Abstract
Members of a family of catecholate siderophores, called salmochelins, were isolated by reversed-phase HPLC from Salmonella enterica serotype Typhimurium and structurally characterized by Fourier transform ion cyclotron resonance–MS/MS and GC–MS. The tentative structure of salmochelin 1 contained two 2,3- dihydroxybenzoylserine moieties bridged by a glucose residue, bound to the serine hydroxyl group of one moiety and the carboxylate of the second moiety. Salmochelin 2 contained in addition a second glucose residue linked to a third 2,3-dihydroxybenzoylserine moiety. Salmochelins were not produced by an iroBC mutant, which indicated that the IroB protein might be responsible for the glucosyl transfer predicted by sequence similarities to known glycosyltransferases. Uptake experiments with radiolabeled 55Fe-salmochelin and growth promotion tests with salmochelins showed that the IroN outer membrane receptor, encoded in the iroA locus of S. enterica and uropathogenic Escherichia coli strains, was the main receptor for ferric salmochelin transport.
In iron-poor environments, many bacteria secrete iron-complexing agents called siderophores to satisfy their iron needs. For some pathogenic bacteria, siderophores are important virulence factors because iron is bound to transferrin and lactoferrin in body fluids. These proteins reduce the free Fe3+ concentration to about one molecule per liter. Enterobacteria, including Escherichia coli and Salmonella enterica, often produce the catecholate siderophore enterochelin (also called enterobactin) (1, 2). It has been postulated that enterochelin is an inferior siderophore in serum because it adsorbs to hydrophobic sites in serum proteins, such as albumin (3). This fact has always been puzzling because iron supply for many pathogens plays a decisive role in the infection process, and enterochelin, a major siderophore synthesized by S. enterica, does not seem to enhance pathogenicity.
Recently, the iroA locus, consisting of the two convergent operons iroN and iroBCDE, has been defined in S. enterica serotype Typhi and also in most other S. enterica serotypes (4, 5). The outer membrane siderophore receptor, IroN, is involved in the transport of several catecholate siderophores in S. enterica (5, 6). In the present investigation, we show that the presence of the iro gene cluster in S. enterica leads to glycosylation of the enterochelin building block 2,3-dihydroxybenzoylserine (DHBS), which makes the hydrophobic enterochelin molecules more hydrophilic, thereby possibly contributing to the observed pathogenicity of Salmonella strains. This siderophore was named salmochelin because it appears to be a characteristic siderophore of Salmonella strains. Interestingly, certain E. coli strains, e.g., the uropathogenic E. coli 563, also possess a very similar iro gene cluster on pathogenicity island III (7). The production and the tentative structural elucidation of salmochelins and their specific uptake via the outer membrane receptor IroN are described. In addition, it is shown that salmochelin is not secreted in an iroBC mutant.
Materials and Methods
Strains and Growth Conditions.
The strains used are described in Table 1. Strains were routinely grown on TY medium containing 8 g/liter tryptone, 5 g/liter yeast extract, and 5 g/liter sodium chloride. Growth response to siderophores was tested on nutrient broth plates (8 g of nutrient broth, 5 g of NaCl, and 15 g of Difco agar per liter) containing 0.2 mM 2,2′-dipyridyl; the plates were seeded with the appropriate strain in 2.5 ml of soft agar in water. Filter-paper discs loaded with 15 μl of siderophore solution were applied, and growth zones were measured after 18 h. Strain AIR50 has been constructed by A. Bäumler and his group (Texas A&M University, College Station) by inserting a kanamycin resistance cassette into the unique BglII site of iroD and introducing this mutation by gene replacement in a similar way as described for AIR49 (4, 5). S. enterica serotype Stanleyville strains H5546 and H5547 were constructed by P22 transduction of the aroA+ gene from S. enterica serotype Typhimurium strain LT2. The WT strain S. enterica serotype Stanleyville strain 207/81, isolated from an infected cow, produces neither enterochelin nor aerobactin (8). Strain S. enterica serotype Paratyphi B IHS 1319 was kindly provided by B. Stocker (Stanford University, Stanford, CA).
Table 1.
