Abstract
Genome sequencing has revolutionized all fields of life sciences. Bioinorganic chemistry is certainly not immune to this influence, which is presenting unprecedented challenges. A new goal for bioinorganic chemistry is the investigation of the linkages between inorganic elements and genomic information. This requires new advancements and/or the development of new expertise in fields such as bioinformatics and genetics but also provides a driving force to push forward the exploitation of traditional analytical techniques and spectroscopic tools. The “case study” of metal homeostasis in cells is discussed to provide a flavor of the current evolution of the field.
Bioinorganic or biological inorganic chemistry is the discipline dealing with the interaction between inorganic substances and molecules of biological interest. It is a rather wide field, because it addresses the role, uptake, and fate of elements essential for life, the response of living organisms to toxic inorganic substances, the function of metal-based drugs, the synthetic production of functional models, the production of MRI contrast agents in medical applications, the development of theoretical models for the above topics, and so on.
The importance of the field stems from the fact that life originated and developed on the earth's crust, i.e., within an inorganic environment. Biological inorganic chemistry has always been part of chemistry and evolved as did the research tools of chemistry. The discovery of iron in the blood and of copper and zinc as essential elements for many physiological reactions may be referred to as fundamental steps in the field. The beginning of the present flourishing may be dated back to the late 1950s to early 1960s, when the x-ray structures of myoglobin and hemoglobin were solved by the illustrious chemists J. C. Kendrew (1) and M. Perutz (2), respectively. Afterward, numerous meetings in the field, also organized by several learned societies, took place. In the 1970s, a significant share of inorganic chemists entered the field, which has been steadily growing since then, as has the flood of publications related to the field. The birth of the Journal of Inorganic Biochemistry in 1971 was quite timely. Eventually, the Society of Biological Inorganic Chemistry was founded in 1995, followed after 1 year by the Journal of Biological Inorganic Chemistry. The mid-1990s were also the time for the birth of genome-based research, which raised the new challenge of matching genomic information with bioinorganic knowledge.
Today's Mission of Bioinorganic Chemistry
Every historical period is characterized by major scientific challenges. For example, at the beginning of the 20th century, the challenge was understanding atomic structure. The challenge of the beginning of this century is the understanding of the molecular bases of living processes. This challenge takes advantage of the availability of genome sequences. In 1995, the first sequence of an entire genome, that of Haemophilus influenzae, was reported (3). At present, genome sequences for 16 archaea, 87 bacteria, and 8 eukaryota are available (see www.ncbi.nlm.nih.gov/Genomes/index.html). In addition, the genome sequences for nine other eukaryota are being completed, six of which are plants. And the amount of data is increasing like an avalanche!
From the genome sequences, it is possible to deduce the primary sequences of essentially all of the proteins that a living organism can produce. The 3D structures of these proteins are being collected at the genome scale within programs of structural genomics financed in North America, Japan, and Europe. These programs are developed around the concept of high-throughput structure determination, which per se does not pay attention to issues such as incorporation of cofactors containing metals. This unprecedented availability of data opens new perspectives for bioinorganic chemists. Particularly, it is well established that a number of metal ions are essential to life. Other metal ions are instead poisonous to living organisms, even when present in the environment at very low concentrations. Sometimes, the same element may be beneficial or noxious depending on speciation. The investigation of the linkages between inorganic elements (essential and noxious) and the information obtainable from genome sequences, as well as of the mechanisms that warrant homeostasis in cell compartments, represent a further challenge for bioinorganic chemists in the so-called postgenomic era.
The Identification of Metal-Binding Proteins in Gene Banks
A number of bioinformatic web servers and databases have been created and updated (or, in some cases, discontinued) before and after the advent of genome sequences with the aim of providing the scientific community with tools for searching gene banks, for the analysis of protein sequences, and for the prediction of a variety of protein properties. Surprisingly enough, very few of these resources are dedicated to the analysis of the interaction between metal ions and proteins. This situation was already pointed out in 2000 (4). Genome browsing in the field of bioinorganic chemistry cannot be limited to the analysis of amino acid identity or similarity in sequences [i.e., as is the case for one of the most popular programs for sequence comparison and genome browsing, blast (5)]. We may take advantage of known consensus sequences, i.e., by taking into account the nature and spacing of amino acids acting as metal ligands and the nature of amino acids present in the metal-binding region. When the consensus sequence is well known, it can thus be used as a further input for genome browsing, together with the primary sequence of one or more proteins evolutionarily related to the one(s) of interest, by using the program phi-blast (6). This poses the problem of having at one's disposal databases of consensus sequences for metal binding. An available database addressing this problem is PROMISE, which collects structural and functional information (including bibliography) on metalloproteins, with emphasis on the properties of the metal site (7). The proteins have been categorized on the basis of the metal cofactor bound. Unfortunately, this service has been discontinued, so the database is no longer updated. A second relevant database publicly available is the Metalloprotein Database, maintained at The Scripps Research Institute (8). This database provides quantitative information on geometrical parameters of metal-binding sites in metalloprotein structures deposited in the Protein Data Bank (PDB), as well as statistical information about the recurrence of consensus sequences (called patterns) within the PDB. An explicitly stated goal of the database is that of becoming a comprehensive source of useful information for metalloprotein design.
