Abstract
Bacteria have aggressive acquisition processes for iron, an essential nutrient. Siderophores are small iron chelators that facilitate cellular iron transport. The siderophore enterobactin is a triscatechol derivative of a cyclic triserine lactone. Studies of the chemistry, regulation, synthesis, recognition, and transport of enterobactin make it perhaps the best understood of the siderophore-mediated iron uptake systems, displaying a lot of function packed into this small molecule. However, recent surprises include the isolation of corynebactin, a closely related trithreonine triscatechol derivative lactone first found in Gram-positive bacteria, and the crystal structure of a ferric enterobactin complex of a protein identified as an antibacterial component of the human innate immune system.
Various aspects of iron regulation, transport, storage, and utilization appear in several articles in this issue of PNAS. As often noted (1–4) iron is needed in organisms in relatively large amounts. A 70-kg adult human has ≈5 g of iron (≈10−3 M for body volume), whereas a bacterial cell of 10−9 cm3 requires 105 to 106 ferric ions per generation to maintain the required 10−6 M internal concentration (5).
Iron: Can't Live With It, Can't Live Without It
While iron's abundance in the Earth's crust, spin state, and redox tuneability makes it the most versatile of the transition elements, the insolubility of ferric hydroxide at pH 7.4 limits the concentration of [Fe3+] (the free aqueous ion) to ≈10−18 M (1). However, even below this concentration, free ferric ion is toxic. To avoid toxicity and regulate iron transport, the human serum iron transport protein, transferrin, maintains the free ferric ion concentration at ≈10−24 M (6–8). Pathogenic bacteria must compete against this thermodynamic limit to obtain iron from the serum or tissues of its human host.
It is difficult to overestimate the significance of iron as a limiting nutrient in microbial growth. Excess iron increases the virulence of organisms as diverse as Escherichia (9), Klebsiella (10), Listeria (11), Neisseria (12), Pasteurella (11), Shigella (13), Salmonella (14, 15), Vibrio (16), and Yersinia (17). Iron dextran injections in children, originally intended to prevent iron deficiencies, enhanced Escherichia coli bacteremia and meningitis (18). As early as the 1850s, Dr. Armand Trousseau, a Parisian professor of clinical medicine, warned his students against administering an iron preparation, then widely used, to tuberculosis patients (19). Nonlethal injections of E. coli in guinea pigs become lethal by the concomitant addition of either heme or enough iron to saturate the transferrin (9, 20). The virulence of Yersinia enterocolitica is enhanced 10 million-fold by the peritoneal injection of ferric desferrioxamine (17, 21). A similar effect is seen if desferrioxamine is supplied during infections of Klebsiella and Salmonella (15), whereas direct correlation between the LD50 of Vibrio vulnificans and iron availability has been demonstrated (22).
Bacteria have consequently evolved aggressive iron acquisition processes. Powerful and selective iron chelators (siderophores) are produced and secreted specifically in response to iron deficiency. The regulator protein Fur (ferric uptake regulation) (23–25) or Fur-like proteins, regulate iron uptake in many bacterial species. Some Gram-positive bacteria (such as streptomyces, corynebacteria, and mycobacteria) use the DtxR (diphtheria toxin regulator) protein (26). Regulation of iron uptake and siderophore production has recently been the subject of an excellent review (27).
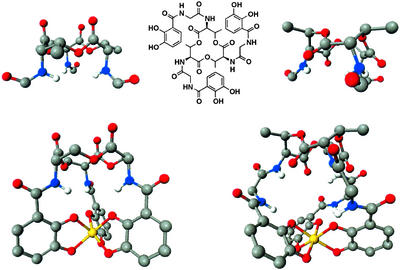
Spectacular advances have taken place in the last 10 years in understanding the recognition and transport processes involved in siderophore-mediated iron acquisition. The structural characterization of proteins and identification of genes have elucidated the steps involved in siderophore-mediated iron transport in several different systems. The general mechanism is initiated when the ferric siderophore complex binds to the receptor protein on the microbial cell surface. Translocation of the complex requires active transport, ending with iron release and metabolism inside the cell. Enterobactin (Fig. 1) coordinates iron through three catecholate functionalities that are linked to a triserine macrocycle. The remarkable stability of the ferric enterobactin (FeEnt) complex, the role of its stereochemistry in recognition and transport, and the significance of the trilactone ring for iron release are all now understood. Although many different siderophores and several types of transport systems are known, it seems timely to take a retrospective look at enterobactin as perhaps the best understood of the siderophore-mediated iron transport systems. How can so much function be packed into this small molecule?