Strains used in this study
| Strain | Relevant genotype | Ref. |
|---|---|---|
| E. coli | ||
| CA 46 | Produces salmochelin and microcins M and H47 (formerly called colicin G) | 11 |
| K-12 H1443 | aroB | 18 |
| K-12 H1876 | aroB cir fiu∷Mud1X fepA∷Tn10 | 18 |
| S. enterica | |
| Typhimurium LT2 | Lab collection |
| Paratyphi B IHS 1319 | B. Stocker |
| Stanleyville207/81 | 8 |
| WR1359 as 207/81 but iroN∷pGP704 | This work |
| WR1361 as 207/81 but cir∷MudJ | This work |
| WR1363 as 207/81 but iroN∷pGP704 cir∷MudJ | This work |
| WR1366 as 207/81 but iroN∷pGP704 fepA∷Tn10 | This work |
| WR1367 as 207/81 but cir∷MudJ fepA∷Tn10 | This work |
| WR1368 as 207/81 but iroN∷pGP704 cir∷MudJ fepA∷Tn10 | This work |
| ATCC14028, serotype Typhimurium | 4, 5 |
| AIR49 as 14028 but aroA∷Tn10 iroBC∷kan | 4, 5 |
| AIR50 as 14028 but aroA∷Tn10 iroD∷kan | A. Bäumler |
| H5546 as AIR49 but aroA+iroBC∷kan | This work |
| H5547 as AIR50 but aroA+iroD∷kan | This work |
Isolation of Salmochelins.
Cells grown to stationary phase in TY medium were used to inoculate [1% (vol/vol) inoculum] M63-glycerol minimal medium (9). After 16–18 h, the cells were sedimented, and 5 mM FeSO4 was added to the supernatant. After 10 min, the precipitate formed was removed by sedimentation, and the clear supernatant was passed slowly through a column containing 100 ml of DE52 DEAE-cellulose; all negatively charged siderophores bound to the DEAE-cellulose. After washing with 2 column vol of water, the iron siderophores were eluted either batch-wise with 2 M ammonium chloride or a water/2 M ammonium chloride gradient. The blue-red fractions containing the catecholate siderophores were pooled, concentrated by evaporation, and purified on a Biogel P2 column by using water as an eluent. The deep-red fractions obtained were subjected to further purification by HPLC. When some batches of culture medium were extracted with ethyl acetate according to published procedures (2), it was observed that salmochelins remained in the water phase (see Results).
HPLC.
Siderophore fractions obtained from DEAE-cellulose and Biogel P2 columns were analyzed by HPLC as described (10). Highly purified salmochelin samples were obtained by preparative HPLC (Shimadzu LC20 pumps) on a reversed-phase Nucleosil C18 column (5 μm, 4 × 250 mm, Grom Herrenberg, Germany) by using a gradient of acetonitrile/water (6–40%) containing 0.1% trifluoroacetic acid over 20 min (flow rate 1 ml/min), followed by 60% acetonitrile for 5 min; detection was at 220 nm. The collected fractions were checked for purity by analytical HPLC, lyophilized, and analyzed by GC–MS, electrospray ionization–MS, and Fourier transform ion cyclotron resonance (FTICR)–MS.
MS.
Electrospray ionization–FTICR–MS was carried out with a passively shielded 4.7 Tesla APEX II-electrospray ionization/matrix-assisted laser desorption ionization–FTICR mass spectrometer (Bruker Daltonik, Bremen, Germany); the data obtained were processed with the MS software xmass version 5.0.10 (Bruker Daltonik) running on a Silicon Graphics O2 Workstation. Substances were dissolved in methanol containing 0.1% triethylamine, and negatively charged ions formed by electrospray were measured over the mass range of m/z 150–2,000. The samples were internally calibrated by adding Hewlett–Packard MS calibration standard to the sample solutions.
For fragmentation studies, high-resolution FTICR–MS/MS was acquired by sustained off-resonance irradiation (SORI)–collision-induced dissociation (CID) by using argon as the collision gas.
GC–MS.
For amino acid analysis, the sample (≈50 μg) was hydrolyzed in 6 M HCl (110°C/24 h), the hydrolysate was dried at 110°C under a stream of nitrogen, and the liberated amino acids were derivatized to their N-(O-)trifluoroacetic acid/ethyl esters and analyzed by GC–MS (Agilent 5973/6890 GC-MS, Waldbronn, Germany) on a homemade chiral capillary column (20 m × 0.25 mm; 30% 2,6-dipentyl-3-butyryl-γ-cyclodextrin/70% PS 255). The above-mentioned hydrolysate was extracted with ethyl acetate, the extract was dried over Na2SO4, and the solvent was evaporated under a gentle stream of nitrogen. The products were trimethylsilylated (60 μl of 1:1 N,O-bis(trimethylsilyl)trifluoroacetamide/acetonitrile; 60°C/60 min) and analyzed by GC–MS as described above on a 15 m × 0.25 mm BD5-MS capillary column.
For methanolysis, samples (≈20 μg) were heated to 110°C for 1 h in 1.5 M HCl/methanol. After removing excess reagent in a stream of nitrogen at room temperature, the products were trimethylsilylated and analyzed by GC–MS (DB5-MS capillary and derivatization as described above).
Results
Sequence Analysis of the Predicted Iro Proteins.
