Abstract
Iron is concentrated in ferritin, a spherical protein with a capacious cavity for ferric nanominerals of <4,500 Fe atoms. Global ferritin structure is very stable, resisting 6 M urea and heat (85°C) at neutral pH. Eight pores, each formed by six helices from 3 of the 24 polypeptide subunits, restrict mineral access to reductant, protons, or chelators. Protein-directed transport of Fe and aqueous Fe3+ chemistry (solubility ≈10−18 M) drive mineralization. Ferritin pores are “gated” based on protein crystals and Fe chelation rates of wild-type (WT) and engineered proteins. Pore structure and gate residues, which are highly conserved, thus should be sensitive to environmental changes such as low concentrations of chaotropes. We now demonstrate that urea or guanidine (1–10 mM), far below concentrations for global unfolding, induced multiphasic rate increases in Fe2+-bipyridyl formation similar to conservative substitutions of pore residues. Urea (1 M) or the nonconservative Leu/Pro substitution that fully unfolded pores without urea both induced monophasic rate increases in Fe2+ chelation rates, indicating unrestricted access between mineral and reductant/chelator. The observation of low-melting ferritin subdomains by CD spectroscopy (melting midpoint 53°C), accounting for 10% of ferritin α-helices, is unprecedented. The low-melting ferritin subdomains are pores, based on percentage helix and destabilization by either very dilute urea solutions (1 mM) or Leu/Pro substitution, which both increased Fe2+ chelation. Biological molecules may have evolved to control gating of ferritin pores in response to cell iron need and, if mimicked by designer drugs, could impact chelation therapies in iron-overload diseases.
The mechanism that controls the rates of Fe transfer from ferritin to other cell molecules or cells in vivo remain poorly understood (1, 2). Iron, which is central to biological catalysis, is concentrated in ferritin from the attomolar solubility in neutral water and air to the micromolar and millimolar ranges used in living cells for respiration, photosynthesis, nitrogen fixation, and DNA synthesis (conversion of ribonucleotides to deoxyribonucleotides) (1). Ferritin, which is nature's only solution to concentrating iron, is found in all known organisms and is a large spherical protein of multiple polypeptide subunits (usually 24) with an internal nanocavity (8-nm diameter) that is the location of a hydrated ferric oxide mineral of up to 4,500 Fe atoms. The biomineral, formed by a combination of protein-based oxidation, and inorganic, aqueous Fe3+ chemistry can be dissolved by adding reductant and chelators (1, 2). Much more is known about Fe2+ entry and oxidation than Fe2+ reduction and exit from ferritin (2, 4–6), in part because biological catalysis is understood better than dissolution of biominerals and in part because the catalytic ferroxidase site in ferritin shares common reaction intermediates with enzymes, exemplified by methane monooxygenase, that provide a frame of reference (7–10). Recovering the iron concentrated in ferritin is crucial for biosynthesis of heme, Fe-S clusters, and diiron cofactors. Safely removing the iron from ferritin is a goal of the chelation therapies used to manage hematological diseases such as β-thalassemia and sickle cell disease, where the aging of red cells in the required, regular transfusions results in the accumulation of massive amounts of iron that saturate the storage capacity of ferritin.
Eight protein pores in ferritin, formed at the threefold axes by junctions of three subunits of the highly symmetric protein, control the access of reductant and chelators to the ferritin mineral. Alterations of pore function can be measured as rates of Fe2+ chelation from mineralized, recombinant ferritin, modified by amino acid substitutions from site-directed mutagenesis (SDM) and triggered by the addition of reductant (NADH/FMN) (3). For example, a 30-fold increase in the rate of dissolving the ferric mineral occurred with substitutions for a conserved leucine residue involved in helix–helix interactions near the pores. The crystal structure of the altered ferritin showed localized unfolding of the pores, with no detectable change in overall subunit folding or assembly elsewhere (3).
Success in unfolding the ferritin pores (measured as increased Fe2+-bipyridyl chelation triggered by NADH/FMN), was achieved in all (10/10) SDM designs based on phylogenetic sequence conservation, location near the pores in three-dimensional space, and lack of any other assigned function (11). All the altered ferritins assembled normally (gel filtration) and produced normal yields after bacterial expression. Three types of conserved structural motifs were altered in the set of ferritins with “open” or unfolded pores: hydrophobic, interhelix packing (Leu-134/Leu-110); ionic loop/helix packing (Asp-122/Arg-72); and helix/helix loop size (lengthening the interhelix loop by five residues). Sensitivity of ferritin-pore function to conservative amino acid changes suggested that the ferritin pores would also be differentially sensitive to solvent molecules that unfold proteins, such as urea (12). We now show that in ferritin, although the global structure is stable to 6 M urea at neutral pH (13, 14) and temperatures up to 85°C (15), ferritin pores are opened by low chaotrope concentrations (1.0 mM to 1 M urea or guanidine), assessed as Fe2+-chelation rates. Moreover, a melting transition (53°C) of a helix subdomain that we observed in ferritin by CD spectroscopy was downshifted 10°C by 1 mM urea, in parallel with the increased rate of Fe2+ chelation, indicating a relationship between the helix subdomain and the pores.