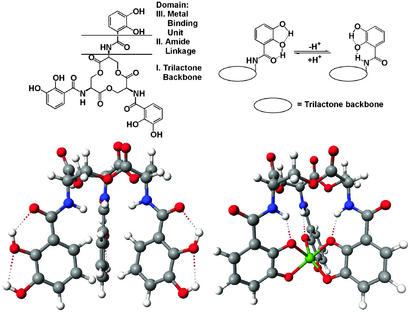
Figure 1.
Schematic and space filling structures of enterobactin and its ferric complex. (Upper) The catechol, amide linkage and triserine ring components of enterobactin, and the conformation charge, driven by hydrogen bonding, following deprotonation (or metal complexation). (Lower Right) The structure of the V(IV) complex (30), considered to be a close model of the Fe(III) complex. (Lower Left) A computer-generated structure of uncomplexed enterobactin based on the trilactone structure of Seebach et al. (52) and appended catecholamide groups as seen in the crystal structure of an enterobactin analog (79). Note how hydrogen bonding locks the catechol group into one of two rigid conformations, the interconversion of which is triggered by deprotonation/metal complexation.
FeEnt Structure
The isolation of enterobactin (or enterochelin) in 1970 resulted in the first of many controversies about this molecule. Pollack and Neilands (28), who isolated the compound from Salmonella typhimurium, named it enterobactin, whereas O'Brien and Gibson (29), who isolated the compound from E. coli, called it enterochelin. Although the O'Brien and Gibson paper was submitted first, the Pollack and Neilands paper was printed first, resulting in the widespread use of both names.
Characterization of enterobactin proved challenging, with many flawed analyses published over the years. The coordination mode of FeEnt involves a hexadentate triscatecholate geometry with a Δ configuration at the metal center, as shown in Fig. 1 Lower Right (30, 31). Iron release is through the action of the cytoplasmic esterase (32, 33). However, synthetic analogs of enterobactin, which are not susceptible to hydrolysis, are ≈5% as effective in delivering iron (34), implying a secondary pathway of iron release. Unfortunately, perhaps because of previous errors, there continues to be confusion on this subject. Even a recent major microbiological review (35) incorrectly assigned the structure of FeEnt and its major route of iron release.
Enterobactin is predisposed for metal binding (31, 36–38). The conformation of the neutral catecholamide free ligand has the ortho-hydroxy proton hydrogen-bonded to the amide oxygen atom. Upon deprotonation this conformation changes to the trans form (Fig. 1 Upper Right), in which the amide proton hydrogen-bonds to the ortho-hydroxy oxygen atom. At physiological pH ≈50% of the three catecholate units have their hydroxy oxygen atoms oriented in the same direction as the amide proton. If we view the siderophores as both iron prospecting and sequestering agents, this dynamic conformation of free enterobactin seems optimally tuned to perform both tasks: the free ligand conformation favors rapid initial binding of an iron atom, whereas the conformation change that results from proton loss promotes full encapsulation of the iron atom.