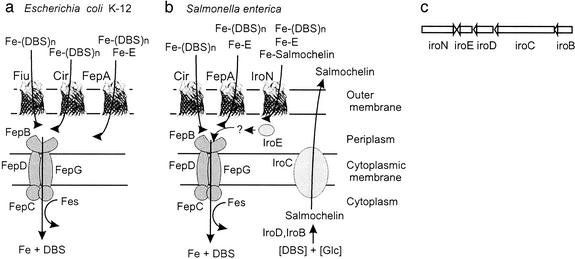
The iroA locus of S. enterica and E. coli contains five genes (Fig. 1). Only the function of the gene product of iroN from E. coli (82% identical to IroN of S. enterica), an outer membrane siderophore receptor with the ability to transport different catechol siderophores, including N-(2,3-dihydroxybenzoyl)-l-serine and enterochelin, has been experimentally defined (5, 6). The second gene of S. enterica related to siderophore function is iroD, whose gene product has low similarity to enterochelin esterase encoded by the fes gene of E. coli (Table 2). The gene products of all other genes in this Fur- and iron-regulated operon (4) are not related to any known protein involved in siderophore biosynthesis or uptake. The IroB protein has similarity to glycosyltransferases. SnoG from Streptomyces nogalater, involved in the glycosylation of an anthracycline antibiotic, is the closest functionally defined and related protein. IroC has similarity to eukaryotic multidrug exporters; in bacteria, no functionally characterized homolog was found. IroE has a signal sequence characteristic for exported proteins and has similarity to hydrolases. Based on these characteristics, we propose that the iroA gene locus is responsible for the glycosylation of the catecholate siderophore enterochelin, which is secreted, loaded with Fe3+, and subsequently transported by IroN into the periplasm. There, the ferric siderophore might be hydrolyzed by IroE to allow Fe-catecholate uptake via the FepBCDG transporter across the cytoplasmic membrane (Fig. 1). However, IroE could also be involved in further modification of salmochelin during export. Fig. 1a provides an overview of our current working model on the functions of the different Iro proteins in S. enterica.
Figure 1.
Tentative scheme of the catecholate siderophore transport systems of E. coli K-12 (a) and S. enterica (b). In both systems, ferric enterochelin (Fe-E) is transported mainly through FepA across the outer membrane, whereas the linear degradation products, ferric DHBS (DHBS)n (n = 1, 2, 3), are taken up mainly via Fiu and FepA in E. coli. In Salmonella, this has not been studied in detail. However, because of the high similarity of the two systems, the same specificities can be expected. Iron complexes are then transported via the ABC transporter consisting of the binding protein FepB, the membrane components FepD and FepG, and the ATPase FepC through the cytoplasmic membrane. Inside the cell, the Fes protein is required for iron release from the ferric enterochelin complex. In Salmonella, salmochelins are synthesized from glucose and DHBS, possibly with the help of IroD and IroB and secreted by the exporter IroC. IroE might degrade the iron–salmochelin complexes to allow their transport via the ABC transporter FepBDGC. (c) The same gene order is found in the iroA gene locus of S. enterica and uropathogenic E. coli strains (5, 7).
Table 2.
Comparison of Salmonella Iro proteins with E. coli Iro proteins and proteins involved in microcin biosynthesis
| iro gene from Salmonella | Homologs | Size of gene product, aa | Putative or demonstrated* function of the proteins | % Amino acid identity |
|---|---|---|---|---|
| iroB | 371 | Glycosyltransferase | ||
| iroB E. coli 536 | 371 | Glycosyltransferase | 72 | |
| mceC Klebsiella pneumoniae | 370 | Maturation of microcin E492 | 77 | |
| mcmL E. coli | 372 | Maturation of microcins M and H | 72 | |
| snoG Streptomyces nogalater | 390 | Glycosyltransferase* | 33 | |
| iroC | 1,261 | ABC exporter | ||
| iroC E. coli 536 | 1,218 | ABC exporter | 76 | |
| SCO2763 Streptomyces coelicolor | 1,243 | ABC transporter | 39 | |
| MDR1 Homo sapiens | 1,280 | ABC exporter, multidrug resistance protein 1* | 28 | |
| iroD | 409 | Similar to enterochelin esterase | ||
| iroD E. coli 536 | 414 | Similar to enterochelin esterase | 68 | |
| mceD K. pneumoniae | 414 | Maturation of microcin E492 | 56 | |
| mcmK E. coli | 424 | Maturation of microcins M and H | 55 | |
| fes E. coli | 374 | Enterochelin esterase* | 27 | |
| iroE | 318 | Periplasmic hydrolase | ||
| iroE E. coli 536 | 311 | Periplasmic hydrolase | 61 | |
| AMY3E Oryza sativa | 437 | Alpha-amylase isozyme 3E* | 26 | |
| iroN | 725 | Catecholate siderophore receptor* | ||
| iroN E. coli 536 | 726 | Catecholate siderophore receptor* | 82 | |
| fepA E. coli | 746 | Enterochelin receptor* | 52 | |
| cir E. coli | 663 | DHBS-catecholate siderophore receptor* | 33 | |
| fiu E. coli | 760 | DHBS-catecholate siderophore receptor* | 22 |
The similarity to some better-defined homologs and the percent identity are given.