Experimental Methods
Recombinant frog ferritin was used as the model ferritin, because the kinetic properties of this ferritin have been particularly favorable for study (3, 7, 16, 18). Frog and human ferritins are very similar structurally (19, 20) and mechanistically as well (6, 7, 16). Of the two types of ferritin subunits, H and L, we chose the H type for study, because functional integrity of protein preparations could be analyzed by specific UV-visible (UV-vis) spectroscopy. The diferric peroxide product of catalytic ferroxidase activity in H ferritin has a spectroscopically distinctive spectrum with an absorbance maximum at 650 nm (7, 16, 17). Note that L subunit ferritin, which is sometimes studied, lacks the catalytic site and the convenient spectroscopic signature of A650. All ferritin preparations used in this study showed maximal formation of the diferric peroxo intermediate (A650) within 25 msec and decay to Fe3+-O-Fe3+ products within 1 sec. Two proteins were compared in the study: H-WT and H-L134P, an open-pore protein for which the crystal structure is known (3). In the H-L134P no electron density was detected in protein crystals from residues 110–134 (represented in Fig. 1 Right) at the turns of three sets of C,D helices around the pores, which contrasts with order in this region for all other known ferritin structures. The local, disordered structure coincides with rapid Fe-chelation rates, thereby defining the exit pores (3). Frog H ferritin sequences encoded in pET-9a and expressed in Escherichia coli BL21-DE3-PLysS (Stratagene) were isolated as the 24-subunit assembly without Fe (<0.5 per subunit) by using the methods described (18). Recombinant ferritin was mineralized by adding a freshly prepared, acidic solution of Fe SO4 (to minimize oxidation and hydrolysis) to apoferritin in buffer and incubating for 1 h at room temperature and then overnight at 4°C to complete hydrolysis of ferric mineral precursors (16). The final solution was 2.08 μM protein with 480 Fe/protein (24 Fe per subunit) in 0.1 M Mops/0.1 M NaCl, pH 7. Sterile solutions of mineralized ferritin are stable for months to years, although catalytic activity can decline after 2 weeks.
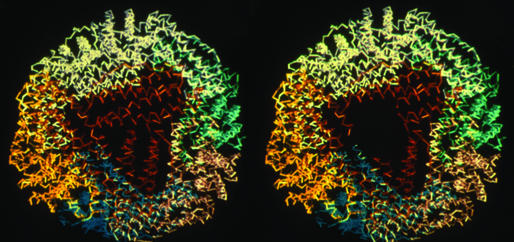
Figure 1.
Structure of a representative ferritin (recombinant frog L subunit) around one of the eight pores. Polypeptide backbones are colored to show the four-α-helix bundle subunits packed into the assembled 24-subunit structure, viewed down the threefold axis, at the pore. The structure, similar in >20 ferritin structures currently in the Protein Data Bank, is based on data from refs. 3 and 19. When the pore is open and the helices that form the pores are unfolded, as in recombinant frog H-L134P, the backbone structure remains superimposable on the WT frog L protein structure except at the pore, where no electron density was detectable in the x-ray diffraction pattern of protein crystals (3). (Right) The absence of pore structure is illustrated as the black or empty area, which was made from Left and the data in ref. 3.
Chelation of Fe2+ in mineralized ferritin, after adding NADH/FMN to reduce the ferric mineral, was monitored as formation of Fe2+-bipyridyl (ΔA522/sec). To start the reaction, equal volumes of mineralized ferritin solution and a fresh solution of 5 mM β-NADH/5 mM FMN/5 mM bipyridyl in 0.1 M Mops/0.1 M NaCl, pH 7, were mixed as first described by Jones et al. (21). The effects of chaotropes on pore function were studied in mineralized ferritin by mixing solutions of protein with unfolding reagents [urea, guanidine hydrochloride (Gdn⋅HCl), and Triton X-100] and incubating at room temperature for 1 h followed by incubating overnight at 4°C before adding reductant and chelator. The final concentrations in the Fe chelation assay were 1.04 μM protein, 2.5 mM FMN, NADH, and bipyridyl, 0.10 M Mops, pH 7, and 0.10 M NaCl. Initial rates of iron release were calculated from the progress curves for the formation of Fe2+-bipyridyl complex, between 0 and 1 min (>40 data points) except for the WT protein, where the window was 0–2 min (>80 data points), by using the Enzyme Kinetics Module from SIGMA PLOT for linear plots and setting the goodness of fit parameter (ratio of the χ2 value to the degrees of freedom in the model) at <6; all nonlinear models tested were rejected by the program. For linear fits P = 0.03–0.04. Data from three independent measurements per protein preparation were averaged for each time point, and the average was used for fitting. The rates thus calculated were compared for two to three independently prepared protein preparations with the error presented as standard deviation. Units are μmol⋅liter−1⋅s−1 Fe2+-bipyridyl complex formed. One-hundred-percent conversion of ferric mineral to Fe2+-bipyridyl, measured under these conditions, has an A522 of 0.8.