Biosynthesis of Enterobactin
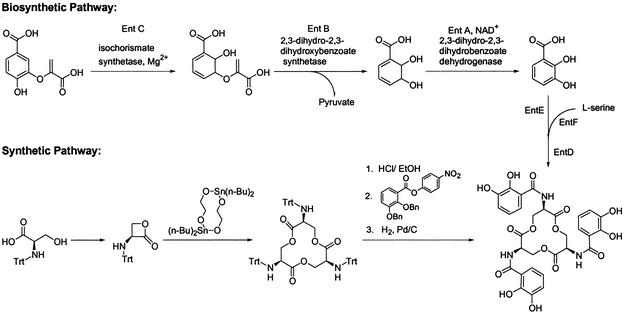
The peptidic assemblage of enterobactin is achieved by nonribosomal peptide synthetases. Walsh and coworkers (39, 40), Earhart and coworkers (41, 42), and McIntosh and coworkers (43, 44) have shown that enterobactin is synthesized from a fork in the aromatic amino acid pathway in a two-step process (40, 41, 45). Chorismic acid, an aromatic amino acid precursor, is converted into isochorismate, then into 2,3-dihydro-2,3-dihydroxybenzoate, and finally into 2,3-dihydroxybenzoic acid (DHB). The amide linkage of DHB and l-serine is catalyzed by EntD, EntE, EntF, and a C-terminal aryl carrier of EntB, which is a bifunctional protein (46). Serine is activated by adenylation and subsequently binds onto a peptidyl carrier protein domain of EntF as an acyl-S-pantetheine intermediate (47). The terminal thioesterase domain of EntF later releases enterobactin after the hydrolysis of three molecules of DHB-Ser by intermolecular cyclization (48) (Scheme S1 Upper).
Scheme 1.
Chemical Syntheses of Enterobactin
The first chemical synthesis of enterobactin was reported by Corey and Bhattacharya (49). Their procedure gave enterobactin with a relatively low yield (≈1%). Subsequent syntheses for both enterobactin and its mirror image, enantioenterobactin (50), have steadily improved the yield (51). A single-step synthesis of the triserine lactone (52) (Scheme S1 Lower) provides an overall yield of ≈50% and also enables the functionalization of the trilactone by attaching chelating groups other than catecholamides (53, 54).
Enterobactin Recognition and Transport
The low concentration and large size of ferric siderophore complexes require active transport (Fig. 2). Recognition and incorporation of the ferric siderophore complex begins at an outer membrane protein receptor. This constitutes the rate-limiting step for the entire siderophore-mediated iron transport process (55, 56). Under iron stress, organisms typically express highly specific receptors capable of binding a very narrow subset of ferric siderophores (27, 57). Several bacteriophages (T1, T5, φ80, UC-1) and bacteriocins (colicin M and microcin 25) opportunistically use these outer membrane receptors for entry into the cell (58).
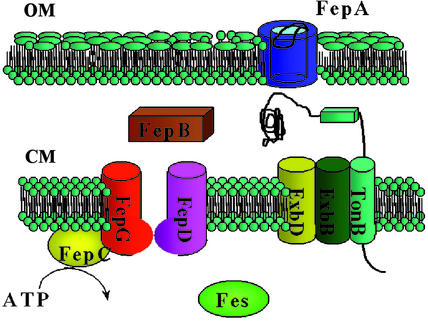
Figure 2.
A pictorial scheme shows the transmembrane topology of the FeEnt uptake proteins and how they function. In an iron-deficient state, iron receptors proliferate among the outer membrane (OM) proteins. FepA is a channel protein composed of a β-barrel and an N-terminal gate protein (see Fig. 3). The FepA receptor is highly specific and recognizes the iron binding domain and amide linkage domains of FeEnt. The gating movement of FepA is transduced by the complex TonB–ExbB–ExbD, which is anchored in the cytoplamic membrane (CM). FepB delivers FeEnt to the cytoplasmic pores formed by FepD and FepG. It appears that the cytoplasmic ATPase, FepC, provides energy to assist the uptake through the inner membrane. FeEnt esterase, which is encoded by the fes gene, catalyzes hydrolytic cleavage of the backbone, leading to the intracellular release of iron.
The octahedral metal center of a tris-bidentate complex is chiral, with either Λ or Δ absolute configuration. In many siderophore systems this chirality is part of the recognition process for siderophore receptors (59). FeEnt has Δ absolute configuration, and although the synthetic mirror image (Λ configuration, enantioenterobactin) does not promote the growth of E. coli (60), subsequent studies have revealed that both enantiomers are transported into the bacterium, showing that chirality of the enterobactin metal center is not required for its recognition by the receptor FepA (61). The trilactone backbone is also not required for recognition; however, an unsubstituted triscatechol iron center and an amide linkage is required (Fig. 1 Upper Left). The early studies of receptor binding and transport of FeEnt using a wide range of synthetic analogs showed that the coordinated catechol amide groups are essential and cannot be significantly changed without disrupting recognition, whereas the triserine macrocycle is not recognized and can be replaced with very different molecular scaffolds (34, 61).
