Abstract
Non-heme iron enzymes catalyze a wide range of O2 reactions, paralleling those of heme systems. Non-heme iron active sites are, however, much more difficult to study because they do not exhibit the intense spectral features characteristic of the porphyrin ligand. A spectroscopic methodology was developed that provides significant mechanistic insight into the reactivity of non-heme ferrous active sites. These studies reveal a general mechanistic strategy used by these enzymes and differences in substrate and cofactor interactions dependent on their requirement for activation by iron. Contributions to O2 activation have been elucidated for non-heme relative to heme ligand sets, and major differences in reactivity are defined with respect to the heterolytic and homolytic cleavage of O—O bonds.
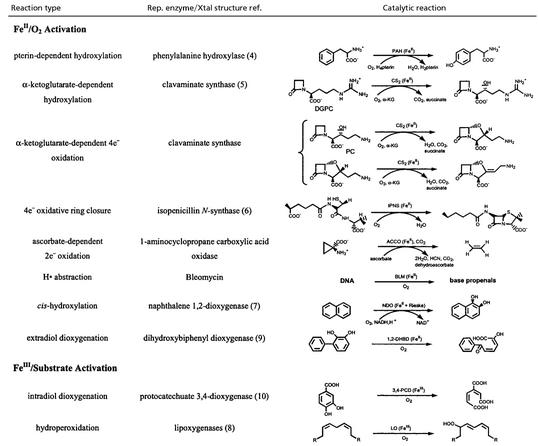
There has been much research effort directed toward determining reaction mechanisms, the nature of intermediates, and structure/function correlations over the heme enzymes involved in dioxygen activation (cytochrome P450, indolamine 2,3- dioxygenase, cytochrome c oxidase, etc.; refs. 1 and 2). However, it can be seen from Table 1 that non-heme iron enzymes also catalyze a wide range of reactions involving dioxygen; those reactions include monooxygenation, dioxygenation, desaturation, and the 4-electron reduction of dioxygen to water. The mononuclear non-heme iron enzymes divide into two categories. The intradiol dioxygenases and the lipoxygenases use FeIII centers to activate substrate for reaction with O2 (see ref. 3 for mechanistic details). The rest of the classes of non-heme iron enzymes in Table 1 use an FeII active site to activate O2, in parallel to heme systems. However, the non-heme iron sites have been much more difficult to study spectroscopically than heme systems in that they do not exhibit the intense π→π* transitions characteristic of the porphyrin ligand. Also, the FeII active sites are even electron (d6), integer spin (high-spin S = 2), non-Kramers ions which generally cannot be studied by traditional EPR-related methods, and ligand-to-FeII charge transfer transitions are usually too high in energy to contribute to the accessible region of the absorption spectrum (<30,000 cm−1).
Table 1.
Mononuclear non-heme iron enzymes
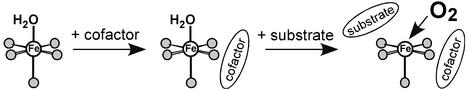
Crystal structures are now available for many of these enzymes in Table 1 (4–10). From these structures, the non-heme FeII active sites generally have two His, one monodentate carboxylate, and two to three water ligands (11). Thus, in contrast to heme sites where the porphyrin macrocycle leaves only one axial position for O2 binding and activation, the non-heme FeII sites have additional, exchangeable positions that can also allow the possibility of substrate and/or cofactor binding to the FeII and new coordination modes for oxygen activation. The His/carboxylate/H2O ligation is also much less covalent, with more limited π-interactions with the iron relative to heme, which can greatly modify the electronic structure of the active site and, hence, its activation of O2.
The next section in this perspective, Spectroscopic Methodology, briefly summarizes the spectroscopic methodology we have developed to study non-heme ferrous active sites. This methodology is applied in the section Structure/Function Correlations to several representative non-heme ferrous enzymes. These studies define a general mechanistic strategy used by these enzymes, as well as significant differences in substrate and cofactor interactions with the FeII which relate to differences in their requirement for activation. Finally, in the section Oxygen Intermediates, we consider how the large covalency differences of a non-heme relative to heme ligand system can contribute to differences in dioxygen activation.
Spectroscopic Methodology (12, 13).