Global ferritin unfolding was monitored as changes in UV-vis absorbance, recorded on a Cary 100 Bio UV-vis spectrophotometer (Varian) over the range of 35–95°C. Ferritin-subdomain unfolding was monitored as changes in the CD spectra recorded on a Pi-180 spectrophotometer (Applied Photophysics, Surrey, U.K.) over the temperature range of 5–65°C. Solutions of protein (2.08 × 10−1 μM in 10 mM Mops buffer, pH 7) were incubated with or without 1 mM urea overnight as described above and equilibrated for 10 min at the selected temperature before recording spectra between 300 and 200 nm, with the resolution set at 0.5 mm. Contributions of the solvent to the protein spectra were subtracted, and the difference spectra for five replicates were averaged. Absorbance values, which represent global protein structure, were plotted against temperature for the values at 280 nm. Molar ellipticities at 220 nm were determined from the optical rotation, and the percentage of α-helix content was calculated as described by Frere et al. (22) and Zhong and Johnson (23): percentage α-helix = Δɛ220 nm × −10. Δɛ220 nm is the CD/residue: Δɛ220 nm = θ220 nm/3,300. Ferritin subunit structure is mainly a four-helix bundle, A–D, plus a short helix, E (24). The data presented for H-L134P ferritin are representative of two sets of measurements on one preparation, and the data for the H-WT protein with and without urea are representative of three independent protein preparations.
Results
Alterations of local structure in ferritin by SDM around the threefold iron-exit pores, which increased rates of formation of Fe2+-bipyridyl in the presence of NADH/FMN as the reductant, defined the residues that “gate” the ferritin pores (11). The alterations were in conserved leucine, aspartate, and arginine residues as well as the insertion of five amino acids to double the length of a short, conserved loop at the pores. The altered rates of Fe-chelator formation were multiphasic (11), indicating limited, albeit increased, access between Fe, chelator, and/or reductant. If the pores and the global structure of ferritin were differentially sensitive to disruption, as suggested by protein crystal structures and the location of the sites sensitive to SDM (3, 11), low concentrations of chaotropes might mimic disruption of the pores by amino acid substitution.
Effects of Chaotropes, Detergent, and NaCl on Chelation of Fe in Ferritin (Formation Rate of Fe2+-Bipyridyl in the Presence of NADH/FMN).
Urea disrupts quaternary structure of the sort stabilized by the Leu-134/110 interhelix hydrogen bonds around the ferritin pores. Because the supramolecular ferritin structure is unusually resistant to urea, maintaining the supramolecular structure in urea concentrations as high as 6–10 M (13, 14, 25), we investigated effects on pore gating as the formation of Fe2+-bipyridyl in the presence of NADH/FMN by using mineralized ferritins with millimolar concentrations of urea that would not affect global ferritin structure (13, 14). Both 1 and 10 mM urea increased rates of Fe2+ chelation significantly (Fig. 2), supporting the notion that the pores would be differentially sensitive to chaotropes. For example, the initial rate of Fe2+-bipyridyl formation was 1.90 ± 0.09 μmol⋅liter−1⋅s−1 with 1 mM urea compared with 0.89 ± 0.06 μmol⋅liter−1⋅s−1 without urea (Table 1). Rates of Fe2+-bipyridyl formation increased with increasing concentrations of urea up to 1 M, where the initial rate was 5.4 times as fast as without urea. The ferric ferritin mineral was dissolved completely and chelated in 3–4 min (Fig. 2) compared with hours in the absence of urea (3). In contrast to the multiphasic kinetics observed in the absence of urea or at lower concentration of urea, the kinetics were linear (see Experimental Methods) at 1 M urea for the removal of 75% of the mineralized Fe (Fig. 2). Thus, only ≈110 Fe atoms were removed at a slower rate and may represent Fe in nucleation sites attached to the protein (4). The kinetics for WT ferritin in 1 M urea and the H-L134P protein without urea were similar. In H-L134P, the pores were fully opened by unfolding between residues 110 and 134 in each of the three subunits that form the pores (3).