Recently, the crystal structure has been described for the outer membrane protein of E. coli that is responsible for the recognition of FeEnt and similar triscatecholate analogs. FepA (Fig. 3) consists of ≈700 residues organized into a C-terminal, 22-stranded β-barrel and an N-terminal plug domain located within the barrel channel (62). The crystal structure and biochemical data have implicated several positively charged and aromatic residues of FepA in the binding of FeEnt (63). The 22-stranded antiparallel β-barrel (residues 154–724) forms a domain that spans the outer membrane, with connecting loops, and functions in FeEnt binding. Several of the extracellular loops in FepA extend 30–40 Å above the membrane bilayer, which may facilitate the initial binding of FeEnt. Although similar to porin proteins, FepA lacks the long loops that fold back into the pore and uses more aromatic residues. The N-terminal globular plug domain (residues 1–153) consists of two long loops, several short β-strands, and single-turn helices that fold into the barrel, plugging the barrel pore. The N domain is firmly anchored to the barrel protein and completely blocks the transport of FeEnt into the periplasm. Transport requires the N-terminal domain to undergo a conformational rearrangement. The unfolded plug domain of FepA binds FeEnt, although at a 100-fold lower affinity than the intact protein, suggesting that the plug domain rearranges to allow the FeEnt to pass through the channel, rather than moving out as the intact plug (56).
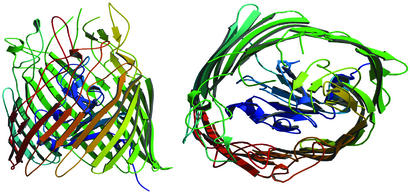
Figure 3.
The crystallographic structure of unliganded FepA (Protein Data Bank code 1FEB). (Left) A view perpendicular to the channel axis. The periplasm side of the outer membrane is at the bottom. (Right) A view down the axis from the extracellular space. FepA is monomeric (porins are trimeric) and composed of an antiparallel β-stranded barrel (571 residues) and an N-terminal plug domain (153 residues, colored blue) anchored within the barrel. The right-handed twist barrel is ≈70 Å in height and the ellipsoidal cross-section dimension is 30 × 40 Å. The β-barrel is tilted ≈45° to the axis of the barrel. The extracellular loops of the β-barrel extend ≈30 Å above the membrane.
Once across the outer membrane, FeEnt is translocated into the cytoplasm via the ATP binding cassette transporter. This transport mechanism of ferric siderophores seems widely conserved and includes: (i) binding of the substrate by the periplasmic binding protein, (ii) interaction of the binding protein with the cytoplasmic membrane spanning protein, and (iii) hydrolysis of ATP via a membrane bound protein to transport the ferric siderophore across the cytoplasmic membrane. Although the structure of the periplasmic enterobactin binding protein, FepB, has not yet been determined, recent binding studies suggest that this protein is far more specific for enterobactin (Kd = 30 nM) (64) than the similar catecholate siderophores vibriobactin (65) or agrobactin (64). Once carried across the periplasm by FepB, FeEnt is delivered to cytoplasmic transmembrane proteins (FepD and FepG). Transport across the cytoplasmic membrane is driven by hydrolysis of ATP, achieved via the ATP binding protein FepC, which is attached to the cytoplasmic membrane. As shown in Fig. 2, FeEnt is a substrate for the cytoplasmic esterase, which is even more effective in hydrolyzing the free enterobactin than the FeEnt complex (32).
Human Defense Against Enterobactin
It has long been known that enterobactin is bound strongly by serum albumin and hence is inactivated in human serum (66). Recently, neutrophil gelatinase-associated lipocalin [NGAL, also called superinducible protein 24 (SIP24), 24p3, neu-related lipocalin (NRL0), and uterocalin] has been proposed as an antibacterial component of the innate immune system (67). NGAL is a human protein secreted by neutrophils (68, 69) and epithelial cells during inflammation and rapid cell growth (70). NGAL, like most lipocalins, is thought to modulate cellular processes by binding to specific cell-surface sites or receptors (71).