The two databases mentioned taken together provide the largest amount of information on metal-binding sites and consensus sequences available from the web. It appears that specific validated bioinformatic tools for the search of metal-binding sequences within gene data banks, possibly building on the information that the above databases contain, are still lacking. The development of such tools will most likely be crucial to extend and fully exploit the success of genome sequencing projects in the field of bioinorganic chemistry. Such tools would not only allow researchers to identify uncharacterized metalloproteins in genome sequences for subsequent experimental studies but would also enable broader applications of other bioinformatic methods, such as molecular modeling. Molecular modeling indeed has been proving extremely powerful in linking structural to functional information (9), also in the case of metalloproteins (10–12); however, available approaches do show some (potential) limitations in the identification and treatment of metal-binding sites, which often can be circumvented only through careful human examination (11).
The Problem of Metal Trafficking in Cells
Being able to browse genomes and gene banks and to identify metalloprotein sequences with a good degree of reliability would be a precious advancement for the field of bioinorganic chemistry but probably would not be sufficient to answer the key question: how do processes involving metal ions occur in cells? To answer this question, i.e., to understand the interaction between cells and metal ions (either essential or noxious) within the context of the global functioning mechanism of the cell, requires synergistic efforts across a number of different disciplines, ranging from genetics to structural biology, from analytical chemistry to biophysics, from enzymology to drug discovery, and beyond. A bioinorganic chemist of the 21st century should be knowledgeable of all of this.
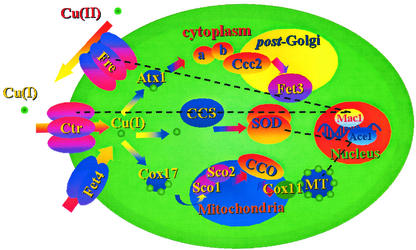
Fig. 1 schematically displays what is currently known about the trafficking pathway of copper in yeast. This is probably the most intensely studied, and thus better understood, “global” scheme of interaction between one metal and a cell. Cu(II) is reduced to Cu(I) at the external surface of the membrane and is then pumped in the cell by Ctr. At this point, in one pathway, Ctr delivers the Cu(I) ion to the Atx1 chaperone, which delivers it to Ccc2 to subsequently reach Fet3, the final target; in a second pathway, CCS (the copper chaperone for superoxide dismutase) gets the Cu(I) ion from Ctr to deliver it to the final target, superoxide dismutase; in the third pathway, Cox17 shuttles Cu(I) from Ctr to Sco1 in the mitochondrion, where it reaches cytochrome c oxidase through Sco2. Regulation of this system is comparatively less clear. It is also very important to note that the trafficking mechanism for a given metal may vary somewhat when comparing bacteria and eukaryota. In addition, an extreme variability is expected and found among bacteria, given their fast evolution processes. Of course, some aspects of these mechanisms can be instead quite well conserved even in different organisms, in which case bioinformatic approaches relying on multiple sequence alignments, phylogenetic trees, and molecular modeling (taking into account all of the caveats discussed in the preceding section) can be extremely informative on the functional features of entire pathways (12). For instance, phylogenetic studies clearly point out that copper chaperones and copper ATPases evolved in parallel but independently from a single ancestor, common to proteins from eukaryota and prokaryota (13). Interestingly, it appears that even in operons coexpressing a copper chaperone and an ATPase, the two proteins have generally evolved independently, suggesting that their coregulation is the result of specific genetic pressure (13).
Figure 1.