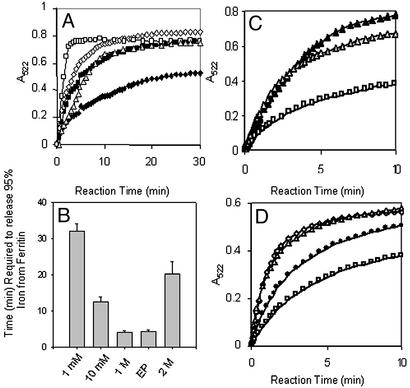
Figure 2.
Chaotropes decrease the time required to chelate Fe in the ferritin biomineral, mimicking the unfolding of ferritin pores associated with amino acid substitution of phylogenetically conserved pore residues. Recombinant ferritin protein (the frog H-WT), isolated with <5 Fe per 24 assembled subunits, was mineralized by mixing a solution of 2.08 μM protein in 0.10 M Mops/0.1 M NaCl, pH 7, with a solution of FeSO4 in 0.001 M HCl (see Experimental Methods). Reduction and chelation were triggered by mixing equal volumes of protein solution and a solution of 5 mM each of NADH/FMN/bipyridyl in 0.10 M Mops/0.1 M NaCl, pH 7 (see Experimental Methods). (A) Progress curve for formation of Fe2+-bipyridyl + urea. Chaotrope concentrations: ♦, 0; ■, 1 mM; ⋄, 10 mM; □, 1 M; ▵, 2 M. (B) Decrease in the time (minutes) required to remove all the Fe from ferritin in the presence of urea. Note that, in the absence of urea, removal of all the Fe required 150 min under these conditions (3). EP, engineered protein H-L134P. (C) Progress curves for formation of Fe2+-bipyridyl with Triton X-100. Chaotrope concentrations: □, 0; ▵, 1%; ▴, 10%. (D) Progress curve for formation of Fe2+-bipyridyl with Gdn⋅HCl. Chaotrope concentrations: □, 0; ●, 0.1 mM; ▵, 1 mM; ⋄, 10 mM.
Table 1.
Millimolar concentrations of urea increase the initial rates of Fe2+-bipyridyl formation from ferritin, triggered by the addition of NADH/FMN to ferritin
| Urea, mol⋅L−1 | Protein | Initial rate, μmol⋅liter−1⋅s−1 (pH 7.0) |
|---|---|---|
| 0 | WT | 0.890 ± 0.063 |
| 0.001 | WT | 1.920 ± 0.088 |
| 0.010 | WT | 2.608 ± 0.246 |
| 1 | WT | 4.848 ± 0.331 |
| 2 | WT | 1.272 ± 0.352 |
| 0 | H-L134P | 3.778 ± 0.410 |
Recombinant ferritin (2.06 μM mineralized with 480 Fe, added as FeSO4 in air) was mixed with an equal volume of NADH, FMN, and chelator each at 5.0 mM as described (3). Initial rates were determined from linear fits (0–2 min) as described in Experimental Methods. The data are means for two to three independent protein preparations with the error as the standard deviation.
The kinetics of Fe removal from the ferritin mineral (formation of Fe2+-bipyridyl formation) changed in 2 M urea. The initial rate decreased (1.02 ± 0.11 μmol⋅liter−1⋅s−1) and a second, linear phase appeared that described the rest of the progress curve (Fig. 2A). In addition, Fe2+ oxidation, a measure of Fe2+ entry and binding to the ferroxidase site, was inhibited slightly, which contrasts with 1 M urea or less. The higher concentration of urea possibly caused the collapse of channels around the pore, destroying the selectivity in control of Fe2+ access to the ferroxidase site and reductant/chelator access to the mineral.
Gdn⋅HCl and the nonionic detergent Triton X-100, two other agents that disrupt hydrophobic interactions in proteins, were also analyzed for effects on the formation rate of Fe2+-bipyridyl with NADH/FMN as the reductant (Fig. 2). The effects of Gdn⋅HCl at 1 and 10 mM and Triton X-100 T at 1% and 10% were comparable to the effects of 1 and 10 mM urea. Gdn⋅HCl increased rates significantly even at 0.1 mM. To determine whether ionic strength might play a role in faster iron release, given that one of the interactions controlling the pore is the conserved ion pair, Arg-72/Asp-122 (11), the effect of the addition of NaCl on the formation rate of Fe2+-bipyridyl was analyzed under the standard conditions. Raising the ionic strength to 1 M NaCl decreased both the initial rate of iron Fe2+-bipyridyl formation and the total amount of Fe iron chelated, indicating that high ionic strength inhibited access of reductant or chelator to Fe, “tightening” protein pores, changing chelator-Fe2+ binding, or both.
Effects of Temperature on Ferritin Protein Stability: Global Structure.