Proteins for which experimental evidence for the function is available.
The iroA locus is not only found in S. enterica, but also in some E. coli strains (7). Homologs of IroB and IroD are also encoded in gene clusters involved in the synthesis of certain microcins, namely microcin E492 from Klebsiella and microcins M and H47 from E. coli (11).
Isolation of Salmochelins from E. coli and S. enterica.
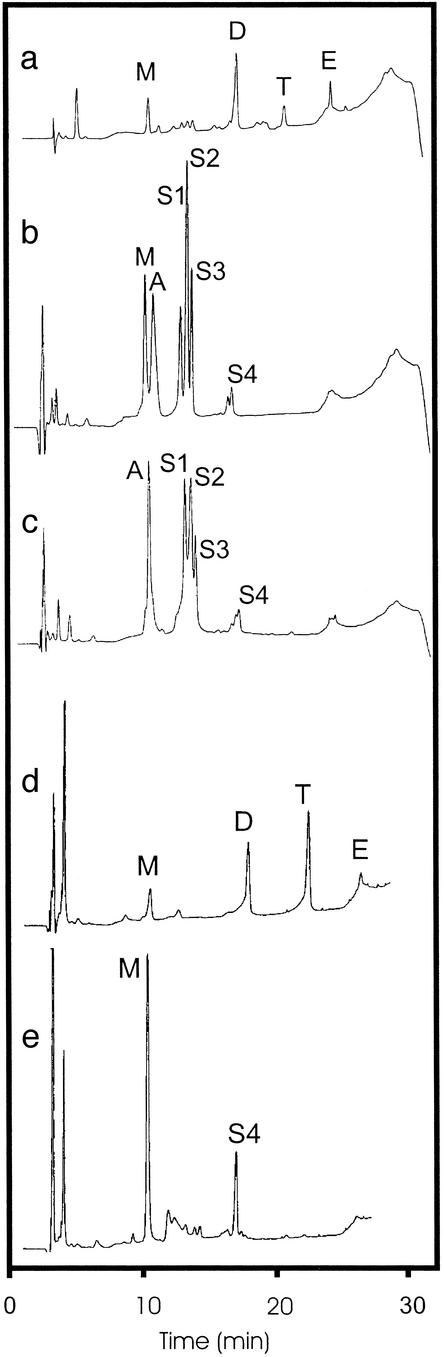
During the study of microcins M and H47 (formerly called colicin G) produced by E. coli CA46, it was observed that large amounts of a siderophore with the typical wine-red color of a catecholate siderophore were produced after addition of iron. E. coli CA46 was grown in M63 minimal medium overnight. Ferric chloride (5 mM) was added to the culture supernatant, which became purple-blue. Following the published protocol for the isolation of enterochelin (2), the siderophore complexes were bound to a DE52 DEAE-cellulose column. After elution of the deep-blue siderophores with 2 M NH4Cl, the eluate was adjusted to pH 2 and extracted with ethyl acetate. The ethyl acetate extract was analyzed by HPLC (12), which allowed the detection of enterochelin and the linear degradation products: monomer of DHBS, dimer (DHBS)2, and trimer (DHBS)3 (Fig. 2a).
Figure 2.
The pooled iron siderophore fraction of E. coli CA46 (a–c) eluted from DE52-cellulose with 2 M ammonium chloride was adjusted to pH 2 with H2SO4, extracted with ethyl acetate, and analyzed by HPLC. (a) The ethyl acetate extract contained the linear degradation products monomer DHBS (M), the dimer (DHBS)2 (D), and the trimer (DHBS)3 (T), and traces of the cyclic enterochelin (E) were identified. In the water phase of the ethyl acetate extract (b), the monomer, aerobactin (A), and salmochelins 1–3 (S1–S3) were found. (c) The ammonium chloride eluate of the DE52-cellulose column contained mainly salmochelins 1–3 and aerobactin. (d) The mutant Salmonella H5546 iroBC∷kan produced only enterochelin and the linear degradation products. (e) The mutant Salmonella H5547 iroD∷kan secreted the monomer and S4, which represents an unidentified compound that awaits further characterization.