Display of the proposed copper trafficking pathways in yeast (adapted from ref. 47). Information on the directionality of copper transport, as well as on the identity of the partners involved in each transfer of the copper ion, is obtained mainly from experimental data but in some instances can be based on speculation or computational predictions. Regulation of the system is maintained at different points: Mac1 regulates the transcription of the genes encoding Fre and Ctr (48), and Ace1 regulates the transcription of the genes encoding superoxide dismutase and metallothionein (MT) (49).
Copper trafficking in cells constitutes an example that may be quite typical of how the field of bioinorganic chemistry may evolve to link genetic information and analytical and spectroscopic methods to unravel the cellular processes at the basis of the role of metal ions in biology. The topic of copper trafficking was initiated by the identification of the chaperone function of the soluble Cu(I) receptor Atx1 (14) and of its 3D structure (15, 16). This and subsequent articles (e.g., ref. 17) clearly demonstrated the existence of a complex molecular machinery in cells controlling uptake, transport, and delivery of copper ions in cells. Genetic studies have been at the basis of the identification of Atx1 as a key factor in metal homeostasis (18), before the identification of its chaperone function. In the subsequent years, a huge amount of data has accumulated on copper trafficking in a variety of organisms, as well as in the various organelles within single eukaryotic cells. Structural biologists have made outstanding contributions in detailing, within the general picture of copper homeostasis, the atomic and molecular determinants of the interaction between the individual proteins and the metal ion (19). The investigation at the atomic level of protein–protein interactions responsible for the key step of transfer of the metal ion along with the delivery (see, e.g., Fig. 1), has proved more difficult, possibly because of the transient nature of these interactions. Here, the bulk of high-resolution structural information on the physiologically relevant interactions has been produced by NMR spectroscopy (20, 21), a technique particularly suitable for the study of this kind of interaction in solution (22). However, through a clever use of site-directed mutagenesis (23) or thanks to repeated trials in a variety of conditions (24), high-resolution information can be gained also through x-ray crystallography. Fig. 2 shows the example of the interaction between two partners in the yeast cell: Atx1 and Ccc2 (20). Molecular modeling (12) and experimental studies (21, 25) indicate that the same mode of interaction is adopted by the homolog proteins in bacteria. However, the directionality of Cu(I) transfer may be species-dependent (21).
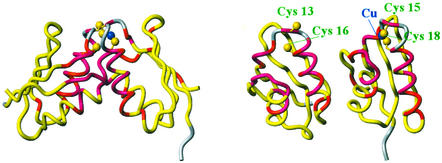
Figure 2.
Display of the complex formed by the first domain of Ccc2 (molecule, Left) and Atx1 (molecule, Right) in solution, as determined by NMR chemical-shift mapping studies (20), based on the independently solved solution structures of the two proteins (16, 50). The configuration of the adduct is supported by the dimerization of Cu(I)Atx1 observed in the crystal structure (24). Atx1 delivers Cu(I) to Ccc2 for subsequent delivery within the post-Golgi apparatus (see Fig. 1). (Left) The proposed structure of the complex. (Right) The two proteins rotated by 90° along the vertical axis, to allow the reader to observe the interaction regions. The figure is color-coded according to the variations in chemical shift of the backbone amide protons experimentally observed: regions in pink feature chemical-shift variations (in absolute value) >0.1 ppm, those in red feature variations between 0.04 and 0.1 ppm, and those in yellow feature variations <0.04 ppm (20). No data could be obtained for regions in gray. The Sγ atoms of the coordinating cysteines and the Cu(I) ion are also shown (gold and blue balls, respectively).
To understand metal homeostasis within the general context of cell metabolism, it is important to address also the topic of how the complex molecular mechanism behind it is regulated. A tight regulation is indeed crucial to maintain the delicate balance of metal partitioning among the various cellular organelles, as well as between the cell and the environment. If we consider again the case of copper homeostasis, here the largest amount of information is probably available for Enterococcus hirae. The cop operon in E. hirae consists of four genes (copY, copZ, copA, and copB) and regulates copper uptake, availability, and export in this bacterium (26). The CopY protein regulates expression of all genes in the operon in dependence of the level of intracellular copper. CopZ is a metallochaperone homologous to Atx1, whose structure has been solved (27), which delivers Cu(I) to CopY, triggering its detachment from the promoter region of the cop operon and thus resulting in expression of the latter (28). In vitro studies coupled to site-directed mutagenesis have provided detailed information about the metal-binding properties in the two proteins, as well as on some details of their mode of interaction, even in the absence of a detailed structural model for CopY (29). It is interesting to note that CopA in E. hirae is responsible for copper uptake under copper-limiting conditions (30), whereas the homolog protein in Bacillus subtilis is most likely responsible for copper efflux from the cell (21, 31).