Ferritin is stable to unfolding by heat. In fact, heating is used to denature contaminating proteins during ferritin isolation from animal tissues (2). The major heat-induced disruption in ferritin structure occurred between 80 and 90°C depending on the protein source (15). To determine whether the global temperature stability of ferritin was altered by changes that enhance mineral dissolution, the effect of SDM on the UV absorbance was measured over the temperature range of 35–95°C, at 280 nm (Fig. 3A), where contributions from aromatic residues dominate. No change in the UV-vis absorbance was observed between 35 and >85°C for proteins representing the helix C/helix D packing interaction (H-L134P and H-L134V) and the ion pair between helix B and the C/D loop (H-R72D and D122R) (Fig. 3A). The results make clear that protein unfolding in ferritin can be localized around the pore with little influence on the global structure of the protein.
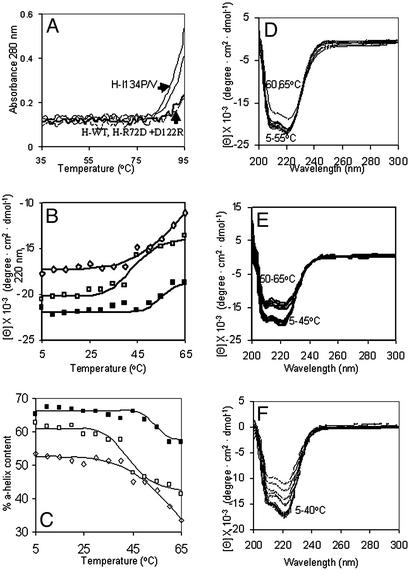
Figure 3.
Subdomain helix-reversible unfolding in the ferritin four-helix bundles. Solutions of recombinant ferritin (frog H-WT, H-L134P, H-R72D+, D122R, and H-WT + 1 mM urea) were analyzed in 0.01 M Mops, pH 7 (see Experimental Methods). CD spectra were recorded in a cuvette with a 1-cm path length at protein concentrations of 2 × 10−7 M except for H-L134P, the protein with fully open pores in protein crystals (Fig. 1), where the concentration was 4 × 10−7 M to compensate on the lower signal at higher temperatures. (A) UV-vis analysis of temperature transitions in global ferritin structure (A280) in WT and pore mutants (L134P, L134V, and R72D+D122R) between 35 and 95°C. (B) CD analysis of subdomain temperature transitions below global melting in ferritin between 5 and 65°C. ■, - H-WT; □, H-WT + 1 mM urea; ⋄, H-L134P. (C) Effect of temperature on % α-helix content in ferritin. ■, H-WT; □, H-WT + 1 mM urea; ⋄, H-L134P. (D–F) CD spectra of ferritin at different temperatures: D, H-WT; E, H-WT + 1 mM urea; F, H-L134P.
Effects of Temperature, Urea, and Amino Acid Substitution on Ferritin Protein: Subdomain Structure.
CD spectroscopy was used to analyze heat-induced structural changes in the α-helix structure of ferritin (Fig. 3 B–D; see Experimental Methods and refs. 22 and 23). Three properties of ferritin were used in designing the experiments: (i) high α-helix content (70%); (ii) pores formed by short sections of six α-helices (two from each of three subunits joined by a short loop) at the junctions of subunit triples; and (iii) pore-function sensitivity to unfolding by millimolar urea compared with 6 M urea for global structure (Figs. 2 A and B; refs. 3, 11, 13, and 15).
CD spectroscopy revealed an unprecedented aspect of ferritin unfolding: subdomains that melted far below the supramolecular assembly with a transition midpoint of 53°C (Fig. 3). The high structural stability of ferritin (1, 4) made detection of the subdomain unexpected, as was the detection of localized unfolding previously observed in protein crystals from H-L134P ferritin (3).
Several features of the low-melting subdomains observed by CD spectroscopy suggest that they are pores. First, the percentage of helix involved in the subdomain was 10 (Fig. 3C), which is comparable to the amount of helix around the pores. Second, 1 mM urea shifted the temperature of subdomain unfolding to 43°C and also increased the rate of forming Fe2+-bipyridyl (Table 1; Fig. 2). Finally, the CD spectrum of H-L134P, which has localized pore unfolding in protein crystals (3), also showed decreased temperature of unfolding in solution (Fig. 3 B, C, and F). The stability of the pore subdomain in H-L134P protein was lower than for WT protein with urea, and loss of helix, measured by CD, continued between 45 and 65°C, although A280 and global structure were unchanged <85°C (Fig. 3A). The local nature of the helix/coil transition induced by urea, observed between 35 and 50°C (Fig. 3 B–F), is emphasized by the small effects on other ferritin functions such as Fe uptake and mineralization, where the rates were 0.67 ± 0.01 and 0.57 ± 0.01 ΔA650 s−1 for 0 and 10 mM urea, respectively, for 480 Fe/protein. Comparisons of temperature effects on global structure (Fig. 3A) and the helical pore subdomain (Fig. 2 B–F) show the relative ease with which pores can be unfolded at or near physiological temperatures.