For NGAL to act as a growth inhibitor of E. coli by binding and sequestering FeEnt, it must have an affinity for FeEnt comparable to FepA. The Kd of FepA for FeEnt has been measured by fluorescence spectroscopy at 50 nM (62) and by filter binding at 0.2 nM (72). Strong and coworkers (67) showed that tryptophan fluorescence quenching analysis of NGAL in the presence of FeEnt gave a Kd of 0.41 ± 0.11 nM at 20°C. Thus NGAL has sufficient affinity to effectively compete with FepA for FeEnt and so to neutralize the use of enterobactin by pathogenic E. coli. Most significantly, the crystal structure of the NGAL:FeEnt adduct shows the specific binding of the ferric siderophore by the protein (67). Because large sections of the NGAL calyx remain unfilled by FeEnt, NGAL may be capable of binding larger catecholate siderophores. Strong and coworkers thus propose that NGAL is a novel component of the innate immune system. Electronic spectra and calculations of metal complexes of enterobactin synthetic analogs have established the highly delocalized nature of the net negative charge (31). It was therefore proposed that the NGAL:FeEnt interactions with lysine and arginine side chains are a hybrid between a simple ionic interaction and a cation–π interaction in that both interacting moieties have a net charge. This is also the case for FeEnt binding to FepA and other FeEnt binding proteins (67).
Corynebactin
Enterobactin was thought unique to Gram-negative bacteria. However, isolation of enterobactin from two Gram-positive Streptomyces species (73) suggests production of this powerful siderophore may be wider than previously thought. It was also thought that the trilactone ring of enterobactin was unique to this siderophore. However, corynebactin (Fig. 4), isolated from Gram-positive Corynebacterium glutamicum (74) and Bacillus subtilis (and then improperly renamed) (75), incorporates a threonine trilactone and glycine spacers, which elongate the three chelating arms as compared with enterobactin. The 12-membered trilactone macrocyclic ring has two stable conformations. If one placed an imaginary carbon atom at the center of the macrocycle, the result would be three fused six-membered rings. The “normal” conformation seen for enterobactin and the “inverted” form found for corynebactin are the two solutions to putting these imaginary six-membered rings all in chair conformations, as was described for enterobactin by Tse and Kishi (76). The result of the two structural changes seems to maintain a very high stability for ferric corynebactin, comparable to that of enterobactin. The structural conformation of ferric corynebactin has been studied by molecular modeling and CD studies (77, 78). Whereas enterobactin forms a Δ-ferric complex, corynebactin is Λ! Both the addition of a glycine spacer and methylation of the trilactone ring (serine to threonine) favor the Λ conformation relative to the Δ conformation of enterobactin. It will be most interesting to see how this stability and the Λ chirality are involved in corynebactin-mediated iron transport in its producing organisms.
Figure 4.
The structures of the two trilactone siderophores: enterobactin (Left) and corynebactin (Right). (Center) The chemical structure of corynebactin (which has a glycine spacer between the catechol ligands and the trilactone ring) is shown (that for enterobactin is in Fig. 1). The conformation for the 12-membered triserine ring of enterobactin places both the lactone carbonyl and the exocyclic amine groups in axial positions. The conformation for the trithreonine ring of corynebactin is inverted, with ester carbonyls equatorial. The consequence is that, while the ferric enterobactin complex is Δ (Lower Left) the ferric corynebactin complex is Λ (Lower Right).
Conclusion
Over a 30-year period enterobactin has provided a merry chase. It was first characterized by microbiologists and biochemists. Its extraordinary iron affinity and structure has been explored by chemists, who have also synthesized the molecule and many analogs. The biosynthesis and regulation has been followed by using the tools of genetics and molecular biology, and the regulatory aspects have provided important insight into what has become a wider field of metal regulation. Finally, elucidating the transport process of FeEnt uptake has provided what today is the clearest picture of siderophore-mediated iron transport. This general topic intersects importantly with a range of applications, most notably bacterial diseases, but also environmental implications of human interest.