HPLC analysis of the water phase of the ethyl acetate extract (Fig. 2b) revealed three additional peaks that contained catechols and did not coincide with known siderophores. The same three peaks were found in the ethyl acetate extract, but only as minor components (Fig. 2a). The finding of catecholates in the water phase suggested that the siderophores are more hydrophilic because of glycosylation. Size fractionation on a P2 column and preparative HPLC yielded three iron-free, colorless compounds, with retention times of 12.9, 13.4, and 13.8 min; these compounds were named salmochelins 1, 2, and 3. The compound S4, obtained only in small amounts, was also observed in a mutant (see below) and remains to be characterized. The same compounds were also identified in extracts from different S. enterica strains by HPLC. Because S. enterica serotype Paratyphi B IHS1319 seemed to be the best producer of salmochelins, this strain was used for further purification experiments.
Salmochelin Production Depends on iro Genes.
To demonstrate that the iro genes take part in the synthesis of salmochelin, S. enterica serotype Typhimurium ATCC14028 and the derived mutants H5546 iroBC∷kan and H5547 iroD∷kan (5) were tested on M63 minimal medium for the production of siderophores. After addition of iron, a deep-purple-red color developed in the spent medium of S. enterica serotype Typhimurium ATCC14028 and mutant H5546 iroBC∷kan, whereas in the medium of mutant H5547 iroD∷kan, a less-intense color developed. HPLC analysis revealed production of salmochelins 1, 2, and 3, and DHBS by S. enterica serotype Typhimurium ATCC14028, similar to that observed in S. enterica serotype Paratyphi B and E. coli CA46. The iroBC mutant H5546 produced the degradation products of enterochelin (DHBS, DHBS2, and DHBS3) and traces of enterochelin, but no salmochelins (Fig. 2d), which shows that iro genes are responsible for salmochelin synthesis. The iroD mutant H5547 produced DHBS and an unidentified peak (S4) (Fig. 2e).
Chemical Characterization of Salmochelin(s).
FTICR–MS of salmochelin 1 revealed an exact m/z for the [M-H]− ion of 625.15343, consistent with an elemental composition of C26H29N2O16, with a relative error between theoretical and experimental mass (Δ) of 1.9 ppm. The double-negatively charged ion [M-2H]2− had an m/z of 312.07281, consistent with an elemental composition of C26H28N2O16 (Δ = 0.6 ppm). Thus, the elemental composition of the neutral molecular species (M = 625) of salmochelin 1 is proposed to be C26H30N2O16.
FTICR–MS of salmochelin 2 revealed an m/z of 1010.2536 for [M-H]−, corresponding to an elemental composition of C42H48N3O26 (Δ = 0.46 ppm). The elemental composition of the neutral molecular species (M = 1011) of salmochelin 2 is proposed to be C42H49N3O26. The difference between salmochelin 1 and 2 is thus C16H20NO10, corresponding to one hexose-DHBS unit.
Amino acid analysis revealed l-serine as the sole amino acid present. 2,3-Dihydroxybenzoic acid was found in the ethyl acetate extract of the hydrolysate. Upon methanolysis and subsequent trimethylsilation, DHBS [OMe/(TMS)3] and glucose [methylglucoside/(TMS)4] were identified by GC–MS, together with 2,3-dihydroxybenzoic acid [OMe/(TMS)3 and (TMS)4].
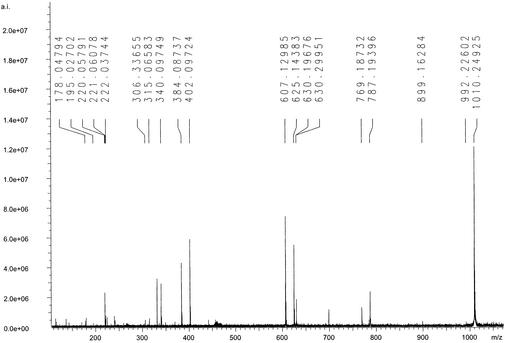
The SORI–CID–MS/MS spectrum of the singly charged ion salmochelin 2 (m/z 1010.25) is shown in Fig. 3, and the spectrum of the singly charged ion of salmochelin 1 (m/z 625.15) is shown in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org. Losses of DHBS (223.05 Da) and a hexose (162.05 and 180.06 Da, depending on loss of H2O) are apparent in both spectra. The similarity of the SORI–CID–MS/MS of the two salmochelin species in a mass range <625 suggests that salmochelin 1 is a structural element of salmochelin 2 (Fig. 4). The sequential loss of DHBS, DHBS-glucose, and DHBS-glucose-DHBS indicated that salmochelins contain alternating DHBS and glucose units.
Figure 3.