Chaotropes Increase Access of both Fe2+ and Fe3+ Chelators.
Desferal (deferoxamine mesylate or DFO), a Fe3+-chelator, is currently the drug of choice to remove Fe from patients with excess tissue iron accumulated from the monthly transfusions used in treating β-globin gene mutations (sickle cell disease and β-thalassemia). DFO is thought to trap Fe2+ during intracellular transfer into and out of ferritin because of the reducing environment in cells, and the more stable Fe3+–DFO forms as oxygen is available. DFO alone removes little Fe from ferritin in solution (26). To determine whether unfolding ferritin pores increased DFO chelation of ferritin Fe, the chelator was substituted for bipyridyl in the aerobic solutions of NADH/FMN used in the previous experiments (Fig. 2; Table 1). Formation of the Fe3+–DFO complex was monitored at 430 nm, which is the absorbance maximum under the conditions used.
Fe3+–DFO was readily detected in aerobic solutions of ferritin, to which NADH and FMN were added (Table 2). DFO was effective in chelating Fe from both H-WT and H-L134P ferritin, the protein in which amino acid substitution Leu/Pro locked pores in an open position (3), although as observed previously (26) only small amounts of Fe were accessible to DFO in the WT ferritin. A lag was observed in the appearance of Fe3+–DFO, which contrasts with the data for Fe3+-bipyridyl. A contribution to the rate from reoxidation of Fe2+ to Fe3+ is suggested by the lag. Only 10% of the Fe could be chelated by DFO in the absence of urea, which corresponds to earlier data obtained with horse spleen ferritin (26), whereas when the pore was open as in H-L134P, 39% of the Fe mineral could be removed with DFO. Fe removal rates from ferritin apparently depend much more on the state of the pores than on the type of chelator or the oxidation state of the Fe-chelator complex (Table 2). Such data emphasize the role of the protein in controlling rates of Fe chelation by pore gating and underscore the complexity of bioinorganic chemistry in current therapies for iron overload.
Table 2.
Accessibility of Fe in ferritin mineral to Fe2+ or Fe3+ chelators with NADH/FMN reduction
| Initial rate for Fe3+-DFO, μmol⋅liter−1⋅s−1 (pH 7.0) | Initial rate for Fe2+-bipyridyl, μmol⋅liter−1⋅s−1 (pH 7.0) | |
|---|---|---|
| H-WT | 0.275 ± 0.041 | 0.972 ± 0.087 |
| H-L134P | 2.443 ± 0.032 | 3.934 ± 0.122 |
Recombinant ferritin (2.06 μM mineralized with 480 Fe, added as FeSO4 in air) was mixed with an equal volume of NADH, FMN, and chelator each at 5.0 mM as described (3). The maximum amount of Fe removed in H-L134P is 39% and 98% for DFP and bipyridyl, respectively. Rates were determined from linear fits (0–2 min, H-L134P; 0–1 min, WT) as described in Experimental Methods. Data are means from two to three independent protein preparations with the error as the standard deviation.
Discussion
Fe in ferritin is used for synthesizing proteins such as hemoglobin (1, 27). Recovering the iron concentrated as the ferritin biomineral under physiological conditions is a process for which the parallels are limited, e.g., turnover or remodeling of bone and resorption of tooth biominerals. Calcium turnover in bone/tooth contrasts with Fe in ferritin in being extracellular and involving many types of macromolecules. Although there may be a number of macromolecules involved in transporting Fe, protons, and oxygen products to and from the ferritin mineral as in bone/tooth minerals, none have been identified.
The multistep process of dissolving the ferritin biomineral under physiological conditions includes (i) hydrolysis of the Fe–O/OH–Fe bridges, (ii) chelation of Fe inside the protein, and/or (iii) transfer of the iron to the chelator outside of the protein. A multiplicity of reactions likely accounts for the complexity of the progress curves for Fe2+-bipyridyl formation with either low chaotrope concentrations (Fig. 2) or conservative amino acid substitutions (3, 11). Experimentally and in vivo where the cellular reducing equivalents should be in excess (28), an additional reaction (4) is the reduction of mineralized Fe3+ inside the protein cavity either directly (29) or indirectly (30) by electron transfer.
Ferritin protein controls access of reductant and/or chelator to Fe3+ in the mineral as demonstrated by the change in rate and complexity of the progress curve for Fe2+-bipyridyl formation (Fig. 2), when pore structure is altered. Changes in ferritin-pore structure can be selectively induced either by millimolar concentrations of urea (Figs. 2 and 3) or by substitution of phylogenetically conserved amino acids around the pores (Fig. 1; refs. 3 and 11). The narrow urea concentration range observed for the induction of maximum rates of Fe2+-bipyridyl formation indicates the structural specificity of pore gating created by phylogenetically conserved amino acids around the ferritin pores.