Finally, it is worth noting that there is yet another very interesting aspect of the topic of metal homeostasis: resistance to toxic metals. Some well documented examples are those of resistance to arsenic (32), mercury (33), and, once again, copper. This last metal is used against pathogens affecting some plants. A strain of Pseudomonas syringae was found to have acquired resistance against copper, the genes responsible for resistance being localized in a copper-inducible operon on a 35-kb plasmid, which contains four genes (copA, -B, -C, and -D) (34). For one of these, CopC (note that the single operon contained in the mentioned plasmid is called cop but does not bear any relationship to the aforementioned cop operon of E. hirae), the solution structure has been recently solved (35). The role of CopC in vivo is most likely that of receiving or donating copper ions from CopA, possibly in cooperation with CopD. It has been proposed that copper binding takes place through an unprecedented consensus sequence (35); furthermore, it appears that the mechanism of interaction between the protein and the metal ion presents peculiar features, shedding new light on the mechanism of copper resistance in P. syringae.
In this section, we have focused on the interaction between cells and copper, both as an essential metal (e.g., in the yeast cell) and as a noxious metal (e.g., for P. syringae). The reason for this choice is that copper is the essential metal for which the largest amount of information is available, ranging from genetic analyses to high-resolution structural information on both individual proteins involved in copper homeostasis and on protein–protein complexes mediating copper transfer. In addition, detailed insights are available on the mechanisms of regulation. Following on the example of copper, several studies addressing some aspects of the regulation and homeostasis of other essential metals are becoming available in the literature. For instance, the tight regulation of the amount of free intracellular zinc has been demonstrated (36), a zinc-specific export pump has been identified and structurally characterized (37), and a few details about the zinc-binding site in the zinc uptake regulator are now known (38). The Ni chaperone (UreE) for Ni-urease has been structurally characterized (39, 40), and a literature is flourishing about iron, manganese, etc. However, we are still far from being able to draw a picture analogous to that of Fig. 1 for the homeostasis of any metal other than copper, Fig. 1 itself representing only a part of the overall mechanism of copper homeostasis. This is an area where bioinorganic chemists will have to be proactive in the coming years. It is also important to mention that metal homeostasis and its regulation are not “isolated” cellular processes but are tightly connected with all of the metabolic pathways in which metal-binding proteins are involved. (Just to give an example with reference to Fig. 1, copper homeostasis is linked to respiration via the enzyme cytochrome c oxidase, which necessitates copper to perform its function, or to iron metabolism via the Fet3 oxidase.) Also, it is often the case that the homeostasis of different metals is interlinked, so that in conditions where there is a shortage of a given metal, a cell can still maintain the functions that would require availability of this metal by producing proteins that use different metal ions [e.g., Monoraphidium braunii can use either plastocyanin or cytochrome c6 as an electron carrier in photosynthesis depending on the availability of copper (41)].
We have not mentioned the hundreds of final users of the metal ions, i.e., the enzymes (and some other metalloproteins) using the metal to perform their catalytic function. Such processes may also control illnesses. At present, there may be a tendency to consider them passive actors, anxiously waiting for their metal to be delivered. It can be anticipated, however, that this picture is not entirely true, and that these systems may actually be active in signaling their need for metal ions (42), thus playing a more relevant role in the broader picture of the cellular processes. Finally, metal ions play a prominent role in protein folding/unfolding (43, 44), as well as neurodegenerative diseases (45, 46).
Concluding Remarks
In perspective, the road for bioinorganic chemists appears to be extremely exciting and challenging, and may yield results capable of driving the development of related areas, e.g., medical sciences, environmental sciences, and nanotechnology. The new concept that the equilibrium between individual cellular components and metal ions is probably never direct but is always mediated by a squad of devoted proteins through an extremely organized and controlled stream of interactions and a careful regulation of protein expression has changed somewhat the way people in the field think. With new bioinformatic tools on the one hand and the new vision of the extent and complexity of cellular processes suggested by genome sequences and genetic studies on the other, the scientific field of biological inorganic chemistry is now bound to a quantum leap linking the classical characterization of the role of essential metals in metal-binding proteins to protein–protein and protein–nucleic acid interactions, up to the unraveling of metabolic pathways and the mechanisms of life.