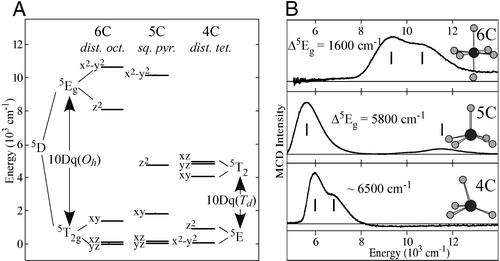
Whereas non-heme ferrous active sites do not have the intense porphyrin π→π* transitions or exhibit ligand to metal charge transfer transitions in the UV/visible spectral region, they are open shell ions and have d→d (i.e., ligand field) transitions in the near-IR region. The 5D ground state for the d6 FeII free ion is split in energy under octahedral symmetry into a 5T2g ground state (dxz, dyz, dxy) and a 5Eg excited state (dz2, dx2−y2), separated by 10Dq ≈ 10,000 cm−1 for the oxygen and nitrogen ligands. The splitting of the 5Eg excited state is sensitive to coordination number and geometry of the FeII site (Fig. 1A). Ligand field theory predicts that six-coordinate (6C) distorted octahedral FeII sites have two transitions centered at ≈10,000 cm−1 with an 5Eg splitting of <2,000 cm−1; 5C sites have these transitions at ≈10,000 cm−1 and ≈5,000 cm−1;, i.e., a large splitting of the 5Eg excited state (≈5,000 cm−1). Upon going to a four-coordinate distorted tetrahedral site, only transitions in the ≈5,000-cm−1 region are expected, because from ligand field theory, 10Dq(Td) ≈ −4/9 10Dq(Oh). Although these d→d transitions are parity-forbidden and weak in absorption spectroscopy, they are intense in the magnetic circular dichroism (MCD) spectrum at low temperatures. Low-temperature MCD spectra have been obtained for a series of 20 structurally defined ferrous model complexes (12), and, as shown in Fig. 1B, the data directly reflect the predictions of ligand field theory.
Figure 1.
Electronic structures/spectra of ferrous sites. (A) Ligand field theory predictions for different geometries. (B) LT MCD spectra of corresponding structures.
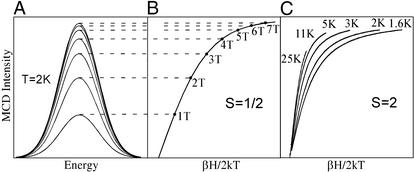
The ligand field splitting of the 5T2g ground state (in Fig. 1A) is sensitive to the π interactions of the ligands with the FeII center. This energy splitting can be obtained experimentally from the temperature and magnetic field dependence of the MCD spectrum [variable-temperature variable-field (VTVH) MCD]. Initially considering the simplest paramagnetic case, an S = 1/2 system, the MCD signal increases rapidly both with increasing magnetic field (shown in Fig. 2A) and decreasing temperature. These data are plotted as a saturation magnetization curve in Fig. 2B (intensity as a function of βH/2kT‡). The MCD signal first increases rapidly with increasing field (and decreasing temperature) and then levels off at the saturation limit. Note from Fig. 2B that, for an S = 1/2 system, MCD data taken at different temperatures lie on the same saturation magnetization curve. However, when VTVH MCD experiments are performed on a non-heme ferrous active site, a spread or nesting of the saturation magnetization curves is found for data taken at different temperatures (Fig. 2C). We have shown that this nesting reflects the non-Kramers (i.e., integer spin) nature of the S = 2 ground state and, thus, can be used to study the ground state as normally would be done by EPR for Kramers (i.e., half-integer spin) ions. An S = 2 ground state is fivefold degenerate in Ms values (0, ±1, ±2), which all split in energy even at zero magnetic field [the zero field splitting (ZFS)] because of the low symmetry of the FeII site in the protein. This ZFS is the explanation for the lack of an EPR signal for the non-Kramers ions; it is also the origin of the nesting of the saturation magnetization curves. Thus, with VTVH MCD, we can use an excited state to probe the ground state and obtain the ZFS parameters for an EPR silent active site. We have further developed the information content of these spin Hamiltonian parameters in terms of the ligand field splittings of the 5T2g ground state in Fig. 1A (see figures 30, 33, and 34 in ref. 13).
Figure 2.
VTVH MCD. (A) MCD signal increases with increasing H. (B) Overlaying saturation magnetization curves for S = 1/2 Kramers system taken at different temperatures. (C) Nesting saturation magnetization curves for S = 2 non-Kramers ions at different temperatures.
From the above summary, VTVH MCD allows the experimental determination of the ligand field splitting of the five d orbitals of an FeII active site (Fig. 1A). We now have a direct probe of active site geometric and electronic structure and have applied this to obtain molecular insight into catalytic mechanisms.
Structure/Function Correlations.
This VTVH MCD methodology has been applied to obtain geometric and electronic structure insight into the reactivity of a wide range of mononuclear and binuclear non-heme ferrous enzymes (3). Here, we focus on the oxygen-activating mononuclear FeII sites in the pterin-dependent hydroxylase, phenylalanine hydroxylase (PAH), and the α-ketoglutarate (α-KG)-dependent dioxygenase, clavaminate synthase (CS2). Spectroscopic studies on these classes of non-heme FeII enzymes illustrate (i) a general mechanistic strategy used by the cofactor-dependent enzymes to control the O2 reaction, and (ii) that because there are potentially three exchange positions on the FeII, there can be very different interactions with the substrate and the cofactor, depending on the requirement for activation.