SORI–CID–MS/MS spectrum of salmochelin 2 and assignment of the resulting fragment ions: m/z 1010.25, [M–H]−; m/z 787.19, [M–H–DHB–Ser]−; m/z 625.14, [M–H–DHB–Ser–glucose]−; m/z 402.10, [M–H–DHB–Ser–glucose–DHB–Ser]−; and m/z 240.04, [M–H–DHB–Ser–glucose–DHB–Ser–glucose]−. The SORI–CID–MS/MS spectrum of salmochelin 1 (see Fig. 6) has the assigned fragment ions m/z 625.12, [M–H]−; m/z 402.09, [M–H–DHB–Ser]−; m/z 315.06, [M–H–DHB–Ser–(Ser–H2O)]− (rearrangement); and m/z 240.05, [M–H–DBH–Ser–glucose]−.
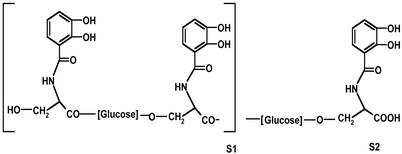
Figure 4.
Tentative structures of salmochelin 1 (S1) and salmochelin 2 (S2).
It was difficult to separate salmochelin 3 in pure form from salmochelin 2. Salmochelin 3 seemed to be an oxidized form of salmochelin 2 (loss of two mass units). Because there was no biological activity detected in growth promotion assays (see below), salmochelin 3 was not chemically characterized further.
Growth Promotion Tests.
To test the siderophore activity of the purified salmochelins, growth promotion assays with different E. coli K-12 test strains were performed. Salmochelins 1 and 2 did not support growth of those E. coli K-12 strains, which do not contain the iro genes. In contrast, S. enterica serotype Stanleyville and the mutants WR1361 cir and WR1367 cir fepA showed good growth; strains WR1359 iroN and WR1366 iroN fepA grew poorly; and strains WR1363 iroN cir and WR1368 iroN cir fepA did not grow (Table 3). These results indicated that IroN is the main outer membrane transport protein for the salmochelin siderophores and that only low amounts are taken up via Cir.
Table 3.
Growth promotion of S. enterica serotype Stanleyville strains by salmochelin
| S. enterica serotype Stanleyville strain | Relevant mutation | Growth zone diameter, mm |
|---|---|---|
| 21 | ||
| WR1361 | cir | 21 |
| WR1359 | iroN | 18 |
| WR1366 | iroN fepA | (17) |
| WR1367 | cir fepA | 20 |
| WR1363 | iroN cir | 0 |
| WR1368 | iroN cir fepA | 0 |
Nutrient broth plates containing dipyridyl to limit available iron were seeded with the indicated strains suspended in 2.5 ml of soft agar in water. Filter-paper discs impregnated with 15 μl of deferri-salmochelin solution (≈1 mg/ml) were applied, and growth zones were measured after 18 h. Numbers in bold indicate good growth; normal numbers and numbers in parentheses indicate poor and very poor growth, respectively.
Salmochelin 3 was also tested, but no growth stimulation of any of these strains was observed, rather a zone of growth inhibition was observed in plates showing background growth of S. enterica serotype Stanleyville.
Uptake Experiments.
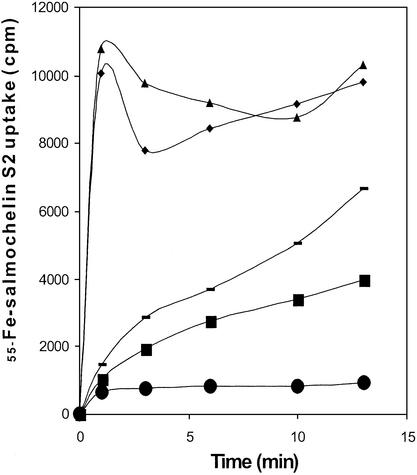
Salmochelin-2-mediated 55Fe uptake was measured in strains S. enterica serotype Stanleyville, WR1361 cir, WR1359 iroN, WR1363 iroN cir, and WR1368 iroN cir fepA. Rapid uptake was observed with the iroN+ strains S. enterica serotype Stanleyville and WR1361 cir (Fig. 5), whereas a gradually reduced uptake was obtained with strains WR1359 iroN and WR1363 iroN cir, no uptake was observed with the triple mutant WR1368 iroN cir fepA, which is unable to make any known iron catecholate outer membrane transport protein. Similar results were obtained with salmochelin 1. From these results, we concluded that IroN is the receptor for salmochelins 1 and 2. Cir and FepA might contribute to salmochelin-mediated iron uptake. Because it is not known how stable the salmochelins are in the transport assay, it cannot be excluded that the residual uptake via Cir and FepA is mediated by small amounts of salmochelin degradation products. It is interesting to note that E. coli K-12 H1443, although not able to use salmochelin 1, bound 10 times more 55Fe-salmochelin1 than mutant H1876 cir fiu fepA, which is devoid of any catecholate receptor.
Figure 5.
