 8-20 proteins were tested for binding from the fluid phase. Wild-type recombinant FH 8-20 showed strong binding to C3d; the FH 8-20/R1210C and also the FH 8-20/R1215G mutants showed rather weak binding to C3d (Figure 4a). Thus, both mutations affect C3d binding.
Figure 4.
Binding of recombinant and purified mutant factor H protein to C3d analyzed by surface plasmon resonance. (a) The wild-type FH8-20 protein (solid line), mutant proteins FH8-20/R1210C (dotted line), and FH8-20/R1215G (dashed line) were injected at a concentration of 2–3 μg/ml into a flow cell coupled with C3d. The binding of the wild-type protein is apparent from the distinct association and dissociation phases of the binding curve, whereas the binding of the mutants is decreased. All recombinant proteins were tested on two different chip surfaces coated with C3d, and representative figures are shown. (b) Binding of purified mutant factor H (FH) (E1172Stop) to C3b was assayed and compared with that of purified native factor H.
In addition, the mutant factor H with the E1172Stop mutation, which was purified from plasma of patient R043, as well as purified factor H, were tested for binding to immobilized C3b (Figure 4b) and C3d (data not shown). Binding of the mutant protein was severely reduced as compared to native factor H.
Binding of recombinant proteins to heparin.
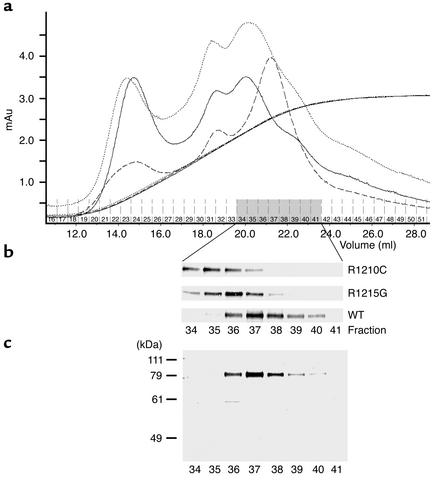
Binding of the two recombinant mutant proteins to heparin was assayed by heparin affinity chromatography. Culture supernatants containing recombinant protein were applied to a heparin column, and after extensive washing, bound proteins were eluted using a linear salt gradient. The elution conditions for the three proteins were identical, as demonstrated by the overlapping conductivity profiles (Figure 5a). Identical amounts of protein were applied to the column, thus the elution profile and the protein concentration in the individual fractions correlate directly with their affinity to the heparin matrix. Eluted fractions were assayed by SDS-PAGE and Western blot analysis. The R1210C mutant (i.e., FH 8-20/R1210C) was predominantly detectable in fractions 34–36, the R1215G mutant (i.e., FH 8-20/R1215G) was present in fractions 35–37, and the recombinant wild-type protein (FH 8-20) was detected in fractions 36–38. Purity of the proteins in the elute fractions of FH 8-20 was confirmed by SDS-PAGE in combination with silver staining, as indicated for the wild-type protein (Figure 5c). This assay shows that both recombinant mutant proteins bind heparin with lower affinity than the wild-type protein and, in addition, reveals a difference between the two mutant forms, with the R1210C mutant demonstrating a lower heparin-binding activity.
Figure 5.
Heparin affinity chromatography binding of recombinant wild-type deletion mutant factor H (SCRs 8-20). (a) Culture supernatant of insect cells infected with recombinant baculovirus coding for the recombinant wild-type protein (FH 8-20) and the two mutant forms, R1210C mutant (i.e., FH 8-20/R1210C) or the R1215G mutant (i.e., FH 8-20/R1215G) was applied to heparin affinity chromatography. After loading, the column was thoroughly washed, and bound proteins were eluted by an NaCl gradient. Absorbencies are indicated for recombinant wild-type protein by the dashed line, the R1210C mutant by the solid line, and the R1215 mutant by the dotted line. The identical conductivity graphs show that elution was performed under identical conditions. mAu, milliampere units. (b) SDS-PAGE and Western blot analysis of fractions 34–41 of the individual proteins. (c) The separation yields pure protein as shown for wild-type protein fractions 34–41 after SDS-PAGE separation in combination with silver staining. The mobility of the marker proteins is indicated on the left.
Binding of recombinant mutant, mutant Factor H (E1172Stop), and wild-type proteins to endothelial cells.