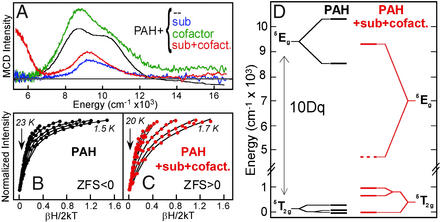
PAH is the non-heme FeII enzyme involved in phenylalanine metabolism. Genetic mutations, often near the catalytic site, result in a build-up of phenylalanine, ultimately leading to severe mental retardation [the disease phenylketonuria (PKU); ref. 14]. The low-temperature MCD spectrum of resting PAH (obtained in collaboration with John Caradonna, Boston University, Boston) is shown in Fig. 3A, black (15). It exhibits two transitions split by <2,000 cm−1 in the ≈10,000 cm−1 region, indicating a distorted six-coordinate (6C) FeII site (see Fig. 1). It was the first insight available into the catalytically active FeII form of this enzyme. Addition of substrate, phenylalanine (blue), slightly shifts the transitions showing that substrate binds near the FeII, perturbing its ligand field. The binding of the pterin cofactor has no significant effect on the MCD spectrum (green), indicating that the pterin does not directly coordinate to the FeII. Importantly, the simultaneous binding of both substrate and cofactor leads to a dramatic change in the spectrum (red) producing one ligand field transition in the ≈5,000-cm−1 region and one in the ≈10,000-cm−1 region, indicating that the site has become 5C (see Fig. 1). VTVH MCD data on the resting and substrate + cofactor bound forms also show dramatic differences (Fig. 3 B and C), where the resting site has the nesting characteristics (fairly small spread in the saturation magnetization curves) of a negative ZFS (Ms = ± 2 lowest in energy), while the substrate + cofactor bound form shows the large nesting of a positive ZFS (Ms = 0 lowest). Analysis of these data using the above methodology leads to the energy level diagram in Fig. 3D, which shows that, upon cofactor and substrate binding, 10Dq dramatically decreases, and both the e and t2 orbitals undergo large energy splittings, all requiring loss of a ligand at the non-heme ferrous active site. From extended x-ray absorption fine structure studies, the ligand lost is a water (16), which is also supported by recent crystallographic results (4). This 6C→5C conversion leads to an open coordination position on the FeII for the O2 reaction and is consistent with the sequential mechanistic order (17) observed for this enzyme, where pterin and cofactor must bind for O2 to react.
Figure 3.
VTVH MCD of PAH. (A) Effects of substrate (blue), cofactor (green), and both substrate + cofactor (red) binding to the resting FeII site. (B and C) Saturation magnetization of resting (black) and substrate + cofactor (red) bound sites. (D) Experimentally determined energy level diagram.
These and parallel VTVH MCD studies on other non-heme FeII enzymes (vide infra) have led to the general mechanistic strategy presented in Scheme S1 (3). The resting site is coordinatively saturated and relatively stable in the presence of O2. Simultaneous binding of cofactor and substrate in the cofactor-dependent enzymes leads to an open coordination position on the FeII and turns on the O2 reaction to generate a reactive oxygen intermediate in the presence of cosubstrates for their coupled hydroxylation. Thus, nature has devised an elegant strategy to turn on the dioxygen chemistry only in the presence of all cosubstrates.
Scheme 1.
General mechanistic strategy.
It should be emphasized from Fig. 3A, green, that the pterin does not bind to the FeII. This lack of cofactor-FeII binding is consistent with the fact that, independently of the iron, reduced pterin will react with dioxygen, and a likely mechanism would involve the initial formation of a peroxy pterin. A peroxopterin bridged to the FeII site would lead to the formation of a ferryl, FeIV⩵O, species (and a 4a-hydroxy-oxidized pterin; ref. 18) suitable for H-atom abstraction from substrate. This mechanism is supported by recent MCD results on PKU mutants which oxidize pterin in a reaction mostly uncoupled from substrate hydroxylation (J. N. Kemsley, E. C. Wasinger, S. Datta, N. Mitic, T. Acharya, J. P. Caradonna, B. Hedman, K. O. Hodgson, and E.I.S., unpublished work).
