Abstract
A rhodium(III) complex, rac-[Rh(bpy)2phzi]3+ (bpy, 2,2′-bipyridine; phzi, benzo[a]phenazine-5,6-quinone diimine) has been designed as a sterically demanding intercalator targeted to destabilized mismatched sites in double-helical DNA. The complex is readily synthesized by condensation of the phenazine quinone with the corresponding diammine complex. Upon photoactivation, the complex promotes direct strand scission at single-base mismatch sites within the DNA duplex. As with the parent mismatch-specific reagent, [Rh(bpy)2(chrysi)]3+ [chrysene-5,6-quinone diimine (chrysi)], mismatch selectivity depends on the helix destabilization associated with mispairing. Unlike the parent chrysi complex, the phzi analogue binds and cleaves with high affinity and efficiency. The specific binding constants for CA, CC, and CT mismatches within a 31-mer oligonucleotide duplex are 0.3, 1, and 6 × 107 M−1, respectively; site-specific photocleavage is evident at nanomolar concentrations. Moreover, the specificity, defined as the ratio in binding affinities for mispaired vs. well paired sites, is maintained. The increase in affinity is attributed to greater stability in the mismatched site associated with stacking by the heterocyclic aromatic ligand. The high-affinity complex is also applied in the differential cleavage of DNA obtained from cell lines deficient in mismatch repair vs. those proficient in mismatch repair. Agreement is found between photocleavage by the mismatch-specific probes and deficiency in mismatch repair. This mismatch-specific targeting, therefore, offers a potential strategy for new chemotherapeutic design.
The targeting of single-base mismatches in DNA represents a challenging problem in molecular recognition. The mismatched site may be of variable sequence and within a variable sequence context. What distinguishes such a site, instead, is the local destabilization in opposing bases because of the absence of Watson–Crick hydrogen bonding (1). Although challenging, however, the development of small molecules targeted to mismatches offers many applications. Site-specific mismatch probes could be used in discovery efforts to identify single-nucleotide polymorphisms. Moreover, such molecules could provide the basis for the development of novel chemotherapeutics. Many cancers are associated with a deficiency in mismatch repair (2, 3). Hence, by directing small molecules to the accumulated mismatches, a cancer-specific targeting strategy could be envisioned.
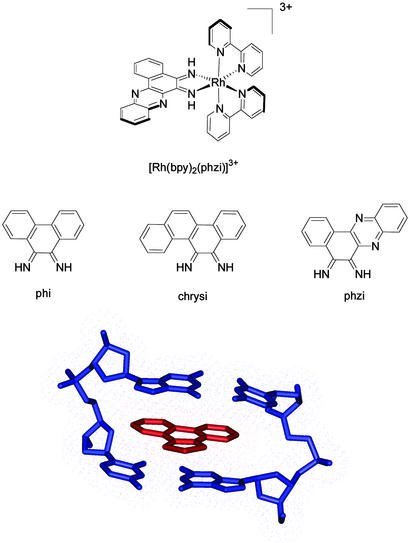
In our laboratory, we have exploited the local helix destabilization associated with mispairing in the design of a metal complex targeted to mismatches (4, 5). Shown in Fig. 1 is a rhodium complex containing the benzo[a]phenazine-5,6-quinone diimine (phzi) ligand, as well as chrysene-5,6-quinone diimine (chrysi) and phenanthrene quinone diimine (phi) ligands (4). Octahedral rhodium(III) complexes containing the phi ligand bind avidly to double-helical DNA by intercalation (6); on photoactivation, direct DNA strand cleavage is also promoted at the bound site (7). By tuning the ancillary ligands, complexes can be prepared that are either site-specific or sequence-neutral. [Rh(bpy)phi2]3+ (bpy, 2,2′-bipyridine), for example, binds to B-DNA with little site selectivity (7), whereas [Rh(S,S-dimethyltrien)(phi)]3+ specifically targets the sequence 5′-TGCA-3′ (8). The high-resolution crystal structure of [Rh(S,S-dimethyltrien)(phi)]3+ bound to a DNA octamer shows site-specific intercalation by the complex from the major groove side of the helix, with site-discrimination determined through an ensemble of hydrogen bonding contacts and methyl–methyl interactions (9). The intercalated phi ligand itself resembles another base pair, stacked at a distance of 3.4 Å between neighboring base pairs, with an expanse across the ligand of dimensions matching that of the base pairs above and below.