For further characterization the recombinant and the purified mutant proteins and wild-type protein were assayed for binding to HUVECs by immunofluorescence staining and FACS analysis.
Immunofluorescence staining.
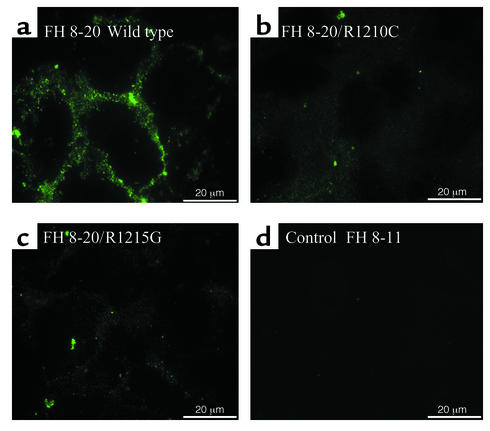
HUVECs cultivated in serum-free medium were incubated in supernatant containing either recombinant wild-type protein (FH 8-20), the FH 8-20/R1210C or the FH 8-20/R1215G mutant, and an additional recombinant control protein (FH 8-11). Cells were stained with factor H antiserum together with FITC-labeled secondary Ab and propidium iodide. The three proteins bound to HUVECs, and FH 8-11 used as a negative control did not bind. Surface staining was more prominent for cells incubated with wild-type protein (Figure 6a) as compared with cells treated with mutant proteins (Figure 6, b and c).
Figure 6.
Binding of recombinant wild-type and mutant factor H proteins to HUVECs: immunofluorescence. HUVECs cultivated in serum-free medium were incubated with cell culture supernatant containing the indicated recombinant proteins, i.e., the recombinant wild-type protein (FH 8-20; WT) (a), the R1210C mutant (i.e., FH 8-20/R1210C) (b), and the R1215G mutant (i.e., FH 8-20/R1215G) (c), and FH 8-11 (d) was used as a control. Unfixed cells were used directly for immunofluorescence analysis by staining with factor H antiserum in combination with an FITC-labeled secondary antiserum. The bars in the lower corner show the length of 20 μm.
FACS analyses.
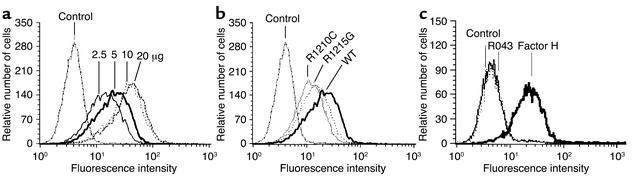
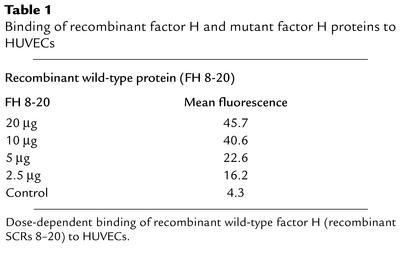
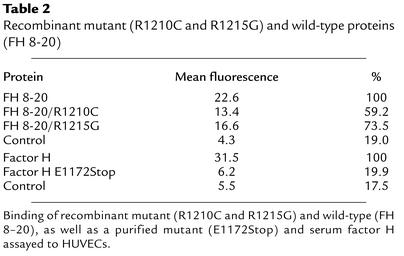
Binding to HUVECs was further assayed by FACS analysis. A dose-dependent binding of the recombinant wild-type protein to endothelial cells was observed (Figure 7a). Mean values ranged from 16.2 to 45.7 mean fluorescence using 3.5 to 20 μg of protein (Table 1). The two mutant proteins bound with lower affinity as compared to wild-type FH 8-20 protein (Figure 7b and Table 2). A comparison of the mean values shows that the binding of FH 8-20/R1210C and FH 8-20/R1215G mutants was reduced to 59% and 73% as compared with the wild-type form (Table 2). Thus, the two mutant proteins bind with a reduced affinity to HUVECs. In addition, binding of purified mutant factor H protein with the E1172Stop was assayed and compared with normal purified factor H. Binding of purified wild-type factor H was almost similar to that of the recombinant wild-type protein. The purified mutant factor H protein with the E1172Stop mutation, however, showed strongly reduced binding to endothelial cells. Binding of this mutant protein was severely reduced (mean 6.2), almost reaching values obtained with control (mean 5.5), and was even lower than that of the recombinant mutant proteins with the single amino acid substitution.