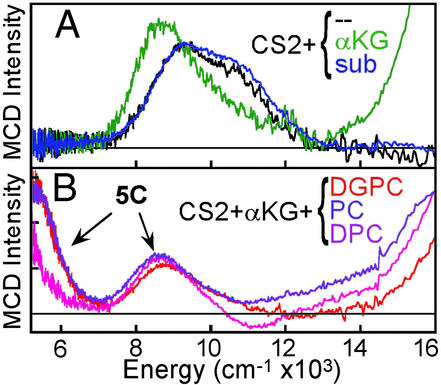
Bacterial resistance to penicillin antibiotics is largely because of the hydrolytic activity of the β-lactamases. CS2 catalyzes the key biosynthetic ring closure step in the formation of clavulanic acid, a potent β-lactamase inhibitor (19). CS2 catalyzes three different reactions (Table 1) depending on substrate [hydroxylation: deoxyguanidinoproclavaminic acid (DGPC); ring closure: proclavaminic acid (PC); desaturation: deoxyproclavaminic acid (DPC); ref. 20]. From the MCD data in Fig. 4A (obtained in collaboration with Craig Townsend, Johns Hopkins University, Baltimore), the resting site is again 6C (black); however, in contrast to the pterin-dependent enzyme PAH, binding of cofactor α-KG to the resting FeII CS2 site greatly perturbs the MCD ligand field transitions (green), indicating a new 6C site (21). The α-KG bound enzyme turns pink because of the presence of new transitions in the UV/visible region (note the intense MCD feature growing in above 15,000 cm−1 in Fig. 4A, green), assigned as a metal-to-ligand charge transfer (MLCT) transition. Because charge-transfer intensity reflects orbital overlap, the α-KG must bind directly to the FeII in a bidentate mode. The presence of MLCT transitions (and a large splitting of the dπ orbitals from the VTVH MCD; ref. 22) reflects back-bonding between the FeII and the α-KG. α-KG coordination to the FeII likely plays an important role in activating the cofactor for O2 reactivity. (Cofactor binding to the FeII is not required in PAH, because pterin is reactive with dioxygen in the absence of FeII).
Figure 4.
LT MCD of CS2. (A) α-KG cofactor (green) or substrate (blue) binding. (B) Both cofactor and substrate (three) binding.
The binding of each of the three substrates to the resting FeII-CS2 site produces no change in the ligand field spectrum (Fig. 4A, blue). Alternatively, binding of cofactor + substrate (Fig. 4B) leads to a large change in the active site. For the FeII-CS2/α-KG/substrate complexes, there are transitions in the ≈5,000-cm−1 region and the ≈9,000-cm−1 region, indicating that the sites have become 5C. Thus, for this large class of non-heme iron enzymes, we find a parallel mechanistic strategy, as in Scheme S1, where the resting site is 6C but simultaneous binding of both cosubstrates opens a coordination position on the FeII for O2 activation for the coupled reaction. (Note that from the charge transfer region, the α-KG remains coordinated in all three active site complexes). There are, however, important differences among the three FeII-CS2/substrate complexes. The MCD band at ≈5,000 cm−1 reduces in intensity with DGPC > PC > DPC. For DPC, there is an additional negative band at ≈11,000 cm−1, and the saturation magnetization curves taken on the low- and high-energy sides of the ≈9,000-cm−1 band show different nesting behaviors (see ref. 21). These show that there is also a 6C component present, which increases from DPGC (5%) < PC (10%) < DPC (50%). Thus, different substrate bindings modulate the affinity of the CS2 FeII site for H2O.
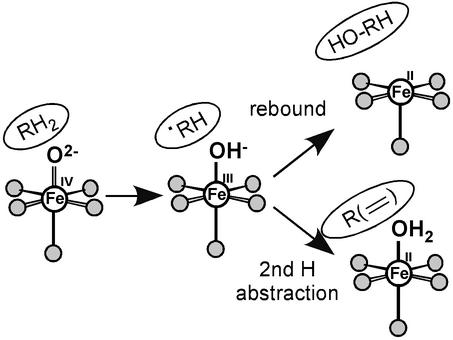
In addition to the substrate orientation relative to the open coordination position on the FeII, the enzyme can affect the type of reactivity (e.g., hydroxylation vs. desaturation) by regulating the binding energy of the water to the FeII site in the enzyme/succinate/product complex. As shown in Scheme S2, reaction of the FeII-CS2/α-KG/substrate complex with O2 is generally thought to generate a ferryl-succinate species initially, which abstracts an H-atom from the substrate. This ferric-hydroxy species can then undergo an OH rebound to generate the hydroxylated product and a 5C FeII site, or abstract the second H-atom to desaturate (or ring close) the substrate, resulting in a 6C FeII—OH2-bound site. The hydroxylation product is more exothermic than the desaturation product + H2O by ≈10 kcal/mol, not including the contribution from the water—FeII bond. An increase in this bond energy would favor the second H-atom abstraction over the rebound reaction in Scheme S2. As presented above, spectroscopy shows the presence of the highest amount of 6C H2O-bound species in the FeII—CS2/α-KG/DPC complex, consistent with the fact that it mostly undergoes a desaturation reaction. Alternatively, for the DPGC complex, a dominant 5C species is present indicating a lower value of the water-FeII interaction energy, which favors rebound and the hydroxylation reaction.
Scheme 2.
FeII⩵OH2 bond contribution to hydroxylation vs. desaturation.