Uptake of ferric salmochelin 2 labeled with 55Fe, measured in M9 minimal medium (22) by using the strains S. enterica serotype Stanleyville (⧫), WR1359 iroN (−), WR1361 cir (▴), WR1363 cir iroN (■), and WR1368 cir fepA iroN (●).
Discussion
Ferric siderophores are specifically recognized by membrane receptors present in the outer membranes of Gram-negative bacteria. Structures and functions of receptors for ferrichromes (FhuA) and enterochelin (FepA) from E. coli have been analyzed and described in detail (reviewed in refs. 13 and 14). Although enterochelin is the major catecholate siderophore in enteric bacteria, the present investigation shows that the iro genes of S. enterica synthesize additional catecholate siderophores together with their cognate receptors.
The function of the iro operon has been puzzling because its identification in 1996 by Bäumler and coworkers (4) in nearly every S. enterica strain tested (5, 15). The specificity of IroN for catecholate-containing siderophores gave the first hint that the other Iro proteins might be responsible for the biosynthesis of a catecholate-type siderophore. Because Salmonella is known to possess the genes for enterochelin biosynthesis, it was tempting to speculate that the Iro proteins also synthesize a modified enterochelin. The sequence similarity of IroB to glycosyltransferases pointed to a sugar-modified enterochelin. By HPLC analysis of low-iron culture supernatants of S. enterica, the predicted salmochelin siderophores were identified. Strain H5546 iroBC produced only DHBS oligomers and traces of enterochelin. Interestingly, strain H5547 iroD seemed to produce mainly DHBS and compound S4, which remains to be characterized.
In 1970, enterochelin was detected in E. coli by O'Brien and Gibson (16), and the same substance, called enterobactin, was found in S. enterica serotype Typhimurium by Pollack and Neilands (17). Our HPLC chromatograms showed that enterochelin represents only a minor fraction of the catecholates synthesized by S. enterica serotype Typhimurium, whereas the salmochelins are a major fraction. The amounts of enterochelin are higher in the supernatants of the E. coli strain, which did not produce salmochelins (16).
As shown in the HPLC chromatograms with C18 reversed-phase columns, the salmochelins eluted between the DHBS monomer and DHBS dimer and consisted of three peaks (S1, S2, and S3) with different ratios depending on the growth conditions, strain, and length of incubation. Whereas S1 was clearly separated, S2 and S3 could not be well separated. Salmochelin 1 and 2 showed high siderophore activity, whereas salmochelin 3 was inactive. The structure elucidation of salmochelins 1 and 2 by MS and GC–MS indicated linear structures consisting of two or three DHBS moieties, respectively, with each DHBS pair bridged by a glucose residue (Fig. 4).
In growth promotion assays, the IroN protein was shown to be the receptor for salmochelin, as was expected from the clustering of the transport and biosynthesis genes in S. enterica and uropathogenic E. coli strains. This was confirmed by uptake experiments with radiolabeled 55Fe-salmochelin. However, low uptake was also observed with the catecholate-specific Salmonella receptors Cir and FepA in feeding assays and uptake experiments. The iron complexes of DHBS, (DHBS)2, and the linear form of enterochelin are transported by Cir (18). In addition, different catechol-cephalosporin derivatives synthesized as antimicrobial agents are taken up via Cir (19). Therefore, it would not be surprising if Cir also transports ferric salmochelin, but, as already pointed out, it is difficult to decide whether this is the case and whether Cir also transports ferric salmochelin fragments to some extent. In E. coli K-12, no growth promotion was observed with salmochelin, which might indicate that the periplasmic hydrolase IroE or the Fes-like IroD protein is necessary for the iron utilization process.
The sequence of IroB clearly suggests a glucosyltransferase function in biosynthesis, and the substrates could be an UDP-activated glucose and DHBS. Although the salmochelins secreted by E. coli CA46 had the same HPLC retention times as those secreted by S. enterica, the sugar and its linkage to DHBS remain to be determined because E. coli IroB and IroD share only 72% and 68% identity, respectively, with their counterparts from S. enterica. A candidate for the esterification of DHBS to glucosyl-DHBS could be IroD, which has low similarity to Fes, the enterochelin esterase. Oligomeric (DHBS)n could be substrates for the transesterification reaction. However, it is also possible that the enterochelin synthase cluster transfers activated DHBS to a hydroxyl group of glucose.
The structure of salmochelin also has implications for the synthesis of microcins E492 (20), H43 (21), and M (11), for which homologs of IroB and IroD have been found (Table 2). These three microcins are peptides of 84, 77, and 60 aa, respectively, and possess a glycine/serine-rich C terminus with a C-terminal serine residue. It is suggested that homologs of IroB and IroD crosslink microcins at their C-terminal serine residue by using a sugar residue in the same way as DHBS is linked by sugars in salmochelins. It is interesting to note that these microcins also use the salmochelin receptor IroN (11).