Figure 7.
Binding of recombinant mutant, mutant factor H (E1172Stop), and wild-type proteins to endothelial cells. (a) Dose-dependent binding of recombinant wild-type factor H (FH 8-20). HUVECs incubated in serum-free medium were incubated with the indicated amounts of purified recombinant wild-type factor H 8-20, and binding was assayed by FACS analysis. (b) Comparison of recombinant mutant R1210C (i.e., FH 8-20/R1210C), or R1215G (i.e., FH 8-20/R1215G), or wild-type factor H (FH 8-20). HUVECs cultivated in serum-free medium were incubated with the indicated proteins, and binding was assayed by FACS analysis. (c) Binding of purified factor H protein with the E1172Stop mutation isolated from serum of patient R043 and of purified wild-type factor H to HUVECs. HUVECs were treated with either 5 μg of mutant factor H protein purified from plasma of patient R043 or with 5 μg purified intact wild-type factor H, and binding was assayed by FACS.
Table 1.
Binding of recombinant factor H and mutant factor H proteins to HUVECs
Table 2.
Recombinant mutant (R1210C and R1215G) and wild-type proteins (FH 8-20)
Discussion
Factor H gene mutations have been reported in a subgroup of patients with familial, recurrent, and even sporadic HUS. Here we demonstrate that three separate single amino acid exchanges (i.e., R1210C, R1215G, and E1172Stop) occurring in the most C-terminal domain of this human immune regulator affect protein function, and we discuss the role of these mutations in the pathophysiology of factor H–associated genetic HUS. In patient F106 the factor H mutant, which has an arginine→cysteine exchange at position 1210 (R1210C), shows an aberrant mobility in SDS-PAGE. Because this patient is heterozygous, the heparin-binding activity of both the mutant and the wild-type factor H could be compared directly in serum. Affinity chromatography shows for the mutant factor H protein a reduced binding to heparin. In addition, recombinant proteins representing either the R1210C or the R1215G mutation, which each have been identified in three independent HUS cases, were created and expressed in the baculovirus system. The C3b/C3d and heparin-binding activity of the two mutant proteins was assayed and compared with that of the recombinant wild-type protein. Both mutants bind with lower affinity to C3d, as assayed by surface plasmon resonance, and similarly both mutant proteins showed reduced binding to heparin. In vitro binding assays, as performed by immunofluorescence staining and FACS analyses, reveal for both mutants reduced binding to intact endothelial HUVECs. Based on the mean fluorescence values, binding of the FH 8-20/R1210C mutant is reduced to 59.2% and for the FH 8-20/R1215G mutant to 73.5%, respectively. In addition the purified mutant factor H protein isolated from patient R043, which lacks the most C-terminal part of SCRs due to an E1172Stop mutation, did not bind to HUVECs at all. These data show for two distinct single amino acid exchanges reduced binding and for the E1172Stop a complete loss of binding. Thus, three mutations that occur in HUS patients show functional defects in terms of C3b/C3d- and heparin-binding as well as attachment to endothelial cells. Particularly, the mutant protein with the E1172Stop did not bind to HUVECs at all.