The above VTVH MCD studies have shown that the resting FeII sites in PAH and CS2 are coordinatively saturated and remain 6C when only one cosubstrate is bound. However, when both cosubstrates are present, the FeII site goes five-coordinate and allows the oxygen reaction for coupled hydroxylation. This general mechanistic strategy, to some extent, parallels heme chemistry, where, in cytochrome P450, substrate binding leads to H2O/OH− ligand loss and reaction with dioxygen. However, this ligand loss occurs at a low spin FeIII site to allow its reduction and does not involve direct coordination of the substrate to the iron. In contrast, there are a number of exchangeable positions on the non-heme FeII sites thus cofactor (for CS2), substrate [for 1-amino-1-cyclopropane carboxylic acid oxidase (ACCO), see ref. 23], or neither (PAH) is found to coordinate to the metal ion dependent on the requirement of the FeII for cosubstrate activation. For the other non-heme FeII/O2 activating enzymes in Table 1, the Rieske center serves the role of the cofactor in the electron donation in the cis-hydroxylating dioxygenases with a similar mechanistic strategy as in Scheme S1, whereas the extradiol dioxygenases have 5C FeII sites in the resting enzymes, which are relatively stable in air (9, 24). For the latter, the one electron reduction of O2 to form an FeIII-O2− complex is thermodynamically unfavorable, and the additional electron required to drive the reaction is only available upon coordination of the catecholate substrate (M. I. Davis, E. C. Wasinger, A.D., M. Y. H. Pau, F. H. Vaillancourt, J. T. Bolin, L. D. Eltis, B. Hedman, K. O. Hodgson, and E.I.S., unpublished work).
Oxygen Intermediates.
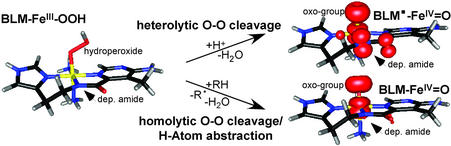
The ferrous enzymes in Table 1 react with dioxygen to generate intermediates, which are active in catalysis. Thus far, the only oxygen intermediate trapped is that for the anti-cancer drug bleomycin (BLM). FeII—BLM reacts with O2 and an exogenous electron to generate activated BLM (ABLM), which is kinetically competent to cleave DNA by H-atom abstraction (25). ABLM has been determined to be a low-spin FeIII—OOH complex (26–29). We have used a combination of spectroscopic techniques and density functional calculations to elucidate the geometric and electronic structure of ABLM and obtain insight into its reactivity (30). The geometry-optimized structure of an ABLM model is given in Fig. 5 Left. The hydroperoxide (with an Fe—O—O angle of 120°) bisects the two chemically interesting ligands of the glycopeptide anti-biotic, the deprotonated amide, and pyrimidine nitrogen. Calculations of the heterolytic cleavage of the O—O bond in ABLM (as is known to occur in heme systems) produce a species analogous to compound I in heme {[FeIV⩵O(Por•)]+}. From the spin density (Fig. 5 Upper Right), a ferryl species (FeIV⩵O) is produced along with oxidation of the ligand (in this case, the deprotonated amide functionality). However, this ligand oxidation is a much higher energy process for the non-heme site in ABLM relative to heme systems.
Figure 5.
Heterolytic and homolytic cleavage of the O—O bond of ABLM.
From the ΔE's in Table 2 Right, heterolytic cleavage of the O—O bond in ABLM (reactant 3 in Table 2) is about 175 kcal/mol higher in energy than for a Fe-porphyrin-thiolate model of cytochrome P450 (reactant 4). From the trend in Table 2, this energy difference reflects the ease of oxidation of the ligand X [ligand X spin density: heme/thiolate (+0.80) > deprotonated amide (+0.6) > OH− (+0.3) > NH3 (0.0)] and the increase in negative charge, which stabilizes the oxidized site. The latter point is reflected in the energetics calculated for a hypothetical hydroperoxo complex of the high-spin non-heme iron enzyme active site of PAH (model 5 with two NH3, one CHOO−, one H2O, and one OH− ligands, where the OH− accounts for the FeIII oxidation state). The heterolytic cleavage of the O—O bond is energetically ≈ 90 kcal/mol more favorable for this PAH model than for model complex 2, which has only one anionic (OH−) ligand. Also, the large solvation effects on the heterolytic cleavage reaction energy should be noted (Table 2), as the charge increases in the reaction, and the change in charge covers a range from +2→+3 to −1→0; this energetic stabilization goes with z2 (Born equation). Experimentally, solvent effects (for 0→±1 reactions) are found to be in the range of 40 kcal/mol (31). Although this value for BLM should be higher, as it involves a +1→+2 reaction, the value calculated with the polarized continuum model (32) of −100 kcal/mol is likely a significant overestimate. In the complex with DNA the effective dielectric experienced by the ABLM molecule will be lower than that of water (ɛ = 78), resulting in a lower solvent stabilization energy. Modeling this system by using a dielectric ɛ = 4.0 raises the free energy by 25 kcal/mol (see Table 2). Additionally, the truncated model of the ABLM molecule used in these calculations§ (shown in Fig. 5 Left) allows excess solvent penetration, artificially increasing the solvent stabilization energy. Finally, zero point vibrational corrections and entropy terms contribute about −7 kcal/mol to the reaction energy. These values total to a calculated free energy estimate for the ABLM heterolysis of ΔG > 13 kcal/mol (Table 2, which does not include the effects of the truncated model of ABLM). Thus, the O—O bond heterolytic cleavage reaction for the non-heme iron site in BLM should be at least 70 kcal/mol less favorable than for the heme site in cytochrome P450.