Both Fe2+ and Fe3+ chelators are effective at removing iron from ferritin when the pores are unfolded. DFO seems to be less efficient than bipyridyl in chelating ferritin Fe even when the ferritin pores are open (30% vs. 98% respectively) (Table 2). However, the apparent difference may relate to rereduction of Fe3+ by the excess NADH/FMN in the reaction mixture, because changing the [NADH/FMN] altered the progress curve for the Fe3+–DFO formation (H. Takagi and E.C.T., unpublished observations). Sorting out the contributions of [O2], NADH, and FMN to the formation of Fe3+–DFO is beyond the scope of this report. However, the combination of reactants used experimentally (Table 2) shares a number of conditions with those in cells of patients with iron overload who are receiving DFO therapy‡ and indicate the importance of understanding the bioinorganic chemistry used in current medical therapies.
Normally the ferritin pore is almost closed in solution, based on rates of dissolving and chelating the ferric mineral solution (Figs. 1 and 2; and refs. 26 and 31), and in protein crystals, based on the apparent pore sizes in WT proteins (19, 20). Evidence that the pores are dynamic and are opening and closing even in solutions without chaotropes or in proteins without amino acid substitution is exemplified by the entrance of solute molecules into the ferritin protein cavity that are too large to pass through the pores observed in protein crystals of WT proteins (32) and by the temperature sensitivity of pore-helix unfolding near physiological temperatures (Fig. 2 B and C). Normally ferritin pores appear to be mostly closed in vivo as well, because the time for iron removal with the therapeutic chelator DFO is very long (8 h/day). However, rapid Fe “mobilization” from ferritin is induced under some physiological conditions, which suggests a change in ferritin-pore gating. Examples are the increased transfer of Fe from the liver to red cells after hemorrhage and from red-cell ferritin to heme when porphyrin biosynthesis rates increase during cell maturation (27, 33). Control of ferritin-pore gating in vivo by regulatory protein–ferritin protein interactions would be analogous to the physiological control of some ion channels (34) and would explain, in part, the high conservation of ferritin-pore structure.
Identification of amino acids, chaotropes, and temperature-sensitive subdomains that alter gating of the ferritin pores is important in understanding recovery of the concentrated Fe from ferritin in health or for removal of the excess iron in disease (Figs. 1–3; refs. 3 and 11). Ferritin-pore gating is a target for therapeutic manipulation and biological regulators. Even before biological regulators of ferritin pore gates are identified, natural or synthetic molecules can be studied that change interactions between the Fe in ferritin and reductants or chelators. Such molecules can include novel chelators for managing Fe overload where removal of excess Fe is crucial to health. Understanding ferritin structure and function illustrate the importance of bioinorganic chemistry in biology and medicine.
Acknowledgments
This work was supported in part by National Institutes of Health Grant DK 20251 and the Children's Hospital of Oakland Research Institute Foundation.
Abbreviations
- SDM
site-directed mutagenesis
- UV-vis
UV-visible
- Gdn⋅HCl
guanidine hydrochloride
- DFO
deferoxamine mesylate
Footnotes
The medical conditions requiring repeated transfusion treatment and iron chelation can be relatively common in populations with genetic origins from areas of endemic malaria; gene frequencies in such populations can be as high as 10%, because carriers are more disease-resistant.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Theil E C. Annu Rev Biochem. 1987;56:289–315. doi: 10.1146/annurev.bi.56.070187.001445. [DOI] [PubMed] [Google Scholar]
- 2.Theil E C. In: Handbook of Metalloproteins. Messerschmidt A, Huber R, Poulos T, Weighart K, editors. Chichester, U.K.: Wiley; 2001. pp. 771–781. [Google Scholar]
- 3.Takagi H, Shi D, Hall Y, Allewell N M, Theil E C. J Biol Chem. 1998;273:18685–18688. doi: 10.1074/jbc.273.30.18685. [DOI] [PubMed] [Google Scholar]