References
- 1.Kendrew J C, Dickerson R E, Strandberg B E, Hart R G, Phillips D C, Shore V C. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 2.Perutz M F, Rossmann M G, Cullis A F, Muirhead H, Will G, North A C T. Nature. 1960;185:416–422. doi: 10.1038/185416a0. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, et al. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 4.Degtyarenko K N. Bioinformatics. 2000;16:851–864. doi: 10.1093/bioinformatics/16.10.851. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S F, Madden T L, Schaeffer A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Degtyarenko K N, North A C T, Findlay J B C. Nucleic Acids Res. 1999;27:233–236. doi: 10.1093/nar/27.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castagnetto J M, Hennessy S W, Roberts V A, Getzoff E D, Tainer J A, Piquet M E. Nucleic Acids Res. 2002;30:379–382. doi: 10.1093/nar/30.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marti-Renom M A, Stuart A C, Fiser A, Sanchez R, Melo F, Sali A. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 10.Banci L, Bertini I, Rosato A, Varani G. J Biol Inorg Chem. 1999;4:824–837. doi: 10.1007/s007750050356. [DOI] [PubMed] [Google Scholar]
- 11.Bertini I, Luchinat C, Provenzani A, Rosato A, Vasos P. Proteins Struct Funct Genet. 2002;46:110–127. doi: 10.1002/prot.10009. [DOI] [PubMed] [Google Scholar]
- 12.Arnesano F, Banci L, Bertini I, Ciofi-Baffoni S, Molteni E, Huffman D L, O'Halloran T V. Genome Res. 2002;12:255–271. doi: 10.1101/gr.196802. [DOI] [PubMed] [Google Scholar]
- 13.Jordan K, Natale D A, Koonin E V, Galperin M Y. J Mol Evol. 2001;53:622–633. doi: 10.1007/s002390010249. [DOI] [PubMed] [Google Scholar]
- 14.Pufahl R A, Singer C P, Peariso K L, Lin S-J, Schmidt P J, Fahrni C J, Cizewski Culotta V, Penner-Hahn J E, O'Halloran T V. Science. 1997;278:853–856. doi: 10.1126/science.278.5339.853. [DOI] [PubMed] [Google Scholar]
- 15.Rosenzweig A C, Huffman D L, Hou M Y, Wernimont A K, Pufahl R A, O'Halloran T V. Struct Folding Des. 1999;7:605–617. doi: 10.1016/s0969-2126(99)80082-3. [DOI] [PubMed] [Google Scholar]
- 16.Arnesano F, Banci L, Bertini I, Huffman D L, O'Halloran T V. Biochemistry. 2001;40:1528–1539. doi: 10.1021/bi0014711. [DOI] [PubMed] [Google Scholar]
- 17.Rae T, Schmidt P J, Pufahl R A, Culotta V C, O'Halloran T V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 18.Lin S J, Culotta V C. Proc Natl Acad Sci USA. 1995;92:3784–3788. doi: 10.1073/pnas.92.9.3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenzweig A C. Acc Chem Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 20.Arnesano F, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Huffman D L, O'Halloran T V. J Biol Chem. 2001;276:41365–41376. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
- 21.Banci L, Bertini I, Ciofi-Baffoni S, Del Conte R, Gonnelli L. Biochemistry. 2003;42:1939–1949. doi: 10.1021/bi027096p. [DOI] [PubMed] [Google Scholar]
- 22.Zuiderweg E R. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 23.Lamb A L, Torres A S, O'Halloran T V, Rosenzweig A C. Nat Struct Biol. 2001;8:751–755. doi: 10.1038/nsb0901-751. [DOI] [PubMed] [Google Scholar]
- 24.Wernimont A K, Huffman D L, Lamb A L, O'Halloran T V, Rosenzweig A C. Nat Struct Biol. 2000;7:766–771. doi: 10.1038/78999. [DOI] [PubMed] [Google Scholar]
- 25.Multhaup G, Strausak D, Bissig K D, Solioz M. Biochem Biophys Res Commun. 2001;288:172–177. doi: 10.1006/bbrc.2001.5757. [DOI] [PubMed] [Google Scholar]