Detailed structure function analyses have localized the complement regulatory domains of factor H within the N-terminal SCRs 1–4 (29–31). Three heparin-binding domains have been mapped to, or close to, SCR 7, SCR 13, and SCR 20 of factor H (16, 18, 27). Similarly, three C3b-binding domains have been localized to SCRs 1–4, SCRs 8–12, and SCRs 19–20, respectively (16, 27). The C terminus of native factor H seems to play an essential role: (a) this domain is highly conserved in all five factor H–related proteins identified so far (32); (b) recent studies show that the C terminus, i.e., SCRs 19 and 20 of native factor H, provides initial contact with target structures (17, 33); and (c) factor H gene mutations observed in HUS patients cluster within SCR 20 (7–9, 34). In two of the mutations, Arg residues are affected, and positively charged residues are considered essential for binding and interaction with heparin. The experimental results presented here show, we believe for the first time, that factor H mutations occurring in HUS patients affect interaction with the ligands C3 and heparin, as well as binding to the surface of endothelial cells (Figures 4, 5, 6, and 7). Similarly, reduced binding to heparin was also observed with the mutated factor H protein derived from patient F106 (Figure 2) and R043 (data not shown). A detailed comparison reveals differences between the three analyzed mutant proteins: the FH 8-20/R1210C mutant binds C3d and heparin, as well as endothelial cells, with lower affinity than the FH 8-20/R1215G mutant, and the purified mutant protein with the E1172Stop mutation binds C3b/C3d and heparin with lower affinity, but did not bind to endothelial cells. Despite this slight difference, all three mutant proteins cause the same disease, indicating that reduced interaction with the surface of endothelial cells is central to the pathophysiology of HUS. The impaired interaction with endothelial cell surface as confirmed by immunofluorescence staining of HUVECs (Figure 6), as well as FACS analyses (Figure 7), suggests reduced binding of the mutated factor H to the damaged endothelium and may favor exposed subendothelial matrix (35–37). This effect may cause a lower density of surface-bound factor H and a diminished complement regulatory and anti-inflammatory activity. The additional missense mutations identified so far in the factor H gene of several HUS patients may have related or even similar biological effects.
The majority of the factor H–associated genetic HUS cases are heterozygous and represent either single amino acid exchanges or premature stop codons within SCR 20 (10, 20). Both factor H alleles are coexpressed, and each allele provides half levels of secreted plasma factor H. In a heterozygous HUS patient the normal allele encodes a functionally intact protein, which provides 50% of plasma factor H. The second mutated allele represents either (a) a null mutant, (b) a truncated protein due to a premature stop codon, or (c) a secreted plasma protein, which is less stable or functionally inactive. Heterozygous mutations are associated with incomplete penetrance of the disease as documented by a number of healthy carriers described within families. It is possible that the genetic change is a predisposing factor and that upon an environmental insult precipitates the disorder. Apparently under normal circumstances half maximal levels of intact factor H are sufficient to maintain tissue integrity. Upon insult, however, e.g. inflammation or infection, complement is activated, initiates inflammatory reactions, and the release of inflammatory mediators cause retraction of endothelial cells, endothelial cell damage, and exposure of the subendothelial matrix. This scenario promotes further activation of the alternative complement pathway, deposition of C3, and formation of C3 convertases, particularly at the damaged site at the exposed subendothelial matrix (37). During such conditions, maximal deposition and activity of factor H as provided by two intact alleles seems essential to control the complement cascade in order to protect damaged cells and tissue. For a HUS patient heterozygous for factor H mutations, the reduced factor H activity seems incapable of efficiently restricting complement activation to protect self cells and the exposed subendothelial matrix. This will allow propagation of endothelial damage and initiation of microvascular thrombosis. In addition, platelet function may be directly affected during these steps. Factor H is stored in the α-granules of platelets and is released upon platelet activation (38), and a loss or modification of factor H activity may directly affect platelet function. The identification of factor H mutations as a cause for endothelial damage and/or platelet activity for progression of HUS allows the design of new approaches of diagnosis, therapy, and disease prevention.
Note added in proof.
After submission of this manuscript, two papers were published. The first paper shows that the C terminus of factor H is relevant for the discriminatory role of factor H (39). The second report characterizes the same mutation R1210C as discussed in this manuscript. The authors show that the mutant protein associates with albumin, and they demonstrate reduced binding of the mutant protein to surface-bound C3b (40).
Acknowledgments
The work of the authors is funded by the Deutsche Forschungsgemeinschaft, the Thüringer Ministerium für Wissenschaft, Forschung und Kunst, and the Foundation for Children with Atypical HUS. M. Jozsi was supported by a research fellowship from the Joseph Eötvös Scholarship Public Foundation, Budapest, Hungary.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: Hemolytic uremic syndrome (HUS); verocytotoxin (VTEC); short consensus repeat (SCR); human umbilical vein endothelial cell (HUVEC); polyethylenglycol (PEG).