Figure 1.
Bulky metallointercalators. (Top) [Rh(bpy)2(phzi)]3+. (Middle) The phi, chrysi, and phzi intercalating ligands. (Bottom) A view of intercalation of the Rh(phi) moiety (red) within a well matched DNA site (blue). The view of intercalation (based upon crystallographic coordinates; ref. 9) shows the snug fit of the phi ligand within its binding site. The bulky chrysi and phzi ligands are precluded sterically from intercalation in a similar fashion.
The chrysi complex represents our first-generation complex targeted to mismatches (4). The site-specificity of the complex is derived from the fact that it is a more bulky intercalator with an expanse exceeding that of the well matched base pair (Fig. 1). Owing to shape selection, the complex is unable to bind to well matched B-form DNA. However, at sites where a base pair is destabilized, intercalation of the chrysi complex can occur. [Rh(bpy)2chrysi]3+ has been shown to be both a general and a remarkably specific mismatch recognition agent (10, 11). Specific DNA cleavage is observed at >80% of mismatch sites in all of the possible single base-pair sequence contexts around the mispaired bases. Moreover, the complex was found to recognize and photocleave at a single base mismatch in a 2,725-bp linearized plasmid heteroduplex.
Here, we describe the synthesis and application of a second generation mismatch recognition agent, [Rh(bpy)2phzi]3+. This complex shares with the chrysi complex a sterically demanding intercalating ligand. However, the phzi ligand, as an aromatic heterocycle, offers the possibility of greater stabilization through stacking within the mismatched site. Indeed, [Rh(bpy)2phzi]3+ binds mismatched sites with a binding affinity increased by two orders of magnitude, without a loss in site-selectivity. This high affinity allows the application of the complex in the differential targeting of DNA in a mismatch-repair-deficient cell line.
Materials and Methods
Materials.
Commercially obtained chemicals were used as received. Bipyridine, triflic acid, 2,3-dichloro-1,4-naphthoquinone, and phenylenediamine were purchased from Aldrich. RhCl3⋅2H2O was obtained from Pressure Chemical (Pittsburgh). Benzo[a]phenazinequinone was synthesized as described (12).
Instrumentation.
Electronic spectra were recorded on a Beckman DU 7400 UV-visible spectrophotometer (Beckman Coulter). Mass spectra [electrospray ionization (ESI)] were measured on an LCQ mass spectrometer. High-performance liquid chromatography was performed on a Waters 996 system using a Vydac C18 column (4.0 ml/min liquid phase, linear gradient over 50 min from 100% 50 mM NH4OAc, pH 7.0, to 100% acetonitrile). NMR spectra were recorded on a Varian Mercury 300 MHz instrument. DNA synthesis was performed on an ABI 392 DNA/RNA synthesizer (Applied Biosystems) by using reagents from Glen Research. DNA was purified by using Glen Research PolyPak II cartridges (Glen Research, Sterling, VA), followed by reverse phase HPLC. Photocleavage reactions were carried out by using a 1,000-W Oriel Hg/Xe arc lamp (Oriel, Stamford, CT) with a monochromator fitted with a 300-nm cutoff filter and an IR filter.