Table 2.
Homolytic vs. heterolytic cleavage of the O—O bond in FeIII—OOH sites*
| Reactant | Spin | Homolysis
|
Heterolysis
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Thermodynamics†
|
Spin densities‡‡
|
Thermodynamics†
|
Spin densities‡‡
|
||||||||||||
| ΔE | +solv | −TΔS‡ | ΔGsolv | Fe | ⩵O | X§ | ΔE | +solv | −TΔS‡ | ΔGsolv | Fe | ⩵O | X§ | ||
| 1. [FeIII(NH3)5OOH]2+ | 1/2 | +40.3 | −10.6¶ | −17.0 | +12.7 | +1.1 | +1.0 | 0.0 | +269.7 | −219.2¶ | −9.4 | +41.0 | +1.8 | +1.4 | 0.0 |
| 2. [FeIII(NH3)4(OH)OOH]+ | 1/2 | +31.5 | −6.5¶ | −15.2 | +9.8 | +1.2 | +0.9 | 0.1 | +136.6 | −132.2¶ | −6.7 | −2.3 | +1.6 | +1.2 | +0.3 |
| 3. [FeIII(BLM)OOH]+ | 1/2 | +29.4 | −2.6‖/−1.8†† | −14.2 | +12.8‖/+13.4†† | +1.1 | +0.9 | 0.0 | +99.3 | −103.4‖/−78.8†† | −7.1 | −11.2‖/+13.3†† | +1.1 | +1.1 | +0.6 |
| 4. [FeIII(Por)(SCH3)OOH]− | 1/2 | +65.2 | −2.4†† | −13.5 | +49.3 | +1.1 | +1.0 | 0.0 | −75.3 | +23.4†† | −6.1 | −58.0 | +1.1 | +1.0 | +0.8 |
| 5. [FeIII(PAH)OOH]** | 5/2 | +37.9 | −3.5†† | −8.0 | +26.4 | +1.2 | +0.9 | 0.0 | +47.0 | −39.9†† | +0.5 | +7.6 | +1.6 | +1.2 | +0.3 |
DFT calculations performed by using B3LYP/TZV.
All values in kcal/mol.
Includes zero point correction energy.
Ligand X = NH3 (1.), OH− (2., 5.), deprotonated amide (3.), heme/SCH3 (4.).
Solvent = CH3CN.
Solvent = H2O.
(PAH): two NH3, H2O, OH−, -OCHO−.
††Solvent with ɛ = 4.0.
‡‡ All FeIV: S = 1, all FeV: S = 3/2.
We have proposed the alternative mechanism of direct H-atom abstraction from the substrate (the C4-H of the deoxyribose), by the low-spin ferric-hydroperoxo ABLM complex (Fig. 5 bottom reaction; ref. 30). We have evaluated this mechanism by means of studies on hydroperoxo- and alkylperoxo-FeIII model systems which undergo the closely related reaction of homolytic cleavage of the O—O bond (33–35). From the homolysis calculations in Table 2 Left, the reaction energy ΔE is much less sensitive to the ligand type, and the solvation effects are small. From the spin densities, the homolysis reaction simply involves oxidation to an FeIV⩵O species, and there is no change in charge. For ABLM, homolytic cleavage of the O—O bond is only endothermic by 29 kcal/mol, which is because of the high stability of the FeIV⩵O species produced. As shown in Fig. 6, there are strong π (two ½ bonds) and σ bonds for this species in both the S = 1 and 2 spin states.¶ These bonding interactions lead to a large donation of charge from the oxo group to the formally FeIV species stabilizing this oxidation state of the iron. This stabilization combined with zero point corrections and entropy (approximately −15 kcal/mol) leads to an O—O bond homolysis free energy of ≈13 kcal/mol for ABLM. This energy is comparable to the lower limit estimate for the heterolysis reaction. H-atom abstraction will be still more favorable than homolysis because of the concerted formation of the H—O bond along the reaction coordinate. From frontier molecular orbital theory, this H—O bond formation requires proper substrate orientation, which seems to be achieved in the ABLM/DNA complex (36). Thus, compared with heme systems, the heterolytic cleavage of the O—O bond in a non-heme environment is energetically much less favorable, due, in part, to the difficulty in oxidizing the non-heme iron ligand. The alternative reaction coordinate of direct H-atom abstraction is favored.