Supplementary Material
Acknowledgments
We are grateful to Andreas Bäumler and Renee Tsolis for constructing strain AIR50. We thank Sandra Frick for technical assistance in HPLC separations, Kerstin Haible for technical assistance in isolating salmochelins, and Volkmar Braun and Karen A. Brune for critical reading of the manuscript. The work was supported by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and Ardeypharm.
Abbreviations
- FTICR
Fourier transform ion cyclotron resonance
- SORI
sustained off-resonance irradiation
- CID
collision-induced dissociation
- DHBS
2,3-dihydroxybenzoylserine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Earhart C F. In: Escherichia coli and Salmonella. Neidhart F C, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1075–1090. [Google Scholar]
- 2.Young I G, Gibson F. Methods Enzymol. 1979;56:394–398. doi: 10.1016/0076-6879(79)56037-6. [DOI] [PubMed] [Google Scholar]
- 3.Konopka K, Neilands J B. Biochemistry. 1984;23:2122–2127. doi: 10.1021/bi00305a003. [DOI] [PubMed] [Google Scholar]
- 4.Bäumler A J, Tsolis R M, van der Velden A W, Stojiljkovic I, Anic S, Heffron F. Gene. 1996;183:207–213. doi: 10.1016/s0378-1119(96)00560-4. [DOI] [PubMed] [Google Scholar]
- 5.Bäumler A J, Norris T L, Lasco T, Voigt W, Reissbrodt R, Rabsch W, Heffron F. J Bacteriol. 1998;180:1446–1453. doi: 10.1128/jb.180.6.1446-1453.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rabsch W, Voigt W, Reissbrodt R, Tsolis R M, Bäumler A J. J Bacteriol. 1999;181:3610–3612. doi: 10.1128/jb.181.11.3610-3612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dobrindt U, Blum-Oehler G, Hartsch T, Gottschalk G, Ron E Z, Fünfstück R, Hacker J. Infect Immun. 2001;69:4248–4256. doi: 10.1128/IAI.69.7.4248-4256.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reissbrodt R, Rabsch W. Zentralbl Bakteriol Mikrobiol Hyg [A] 1988;268:306–317. doi: 10.1016/s0176-6724(88)80015-4. [DOI] [PubMed] [Google Scholar]
- 9.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 10.Winkelmann G, Schmid D G, Nicholson G, Jung G, Colquhoun D J. Biometals. 2002;15:153–160. doi: 10.1023/a:1015206419613. [DOI] [PubMed] [Google Scholar]
- 11.Braun V, Patzer S I, Hantke K. Biochimie. 2002;84:365–380. doi: 10.1016/s0300-9084(02)01427-x. [DOI] [PubMed] [Google Scholar]
- 12.Winkelmann G, Cansier A, Beck W, Jung G. Biometals. 1994;7:149–154. doi: 10.1007/BF00140485. [DOI] [PubMed] [Google Scholar]
- 13.Van der Helm D, Chakraborty R. In: Microbial Transport Systems. Winkelman G, editor. Weinheim, Germany: Wiley-VCH; 2001. pp. 261–287. [Google Scholar]
- 14.Braun V, Hantke K. In: Microbial Transport Systems. Winkelman G, editor. Weinheim, Germany: Wiley-VCH; 2001. pp. 289–311. [Google Scholar]
- 15.Bäumler A J, Heffron F, Reissbrodt R. J Clin Microbiol. 1997;35:1224–1230. doi: 10.1128/jcm.35.5.1224-1230.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Brien I G, Gibson F. Biochim Biophys Acta. 1970;215:393–402. doi: 10.1016/0304-4165(70)90038-3. [DOI] [PubMed] [Google Scholar]
- 17.Pollack J R, Neilands J B. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 18.Hantke K. FEMS Microbiol Lett. 1990;67:5–8. doi: 10.1016/0378-1097(90)90158-m. [DOI] [PubMed] [Google Scholar]
- 19.Curtis N A, Eisenstadt R L, East S J, Cornford R J, Walker L A, White A J. Antimicrob Agents Chemother. 1988;32:1879–1886. doi: 10.1128/aac.32.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagos R, Baeza M, Corsini G, Hetz C, Strahsburger E, Castillo J A, Vergara C, Monasterio O. Mol Microbiol. 2001;42:229–243. doi: 10.1046/j.1365-2958.2001.02630.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez E, Gaggero C, Lavina M. Antimicrob Agents Chemother. 1999;43:2176–2182. doi: 10.1128/aac.43.9.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hantke K. Mol Gen Genet. 1983;191:301–306. doi: 10.1007/BF00334830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.