Synthesis and Characterization: rac-[Rh(bpy)2phzi]Cl3.
rac-[Rh(bpy)2(phzi)]Cl3 was synthesized by using the o-quinone condensation method previously used in the synthesis of [Rh(bpy)2(chrysi)]Cl3 (13). [Rh(bpy)2(NH3)2]OTf3 (100 mg, 0.1 mmol) and benzo[a]phenazinequinone (35 mg, 0.125 mmol) were prepared according to the literature procedure (12, 14). These were dissolved in dry acetonitrile (50 ml). Aqueous sodium hydroxide (2 ml, 0.2 M) was added, and the solution was rapidly stirred for 30 min at ambient temperature. The reaction was quenched with the addition of 35 ml of water and 0.2 N HCl to neutralize. The acetonitrile was removed in vacuo. The remaining solution was filtered from the brown solid and loaded on a CM-C25 Sephadex cation exchange column (resin equilibrated with 0.05 M MgCl2). A yellow compound, likely the metal-free ligand, eluted with the initial volume. After rinsing with water, 0.05 M MgCl2, a green fraction, elutes. It was collected and concentrated on a Seppac-C18 column, eluted with a mixture of acetonitrile, water, and TFA (1:1:0.001), and lyophilized to dryness. The yield was 23 mg (30% yield). Microanalysis found (calculated): C 54.8 (55.4), H 3.6 (3.3), N 15.3 (14.8). (1)H NMR (d6-DMSO, 300 MHz): δ 14.88 (s), 14.70 (s), 9.03 (m, 4H), 8.90 (d, 2H, 7.2 Hz), 8.72 (d, 1H, 6.1 Hz), 8.60 (t, 2H, 8.5 Hz), 8.54 (d, 1H, 6.1 Hz), 8.47 (t, 2H, 8.5 Hz), 8.32 (d, 1H, 8.5 Hz), 8.20 (d, 1H, 8.5 Hz), 8.11 (t, 1H, 7.3 Hz), 8.03 (m, 3H), 7.94 (t, 1H, 6.1Hz), 7.84 (t, 1H, 7.3Hz), 7.75 (m, 3H), 7.69 (d, 1H, 6.1 Hz) ppm. UV/vis (H2O, pH 5): 245 nm (89,800 M−1·cm−1), 304 nm (65,800 M−1cm−1), 314 nm (67,300 M−1·cm−1), 343 nm (39,300 M−1·cm−1). ESI-MS-(cation) 671(M-2H+) observed, 671 calculated.
DNA Preparation and Photocleavage Experiments.
The oligonucleotides, 5′-GAT GTC GGT CCC ACG ATG GTG ACG GAT TAC C-3′ and 5′-GAG TTG GTA ATC CGT CAC CAT CGT GCG ACC GAC ATC ATG CG-3′, where C denotes the position of the mismatch, were synthesized on an ABI 392 DNA/RNA synthesizer (Applied Biosystems) by using standard phosphoramidite solid-phase synthesis; they were initially purified on PolyPak II cartridges and further purified by HPLC (98% 100 mM NH4OAc/2% acetonitrile to 70% 100 mM NH4OAc/30% acetonitrile over 30 min). The single strands were then 5′-labeled with [γ-32P]ATP and T4 polynucleotide kinase. The labeled strands were further purified by gel electrophoresis (20% denaturing polyacrylamide gel), eluted from the gel by soaking in triethylammonium acetate (100 mM, pH 7.0), ethanol precipitated, and annealed in the presence of unlabeled DNA. To the labeled 31/41-mer duplex (2 μM in 50 mM NaCl/10 mM Tris⋅HCl, pH 8.5) was added either [Rh(bpy)2phzi]Cl3 or [Rh(bpy)2chrysi]Cl3, and the sample was irradiated. After irradiation, all samples were lyophilized, denaturing formamide loading dye was added, and the samples were electrophoresed on a 20% polyacrylamide denaturing gel. The photocleavage results were quantitated by phosphorimagery (PhosphorImager, Molecular Dynamics).
Mismatch-Repair-Proficient and -Deficient Cell Lines and Photocleavage of Genomic DNA.