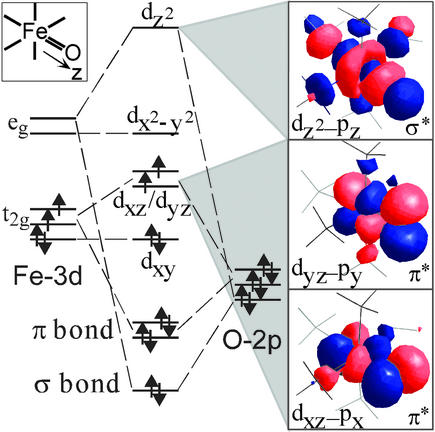
Figure 6.
FeIV⩵O bonding scheme.
With respect to the non-heme ferrous enzymes in Table 1, most of these enzymes have a cofactor or substrate (α-KG, pterin, etc.) that can donate two additional electrons toward O2 reduction and directly allow the formation of an FeIV⩵O intermediate for H-atom abstraction (Scheme S2). However, in the cis-hydroxylating dioxygenases, the Rieske center can only provide one electron; thus, an FeIII—OOH, FeV⩵O, or a redox equivalent species must be invoked in this reaction (37). From the calculations in Table 2 for the non-heme ligand set of PAH, the energetics are reasonable for O—O heterolysis (because of the negatively charged carboxylate and hydroxide ligands), as has recently been calculated for a low-spin FeIII model complex (38). However for the high-spin enzyme case, there is an additional energy barrier (≈10 kcal/mol), at least for O—O bond homolysis, because of an allowed crossing of energy levels (34). For these non-heme ferrous enzymes, it is now of critical importance to trap and define the nature of oxygen intermediates to distinguish among possible mechanisms and experimentally elucidate these reaction coordinates.
Acknowledgments
We greatly appreciate the contributions of our coworkers and collaborators listed in the references. This research was supported by National Institutes of Health Grant GM-40392.
Abbreviations
- 6C
six-coordinate
- MCD
magnetic circular dichroism
- VTVH
variable-temperature variable-field
- PAH
phenylalanine hydroxylase
- α-KG
α-ketoglutarate
- CS2
clavaminate synthase
- BLM
bleomycin
Footnotes
The complete bleomycin ligand also includes a sugar moiety, a β-aminoalanine fragment, and a bithiazole tail.
The S = 2 spin state differs from S = 1 only by the promotion of an electron from dxy to dx2−y2, which does not affect the Fe⩵O bond (along the z-axis).
β = Bohr magneton and k = Boltzmann constant.
References
- 1.Sono M, Roach M P, Coulter E D, Dawson J H. Chem Rev. 1996;96:2841–2887. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson-Miller S, Babcock G T. Chem Rev. 1996;96:2889–2907. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 3.Solomon E I, Brunold T C, Davis M I, Kemsley J N, Lee S K, Lehnert N, Neese F, Skulan A J, Yang Y S, Zhou J. Chem Rev. 2000;100:235–349. doi: 10.1021/cr9900275. [DOI] [PubMed] [Google Scholar]
- 4.Anderson O A, Flatmark T, Hough E. J Mol Biol. 2002;320:1095–1108. doi: 10.1016/s0022-2836(02)00560-0. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Z H, Ren J, Stammers D K, Baldwin J E, Harlos K, Schofield C J. Nat Struct Biol. 2000;7:127–133. doi: 10.1038/72398. [DOI] [PubMed] [Google Scholar]
- 6.Roach P L, Clifton I J, Hensgens C M H, Shibata N, Schofield C J, Hajdu J, Baldwin J E. Nature. 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 7.Carredano E, Karlsson A, Kauppi B, Choudhury D, Parales R E, Parales J V, Lee K, Gibson D T, Eklund H, Ramaswamy S. J Mol Biol. 2000;296:701–712. doi: 10.1006/jmbi.1999.3462. [DOI] [PubMed] [Google Scholar]
- 8.Tomchick D R, Phan P, Cymborowski M, Minor W, Holman T R. Biochemistry. 2001;40:7509–7517. doi: 10.1021/bi002893d. [DOI] [PubMed] [Google Scholar]
- 9.Han S, Eltis L D, Timmis K N, Muchmore S W, Bolin J T. Science. 1995;270:976–980. doi: 10.1126/science.270.5238.976. [DOI] [PubMed] [Google Scholar]
- 10.Orville A M, Lipscomb J D, Ohlendorf D H. Biochemistry. 1997;36:10052–10066. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