Acknowledgments
We thank our many previous coworkers whose names appear in our referenced work and the National Institutes of Health for continuing support through Grant AI11744.
References
- 1.Raymond K N, Carrano C J. Acc Chem Res. 1979;12:183–190. [Google Scholar]
- 2.Neilands J B. J Biol Chem. 1995;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 3.Aisen P, Enns C, Wessling-Resnick M. Int J Biochem Cell Biol. 2001;33:940–959. doi: 10.1016/s1357-2725(01)00063-2. [DOI] [PubMed] [Google Scholar]
- 4.De Freitas J M, Meneghini R. Mutat Res. 2001;475:153–159. doi: 10.1016/s0027-5107(01)00066-5. [DOI] [PubMed] [Google Scholar]
- 5.Braun V, Killmann H. Trends Biochem Sci. 1999;24:104–109. doi: 10.1016/s0968-0004(99)01359-6. [DOI] [PubMed] [Google Scholar]
- 6.Aisen P, Leibman A, Zweier J. J Biol Chem. 1978;253:1930–1937. [PubMed] [Google Scholar]
- 7.Martin R B, Savory J, Brown S, Bertholf R L, Wills M R. Clin Chem. 1987;33:405–407. [PubMed] [Google Scholar]
- 8.Kretchmar S A, Reyes Z E, Raymond K N. Biochim Biophys Acta. 1988;956:85–94. doi: 10.1016/0167-4838(88)90301-9. [DOI] [PubMed] [Google Scholar]
- 9.Bullen J J, Leigh L C, Rogers H J. Immunology. 1968;15:581–588. [PMC free article] [PubMed] [Google Scholar]
- 10.Ward C G, Hammond J S, Bullen J J. Infect Immun. 1986;51:723–730. doi: 10.1128/iai.51.3.723-730.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez J L, Delgado-Iribarren A, Baquero F. FEMS Microbiol Rev. 1990;75:45–56. doi: 10.1111/j.1574-6968.1990.tb04085.x. [DOI] [PubMed] [Google Scholar]
- 12.Bullen J J, Rogers H J, Lewin J E. Immunology. 1971;20:391–406. [PMC free article] [PubMed] [Google Scholar]
- 13.Payne S M. Mol Microbiol. 1989;3:1301–1306. doi: 10.1111/j.1365-2958.1989.tb00281.x. [DOI] [PubMed] [Google Scholar]
- 14.Griffiths E. Biol Metals. 1991;4:7–13. doi: 10.1007/BF01135551. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths E, Williams P. In: Iron and Infection: Molecular, Physiological and Clinical Aspects. Bullen J J, Griffiths E, editors. Chichester, U.K.: Wiley; 1987. pp. 87–212. [Google Scholar]
- 16.Chart H, Griffiths E. FEMS Microbiol Lett. 1985;26:226. [Google Scholar]
- 17.Robins-Browne R M, Prpic J K. Infect Immun. 1985;47:774–779. doi: 10.1128/iai.47.3.774-779.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barry D M J, Reeve A W. Pediatrics. 1977;60:908–912. [PubMed] [Google Scholar]
- 19.Trousseau A. Lect Clin Med. 1872;5:95–117. [Google Scholar]
- 20.Bornside G H, Bouis P J, Cohn I. J Bacteriol. 1968;95:1567–1571. doi: 10.1128/jb.95.5.1567-1571.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bullen J J, Ward C G, Rogers H J. Eur J Microbiol Infect Dis. 1991;10:613–617. doi: 10.1007/BF01975810. [DOI] [PubMed] [Google Scholar]
- 22.Wright A C, Simpson L M, Oliver J D. Infect Immun. 1981;34:503–507. doi: 10.1128/iai.34.2.503-507.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst J F, Bennett R L, Rothfield L I. J Bacteriol. 1978;135:928–934. doi: 10.1128/jb.135.3.928-934.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hantke K. Mol Gen Genet. 1984;197:337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 25.Bagg A, Neilands J B. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 26.Tao X, Schiering N, Zeng H Y, Ringe D, Murphy J R. Mol Microbiol. 1994;14:191–197. doi: 10.1111/j.1365-2958.1994.tb01280.x. [DOI] [PubMed] [Google Scholar]