- 26.Odermatt A, Solioz M. J Biol Chem. 1995;270:4349–4354. doi: 10.1074/jbc.270.9.4349. [DOI] [PubMed] [Google Scholar]
- 27.Wimmer R, Herrmann T, Solioz M, Wüthrich K. J Biol Chem. 1999;274:22597–22603. doi: 10.1074/jbc.274.32.22597. [DOI] [PubMed] [Google Scholar]
- 28.Cobine P, Wickramasinghe W A, Harrison M D, Weber T, Solioz M, Dameron C T. FEBS Lett. 1999;445:27–30. doi: 10.1016/s0014-5793(99)00091-5. [DOI] [PubMed] [Google Scholar]
- 29.Cobine P A, George G N, Jones C E, Wickramasinghe W A, Solioz M, Dameron C T. Biochemistry. 2002;41:5822–5829. doi: 10.1021/bi025515c. [DOI] [PubMed] [Google Scholar]
- 30.Odermatt A, Suter H, Krapf R, Solioz M. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 31.Banci L, Bertini I, Ciofi-Baffoni S, D'Onofrio M, Gonnelli L, Marhuenda-Egea F C, Ruiz-Dueñas F J. J Mol Biol. 2002;317:415–429. doi: 10.1006/jmbi.2002.5430. [DOI] [PubMed] [Google Scholar]
- 32.Rosen B P. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- 33.Foster T J. CRC Crit Rev Microbiol. 1987;15:117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- 34.Cooksey D A. Mol Microbiol. 1993;7:1–5. doi: 10.1111/j.1365-2958.1993.tb01091.x. [DOI] [PubMed] [Google Scholar]
- 35.Arnesano F, Banci L, Bertini I, Thompsett A R. Structure (Cambridge, UK) 2002;10:1337–1347. doi: 10.1016/s0969-2126(02)00858-4. [DOI] [PubMed] [Google Scholar]
- 36.Outten C E, O'Halloran T V. Science. 2001;292:2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 37.Banci L, Bertini I, Ciofi-Baffoni S, Finney L A, Outten C E, O'Halloran T V. J Mol Biol. 2002;323:883–897. doi: 10.1016/s0022-2836(02)01007-0. [DOI] [PubMed] [Google Scholar]
- 38.Outten C E, Tobin D A, Penner-Hahn J E, O'Halloran T V. Biochemistry. 2001;40:10417–10423. doi: 10.1021/bi0155448. [DOI] [PubMed] [Google Scholar]
- 39.Remaut H, Safarov N, Ciurli S, Van Beeumen J J. J Biol Chem. 2001;276:49365–49370. doi: 10.1074/jbc.M108304200. [DOI] [PubMed] [Google Scholar]
- 40.Song H K, Mulrooney S B, Huber R, Hausinger R P. J Biol Chem. 2001;276:49359–49364. doi: 10.1074/jbc.M108619200. [DOI] [PubMed] [Google Scholar]
- 41.Ho K K, Krogmann D W. Biochim Biophys Acta. 1984;766:310–316. [Google Scholar]
- 42.Wei J, Srinivasan C, Han H, Valentine J, Gralla E B. J Biol Chem. 2001;276:44798–44803. doi: 10.1074/jbc.M104708200. [DOI] [PubMed] [Google Scholar]
- 43.Telford J, Wittung-Stafshede P, Gray H B, Winkler J R. Acc Chem Res. 1998;31:755–763. [Google Scholar]
- 44.Bentrop D, Bertini I, Iacoviello R, Luchinat C, Niikura Y, Piccioli M, Presenti C, Rosato A. Biochemistry. 1999;38:4669–4680. doi: 10.1021/bi982647q. [DOI] [PubMed] [Google Scholar]
- 45.Lyons T J, Gralla E B, Valentine J S. Metal Ions Biol Syst. 1999;36:125–177. [PubMed] [Google Scholar]
- 46.Lehmann S. Curr Opin Chem Biol. 2002;6:187–192. doi: 10.1016/s1367-5931(02)00295-8. [DOI] [PubMed] [Google Scholar]
- 47.O'Halloran T V, Culotta V C. J Biol Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 48.Dancis A, Haile D, Yuan D S, Klausner R D. J Biol Chem. 1994;269:25660–25667. [PubMed] [Google Scholar]
- 49.Gralla E B, Thiele D J, Silar P, Valentine J S. Proc Natl Acad Sci USA. 1991;88:8558–8562. doi: 10.1073/pnas.88.19.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Banci L, Bertini I, Ciofi-Baffoni S, Huffman D L, O'Halloran T V. J Biol Chem. 2001;276:8415–8426. doi: 10.1074/jbc.M008389200. [DOI] [PubMed] [Google Scholar]