- 4.Chasteen N D, Harrison P M. J Struct Biol. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- 5.Jameson G N L, Jin W, Krebs C, Perreira A S, Tavares P, Liu X, Theil E C, Huynh B H. Biochemistry. 2002;41:13435–13443. doi: 10.1021/bi026478s. [DOI] [PubMed] [Google Scholar]
- 6.Bou-Abdallah F, Papaefthymiou G C, Scheswohl D M, Stanga S D, Arosio P, Chasteen N D. Biochem J. 2002;364:57–63. doi: 10.1042/bj3640057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang J, Krebs C, Huynh B H, Edmondson D E, Theil E C, Penner-Hahn J E. Science. 2000;287:122–125. doi: 10.1126/science.287.5450.122. [DOI] [PubMed] [Google Scholar]
- 8.Rosenzweig A C, Brandstetter H, Whittington D A, Nordlund P, Lippard S J, Frederick C A. Proteins. 1997;29:141–152. [PubMed] [Google Scholar]
- 9.Shu L, Nesheim J C, Kauffmann K, Munck E, Lipscomb J D, Que L., Jr Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 10.Broadwater J A, Ai J, Loehr T M, Sanders-Loehr J S, Fox B G. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 11.Jin W, Takagi H, Pancorbo N M, Theil E C. Biochemistry. 2001;40:7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- 12.Shortle D, Wand Y, Gillespie J R, Wrabl J O. Protein Sci. 1996;5:991–1000. doi: 10.1002/pro.5560050602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavoie D J, Marcus D M, Otsuka S, Listowsky I. Biochim Biophys Acta. 1979;579:359–366. doi: 10.1016/0005-2795(79)90063-1. [DOI] [PubMed] [Google Scholar]
- 14.Otsuka S, Listowsky I, Niitsu Y, Urushizaki I. J Biol Chem. 1980;255:6234–6237. [PubMed] [Google Scholar]
- 15.Stefanini S, Cavallo S, Wang C Q, Tataseo P, Vecchini P, Giartosio A, Chiancone E. Arch Biochem Biophys. 1996;325:58–64. doi: 10.1006/abbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 16.Pereira A, Small G S, Krebs C, Tavares P, Edmondson D E, Theil E C, Huynh B H. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 17.Moënne-Loccoz P, Krebs C, Herlihy K, Edmondson D E, Theil E C, Huynh B H, Loehr T M. Biochemistry. 1999;38:5290–5295. doi: 10.1021/bi990095l. [DOI] [PubMed] [Google Scholar]
- 18.Waldo G S, Theil E C. Biochemistry. 1993;32:13262–13269. doi: 10.1021/bi00211a039. [DOI] [PubMed] [Google Scholar]
- 19.Trikha J, Theil E C, Allewell N M. J Mol Biol. 1995;248:949–967. doi: 10.1006/jmbi.1995.0274. [DOI] [PubMed] [Google Scholar]
- 20.Hempstead P D, Yewdall S J, Fernie A R, Lawson D M, Artymiuk P J, Rice D W, Ford G C, Harrison P M. J Mol Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 21.Jones T, Spencer R, Walsh C. Biochemistry. 1978;17:4011–4017. doi: 10.1021/bi00612a021. [DOI] [PubMed] [Google Scholar]
- 22.Frere V, Sourgen F, Monnot M, Troalen F, Fermandjian S. J Biol Chem. 1995;270:17502–17507. doi: 10.1074/jbc.270.29.17502. [DOI] [PubMed] [Google Scholar]
- 23.Zhong L, Johnson W C J. Proc Natl Acad Sci USA. 1992;89:4462–4465. doi: 10.1073/pnas.89.10.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banyard S H, Stammers D K, Harrison P M. Nature. 1978;271:282–284. doi: 10.1038/271282a0. [DOI] [PubMed] [Google Scholar]
- 25.Otsuka S, Maruyama H, Listowsky I. Biochemistry. 1981;20:5226–5232. doi: 10.1021/bi00521a020. [DOI] [PubMed] [Google Scholar]
- 26.Crichton R R, Roman F, Roland F, Paques E, Paques A, Vandamme E. J Mol Catal. 1980;7:267–276. [Google Scholar]
- 27.Peto T E A, Thompson J L. Biochim Biophys Acta. 1986;881:79–86. doi: 10.1016/0304-4165(86)90099-1. [DOI] [PubMed] [Google Scholar]
- 28.Carmel-Harel O, Storz G. Annu Rev Microbiol. 2000;54:439–461. doi: 10.1146/annurev.micro.54.1.439. [DOI] [PubMed] [Google Scholar]
- 29.Yang X, Arosio P, Chasteen N D. Biophys J. 2000;78:2049–2059. doi: 10.1016/S0006-3495(00)76752-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watt G D, Jacobs D, Frankel R B. Proc Natl Acad Sci USA. 1988;85:7457–7461. doi: 10.1073/pnas.85.20.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ihara K, Maeguchi K, Young C T, Theil E C. J Biol Chem. 1984;259:278–283. [PubMed] [Google Scholar]
- 32.Chasteen N D. In: Metal Ions in Biological Systems. Sigel H, Sigel M, editors. Vol. 35. New York: Dekker; 1998. pp. 479–514. [PubMed] [Google Scholar]
- 33.Lipschitz D A, Simon M O, Lynch S R, Dugard J, Bothwell T H, Charlton R W. Br J Haematol. 1971;21:2889–2303. doi: 10.1111/j.1365-2141.1971.tb03441.x. [DOI] [PubMed] [Google Scholar]
- 34.Dascal N. Trends Endocrinol Metab. 2001;2:391–398. doi: 10.1016/s1043-2760(01)00475-1. [DOI] [PubMed] [Google Scholar]