- 27.Hantke K. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 28.Pollack J R, Neilands J B. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 29.O'Brien I G, Gibson F. Biochim Biophys Acta. 1970;215:393–397. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 30.Karpishin T B, Raymond K N. Angew Chem Int Ed Engl. 1992;31:466–468. [Google Scholar]
- 31.Karpishin T B, Dewey T M, Raymond K N. J Am Chem Soc. 1993;115:1842–1851. [Google Scholar]
- 32.Greenwood K T, Luke R K J. Biochim Biophys Acta. 1978;525:209–218. doi: 10.1016/0005-2744(78)90216-4. [DOI] [PubMed] [Google Scholar]
- 33.Brickman T J, McIntosh M A. J Biol Chem. 1992;267:12350–12355. [PubMed] [Google Scholar]
- 34.Ecker D J, Matzanke B F, Raymond K N. J Bacteriol. 1986;167:666–673. doi: 10.1128/jb.167.2.666-673.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ratledge C, Dover L G. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 36.Scarrow R C, Ecker D J, Ng C, Liu S, Raymond K N. Inorg Chem. 1991;30:900–906. [Google Scholar]
- 37.Stack T D P, Karpishin T B, Raymond K N. J Am Chem Soc. 1992;114:1512–1514. [Google Scholar]
- 38.Hou Z G, Stack T D P, Sunderland C J, Raymond K N. Inorg Chim Acta. 1997;263:341–355. [Google Scholar]
- 39.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C T. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 40.Walsh C T, Liu J, Rusnak F, Sakaitani M. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 41.Earhart C. In: Iron Transport in Microbes, Plants, and Animals. Winkelmann G, van der Helm D, Neilands J, editors. Weinheim, Germany: VCH; 1987. pp. 67–84. [Google Scholar]
- 42.Chenault S S, Earhart C F. Mol Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 43.Nahik M S, Fleming T P, McIntosh M A. J Bacteriol. 1987;169:4163–4170. doi: 10.1128/jb.169.9.4163-4170.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ozenberger B A, Brickman T J, McIntosh M A. J Bacteriol. 1989;171:775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Braun V, Hantke K. In: CRC Handbook of Microbial Iron Chelators. Winkelmann G, editor. Boca Raton, FL: CRC; 1991. pp. 107–138. [Google Scholar]
- 46.Gehring A M, Bradley K A, Walsh C T. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 47.Reichert J, Sakaitani M, Walsh C T. Protein Sci. 1992;1:549–556. doi: 10.1002/pro.5560010410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw-Reid C A, Kelleher N L, Losey H C, Gehring A M, Berg C, Walsh C T. Chem Biol. 1999;6:385–400. doi: 10.1016/S1074-5521(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 49.Corey E J, Bhattacharyya S. Tetrahedron Lett. 1977;45:3919–3922. [Google Scholar]
- 50.Rastetter W H, Erickson T J, Venuti M C. J Org Chem. 1981;46:3579–3590. [Google Scholar]
- 51.Shanzer A, Libman J. J Chem Soc Chem Commun. 1983;15:846–847. [Google Scholar]
- 52.Seebach D, Muller H M, Burger H M, Plattner D A. Angew Chem Int Ed Engl. 1992;31:434–435. [Google Scholar]
- 53.Meyer M, Telford J R, Cohen S M, White D J, Xu J, Raymond K N. J Am Chem Soc. 1997;119:10093–10103. [Google Scholar]
- 54.Ramirez R J A, Karamanukyan L, Ortiz S, Gutierrez C G. Tetrahedron Lett. 1997;38:749–752. [Google Scholar]
- 55.Braun V, Hantke K. In: Molecular and Cellular Iron Transport. Templeton D M, editor. New York: Dekker; 2002. pp. 395–426. [Google Scholar]