References
- 1.Kaplan BS, Meyers KE, Schulman SL. The pathogenesis and treatment of hemolytic uremic syndrome. J. Am. Soc. Nephrol. 1998;9:1126–1133. doi: 10.1681/ASN.V961126. [DOI] [PubMed] [Google Scholar]
- 2.Remuzzi G, Ruggenenti P. The hemolytic uremic syndrome. Kidney Int. Suppl. 1998;66:S54–S57. [PubMed] [Google Scholar]
- 3.Kaplan BS, Drummond KN. The hemolytic-uremic syndrome is a syndrome. N. Engl. J. Med. 1978;298:964–966. doi: 10.1056/NEJM197804272981710. [DOI] [PubMed] [Google Scholar]
- 4.Thompson RA, Winterborn MH. Hypocomplementaemia due to a genetic deficiency of beta 1H globulin. Clin. Exp. Immunol. 1981;46:110–119. [PMC free article] [PubMed] [Google Scholar]
- 5.Buddles MR, Donne RL, Richards A, Goodship J, Goodship TH. Complement factor H gene mutation associated with autosomal recessive atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2000;66:1721–1722. doi: 10.1086/302877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieson P. Complement factor H and haemolytic uraemic syndrome. Lancet. 2002;359:801–802. doi: 10.1016/S0140-6736(02)07866-2. [DOI] [PubMed] [Google Scholar]
- 7.Caprioli J, et al. The molecular basis of familial hemolytic uremic syndrome: mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J. Am. Soc. Nephrol. 2001;12:297–307. doi: 10.1681/ASN.V122297. [DOI] [PubMed] [Google Scholar]
- 8.Perez-Caballero D, et al. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards A, et al. Factor H mutations in hemolytic uremic syndrome cluster in exons 18-20, a domain important for host cell recognition. Am. J. Hum. Genet. 2001;68:485–490. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor CM. Complement factor H and the haemolytic uraemic syndrome. Lancet. 2001;358:1200–1202. doi: 10.1016/s0140-6736(01)06339-5. [DOI] [PubMed] [Google Scholar]
- 11.Pangburn MK, Muller-Eberhard HJ. Kinetic and thermodynamic analysis of the control of C3b by the complement regulatory proteins factors H and I. Biochemistry. 1983;22:178–185. doi: 10.1021/bi00270a026. [DOI] [PubMed] [Google Scholar]
- 12.Whaley K, Ruddy S. Modulation of the alternative complement pathways by beta 1 H globulin. J. Exp. Med. 1976;144:1147–1163. doi: 10.1084/jem.144.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zipfel PF, et al. Factor H and disease: a complement regulator affects vital body functions. Mol. Immunol. 1999;36:241–248. doi: 10.1016/s0161-5890(99)00038-3. [DOI] [PubMed] [Google Scholar]
- 14.Fedarko NS, Fohr B, Robey PG, Young MF, Fisher LW. Factor H binding to bone sialoprotein and osteopontin enables tumor cell evasion of complement-mediated attack. J. Biol. Chem. 2000;275:16666–16672. doi: 10.1074/jbc.M001123200. [DOI] [PubMed] [Google Scholar]
- 15.DiScipio RG, Daffern PJ, Schraufstatter IU, Sriramarao P. Human polymorphonuclear leukocytes adhere to complement factor H through an interaction that involves alphaMbeta2 (CD11b/CD18) J. Immunol. 1998;160:4057–4066. [PubMed] [Google Scholar]
- 16.Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on complement factor H interacts with a distinct site on C3b. J. Biol. Chem. 2000;275:27657–27662. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- 17.Prodinger WM, Hellwage J, Spruth M, Dierich MP, Zipfel PF. The C-terminus of factor H: monoclonal antibodies inhibit heparin binding and identify epitopes common to factor H and factor H-related proteins. Biochem. J. 1998;331:41–47. doi: 10.1042/bj3310041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blackmore TK, et al. Identification of the second heparin-binding domain in human complement factor H. J. Immunol. 1998;160:3342–3348. [PubMed] [Google Scholar]
- 19.Ault BH, et al. Human factor H deficiency. Mutations in framework cysteine residues and block in H protein secretion and intracellular catabolism. J. Biol. Chem. 1997;272:25168–25175. doi: 10.1074/jbc.272.40.25168. [DOI] [PubMed] [Google Scholar]
- 20.Zipfel PF, et al. Complement factor H and hemolytic uremic syndrome. Int. Immunopharmacol. 2001;1:461–468. doi: 10.1016/s1567-5769(00)00047-3. [DOI] [PubMed] [Google Scholar]