- 11.Hegg E L, Que L., Jr Eur J Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 12.Pavel E G, Kitajima N, Solomon E I. J Am Chem Soc. 1998;120:3949–3962. [Google Scholar]
- 13.Solomon E I, Pavel E G, Loeb K E, Campochiaro C. Coord Chem Rev. 1995;144:369–460. [Google Scholar]
- 14.Kaufman S. Science. 1958;128:1506–1508. doi: 10.1126/science.128.3337.1506. [DOI] [PubMed] [Google Scholar]
- 15.Kemsley J N, Mitic N, Zaleski K L, Caradonna J P, Solomon E I. J Am Chem Soc. 1999;121:1528–1536. [Google Scholar]
- 16.Wasinger E C, Mitic N, Hedman B, Caradonna J P, Solomon E I, Hodgson K O. Biochemistry. 2002;41:6211–6217. doi: 10.1021/bi0121510. [DOI] [PubMed] [Google Scholar]
- 17.Xia T, Gray D W, Shiman R. J Biol Chem. 1994;269:24657–24665. [PubMed] [Google Scholar]
- 18.Lazarus R A, DeBrosse C W, Benkovic S J. J Am Chem Soc. 1982;104:6869–6871. [Google Scholar]
- 19.Baldwin J E, Merritt K D, Schofield C J, Elson S W, Baggaley K H. J Chem Soc Chem Commun. 1993;16:1301–1302. [Google Scholar]
- 20.Busby R W, Townsend C A. Bioorg Med Chem. 1996;4:1059–1064. doi: 10.1016/0968-0896(96)00088-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhou J, Kelly W L, Bachman B O, Gunsior M, Townsend C A, Solomon E I. J Am Chem Soc. 2001;123:7388–7398. doi: 10.1021/ja004025+. [DOI] [PubMed] [Google Scholar]
- 22.Pavel E G, Zhou J, Busby R W, Gunsior M, Townsend C A, Solomon E I. J Am Chem Soc. 1998;120:743–753. [Google Scholar]
- 23.Zhou J, Rocklin A M, Lipscomb J D, Que L, Jr, Solomon E I. J Am Chem Soc. 2001;124:4602–4609. doi: 10.1021/ja017250f. [DOI] [PubMed] [Google Scholar]
- 24.Mabrouk P A, Orville A M, Lipscomb J D, Solomon E I. J Am Chem Soc. 1991;113:4053–4061. [Google Scholar]
- 25.Burger R M. Struct Bonding. 2000;97:287–303. [Google Scholar]
- 26.Westre T E, Loeb K E, Zaleski J M, Hedman B, Hodgson K O, Solomon E I. J Am Chem Soc. 1995;117:1309–1313. [Google Scholar]
- 27.Veselov A, Sun H, Sienkiewicz A, Taylor H, Burger R M, Scholes C P. J Am Chem Soc. 1995;117:7508–7512. [Google Scholar]
- 28.Sam J W, Tang X-J, Peisach J. J Am Chem Soc. 1994;116:3250–5256. [Google Scholar]
- 29.Burger R M, Kent T A, Horwitz S B, Münck E, Peisach J. J Biol Chem. 1983;258:1559–1564. [PubMed] [Google Scholar]
- 30.Neese F, Zaleski J M, Loeb Zaleski K, Solomon E I. J Am Chem Soc. 2000;122:11703–11724. [Google Scholar]
- 31.Sharpe P, Alameddin N G, Richardson D E. J Am Chem Soc. 1994;116:11098–11108. [Google Scholar]
- 32.Cramer C J, Truhlar D G. Chem Rev. 1999;99:2161–2200. doi: 10.1021/cr960149m. [DOI] [PubMed] [Google Scholar]
- 33.Lehnert N, Ho R Y N, Que L, Jr, Solomon E I. J Am Chem Soc. 2001;123:8271–8290. doi: 10.1021/ja010165n. [DOI] [PubMed] [Google Scholar]
- 34.Lehnert N, Ho R Y N, Que L, Jr, Solomon E I. J Am Chem Soc. 2001;123:12802–12816. doi: 10.1021/ja011450+. [DOI] [PubMed] [Google Scholar]
- 35.Lehnert N, Ho R Y N, Neese F, Que L, Jr, Solomon E I. J Am Chem Soc. 2002;124:10810–10822. doi: 10.1021/ja012621d. [DOI] [PubMed] [Google Scholar]
- 36.Wu W, Vanderwall D E, Lui S M, Tang X-J, Turner C J, Kozarich J W, Stubbe J. J Am Chem Soc. 1996;118:1268–1280. [Google Scholar]
- 37.Wolfe M D, Parales J V, Gibson D T, Lipscomb J D. J Biol Chem. 2001;276:1945–1953. doi: 10.1074/jbc.M007795200. [DOI] [PubMed] [Google Scholar]
- 38.Bassan A, Blomberg M R A, Siegbahn P E M, Que L., Jr J Am Chem Soc. 2002;124:11056–11063. doi: 10.1021/ja026488g. [DOI] [PubMed] [Google Scholar]