- 56.Usher K C, Ozkan E, Gardner K H, Deisenhofer J. Proc Natl Acad Sci USA. 2001;98:10676–10681. doi: 10.1073/pnas.181353398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Braun V. Int J Med Microbiol. 2001;291:67–79. doi: 10.1078/1438-4221-00103. [DOI] [PubMed] [Google Scholar]
- 58.Braun V, Hantke K, Koster W. In: Metal Ions in Biological Systems: Iron Transport and Storage in Microorganisms, Plants, and Animals. Sigel A S D, editor. Vol. 35. New York: Dekker; 1998. pp. 67–145. [PubMed] [Google Scholar]
- 59.Matzanke B F, Müller-Matzanke G, Raymond K N. In: Iron Carriers and Iron Proteins. Loehr T M, editor. New York: VCH; 1989. pp. 1–121. [Google Scholar]
- 60.Neilands J B, Erickson T J, Rastetter W H. J Biol Chem. 1981;26:3831–3832. [PubMed] [Google Scholar]
- 61.Thulasiraman P, Newton S M C, Xu J D, Raymond K N, Mai C, Hall A, Montague M A, Klebba P E. J Bacteriol. 1998;180:6689–6696. doi: 10.1128/jb.180.24.6689-6696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buchanan S K, Smith B S, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 63.Cao Z H, Qi Z B, Sprencel C, Newton S M C, Klebba P E. Mol Microbiol. 2000;37:1306–1317. doi: 10.1046/j.1365-2958.2000.02093.x. [DOI] [PubMed] [Google Scholar]
- 64.Sprencel C, Cao Z H, Qi Z B, Scott D C, Montague M A, Ivanoff N, Xu J, Raymond K N, Newton S M C, Klebba P E. J Bacteriol. 2000;182:5359–5364. doi: 10.1128/jb.182.19.5359-5364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wyckoff E E, Valle A M, Smith S L, Payne S M. J Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Konopka K, Neilands J B. Biochemistry. 1984;23:2122–2127. doi: 10.1021/bi00305a003. [DOI] [PubMed] [Google Scholar]
- 67.Goetz D H, Holmes M A, Borregaard N, Bluhm M E, Raymond K N, Strong R K. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 68.Goetz D H, Willie S T, Armen R S, Bratt T, Borregaard N, Strong R K. Biochem. 2000;39:1935–1941. doi: 10.1021/bi992215v. [DOI] [PubMed] [Google Scholar]
- 69.Kjeldsen L, Johnsen A H, Sengelov H, Borregaard N. J Biol Chem. 1993;268:10425–10432. [PubMed] [Google Scholar]
- 70.Nielsen B S, Borregaard N, Bundgaard J R, Timshel S, Sehested M, Kjeldsen L. Gut. 1996;38:414–420. doi: 10.1136/gut.38.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devireddy L R, Teodoro J G, Richard F A, Green M R. Science. 2001;293:829–834. doi: 10.1126/science.1061075. [DOI] [PubMed] [Google Scholar]
- 72.Newton S M C, Igo J D, Scott D C, Klebba P E. Mol Microbiol. 1999;32:1153–1165. doi: 10.1046/j.1365-2958.1999.01424.x. [DOI] [PubMed] [Google Scholar]
- 73.Fiedler H P, Krastel P, Muller J, Gebhardt K, Zeeck A. FEMS Microbiol Lett. 2001;196:147–151. doi: 10.1111/j.1574-6968.2001.tb10556.x. [DOI] [PubMed] [Google Scholar]
- 74.Budzikiewicz H, Boessenkamp A, Taraz K, Pandey A, Meyer J M. Z Naturforsch. 1997;52:551–554. [Google Scholar]
- 75.May J J, Wendrich T M, Marahiel M A. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- 76.Tse B, Kishi Y. J Org Chem. 1994;59:7807–7814. [Google Scholar]
- 77.Bluhm M E, Hay B P, Kim S S, Dertz E A, Raymond K N. Inorg Chem. 2002;41:5475–5478. doi: 10.1021/ic025531y. [DOI] [PubMed] [Google Scholar]
- 78.Bluhm M E, Kim S S, Dertz E A, Raymond K N. J Am Chem Soc. 2002;124:2436–2437. doi: 10.1021/ja016651s. [DOI] [PubMed] [Google Scholar]
- 79.Stack T D P, Hou Z G, Raymond K N. J Am Chem Soc. 1993;115:6466–6467. [Google Scholar]