- 21.Warwicker P, et al. Familial relapsing haemolytic uraemic syndrome and complement factor H deficiency. Nephrol. Dial. Transplant. 1999;14:1229–1233. doi: 10.1093/ndt/14.5.1229. [DOI] [PubMed] [Google Scholar]
- 22.Monnens L, Molenaar J, Lambert PH, Proesmans W, van Munster P. The complement system in hemolytic-uremic syndrome in childhood. Clin. Nephrol. 1980;13:168–171. [PubMed] [Google Scholar]
- 23.Kaplan BS, Chesney RW, Drummond KN. Hemolytic uremic syndrome in families. N. Engl. J. Med. 1975;292:1090–1093. doi: 10.1056/NEJM197505222922102. [DOI] [PubMed] [Google Scholar]
- 24.Noris M, et al. Hypocomplementemia discloses genetic predisposition to hemolytic uremic syndrome and thrombotic thrombocytopenic purpura: role of factor H abnormalities. Italian Registry of Familial and Recurrent Hemolytic Uremic Syndrome/Thrombotic Thrombocytopenic Purpura. J. Am. Soc. Nephrol. 1999;10:281–293. doi: 10.1681/ASN.V102281. [DOI] [PubMed] [Google Scholar]
- 25.Warwicker P, et al. Genetic studies into inherited and sporadic hemolytic uremic syndrome. Kidney Int. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 26.Jokiranta TS, et al. Complement C3b interactions studied with surface plasmon resonance technique. Int. Immunopharmacol. 2001;1:495–506. doi: 10.1016/s1567-5769(00)00042-4. [DOI] [PubMed] [Google Scholar]
- 27.Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10996–11001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn S, Zipfel PF. The baculovirus expression vector pBSV-8His directs secretion of histidine-tagged proteins. Gene. 1995;162:225–229. doi: 10.1016/0378-1119(95)00360-i. [DOI] [PubMed] [Google Scholar]
- 29.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J. Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 30.Kuhn S, Zipfel PF. Mapping of the domains required for decay acceleration activity of the human factor H-like protein 1 and factor H. Eur. J. Immunol. 1996;26:2383–2387. doi: 10.1002/eji.1830261017. [DOI] [PubMed] [Google Scholar]
- 31.Kuhn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J. Immunol. 1995;155:5663–5670. [PubMed] [Google Scholar]
- 32.Zipfel PF, Jokiranta TS, Hellwage J, Koistinen V, Meri S. The factor H protein family. Immunopharmacology. 1999;42:53–60. doi: 10.1016/s0162-3109(99)00015-6. [DOI] [PubMed] [Google Scholar]
- 33.Perkins SJ, Goodship TH. Molecular modelling of the C-terminal domains of factor H of human complement: a correlation between haemolytic uraemic syndrome and a predicted heparin binding site. J. Mol. Biol. 2002;316:217–224. doi: 10.1006/jmbi.2001.5337. [DOI] [PubMed] [Google Scholar]
- 34.Warwicker P, Goodship JA, Goodship TH. Factor H—US? Nephrol. Dial. Transplant. 1998;13:1921–1923. doi: 10.1093/ndt/13.8.1921. [DOI] [PubMed] [Google Scholar]
- 35.Zipfel PF. Hemolytic uremic syndrome: how do factor H mutants mediate endothelial damage? Trends Immunol. 2001;22:345–348. doi: 10.1016/s1471-4906(01)01972-x. [DOI] [PubMed] [Google Scholar]
- 36.Zoja C, Morigi M, Remuzzi G. The role of the endothelium in hemolytic uremic syndrome. J. Nephrol. 2001;4(Suppl.):S58–S62. [PubMed] [Google Scholar]
- 37.Hindmarsh EJ, Marks RM. Complement activation occurs on subendothelial extracellular matrix in vitro and is initiated by retraction or removal of overlying endothelial cells. J. Immunol. 1998;160:6128–6136. [PubMed] [Google Scholar]
- 38.Devine DV, Rosse WF. Regulation of the activity of platelet-bound C3 convertase of the alternative pathway of complement by platelet factor H. Proc. Natl. Acad. Sci. U. S. A. 1987;84:5873–5877. doi: 10.1073/pnas.84.16.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pangburn MK. Localization of the host recognition functions of complement factor H at the carboxyl-terminal: implications for hemolytic uremic syndrome. J. Immunol. 2002;169:4702–4706. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez-Corral P, et al. Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2002;71:1285–1295. doi: 10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]