Cell lines were obtained from the National Cancer Institute cell repository (National Cancer Institute-Frederick Cancer Research and Development Center, Frederick, MD). DNA was isolated according to standard methods (15). Isolated DNA was digested with EcoRI, dephosphorylated with shrimp alkaline phosphatase (SAP), and radiolabeled with [γ-32P]ATP, as follows: 100 μl of 100 μM solution of DNA from each cell line contained 10 μl of 10× EcoRI buffer, to which 5 μl of EcoRI (NEB, Beverly, MA) was added. The reactions were allowed to incubate at 37°C for ≈8 h. The solutions then were put through Bio-Rad microfuge exchange spin columns and subsequently treated with 10 μl of 10× SAP buffer and 7 μl of SAP. The solutions were allowed to incubate at 37°C for at least 4 h before deactivation by heating at 65°C for 15 min. The solutions then were put through Bio-Rad exchange columns once again and subsequently treated with 10 μl of 10× T4 polynucleotide kinase (PNK buffer: 70 mM Tris·HCl, pH 7.6/10 mM MgCl2,/5 mM DTT), 3 μl of [γ-32P]ATP (ICN) and 5 μl of PNK (NEB). The reactions were allowed to incubate at 37°C overnight, after which time, excess ATP was removed with Bio-Rad microfuge exchange spin columns. Samples were then lyophilized and extracted into 100 μl 10 mM TE buffer (10 mM Tris·HCl/1 mM EDTA, pH 7.2).
DNA was then treated with [Rh(bpy)2(chrysi)]3+ or [Rh(bpy)2(phzi)]3+ and irradiated. For each cell line, control samples, containing either metal complex without irradiation or irradiation in the absence of metal complex, were also prepared and analyzed. Samples being photolyzed were exposed to 440 nm light for 5 min. All samples were then lyophilized and extracted into 10 μl of 8 M urea loading dye and heated at 90°C for 2 min. The samples were then treated with 10 μl of 2× alkaline gel buffer and loaded on an alkaline denaturing 1% agarose gel and electrophoresed.
The gels were visualized and quantitated by using IMAGEQUANT software. Line integration down the center of the lanes was carried out to give a quantitative distribution of fragment intensity as a function of gel mobility. The distribution curves were then normalized by using ORIGIN. The dark control data were plotted for each cell line and fit to a Gaussian curve to obtain a midpoint. The raw, normalized intensity data up to the control midpoint for each cell line were then subtracted from the raw, normalized intensity data for the control. The resulting areas were then integrated to obtain relative values of shift in size distribution. These values were then averaged over several trials.
Results
Synthesis and Spectral Characterization of [Rh(bpy)2phzi]Cl3.
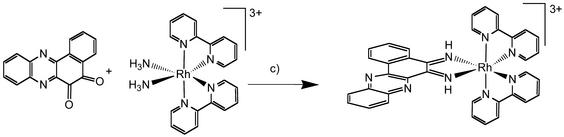
[Rh(bpy)2phzi]Cl3 was prepared as illustrated in Fig. 2. The complex may be readily synthesized by using the condensation method (13). The benzo[a]phenazine quinone is made by the reaction of 2,3-dichloro-1,4-napthoquinone with o-phenylenediamine in pyridine, followed by oxidation with nitric acid (12). Separately, [Rh(bpy)2(NH3)2](OTf)3 is synthesized (14), and then the condensation reaction is carried out in an H2O/acetonitrile solution-containing base. The reaction, as monitored by HPLC, is essentially complete after 30 min at ambient temperatures. The condensation method is significantly less cumbersome than reaction involving direct coordination of either the quinone diamine or diimine. Moreover, the reaction is also more facile than that with the chrysine quinone, given the lower extent of steric crowding around the condensation sites.
Figure 2.
Synthetic scheme for rac-[Rh(bpy)2(phzi)]3+ using the condensation strategy.
Spectral characterization for [Rh(bpy)2phzi]3+ resembles that for the parent phi and chrysi complexes. The spectrum consists of contributions from ligand to metal charge-transfer states near 310 nm and two ligand-centered transitions near 350 and 400 nm. Changes in the spectrum as a function of pH are consistent with deprotonation of the diimmine at pH 5, similar to that for the chrysi complex.
DNA Photocleavage of a Single Base Mismatch.
As is evident in Fig. 3, photoactivation of [Rh(bpy)2phzi]3+ bound to a mismatched site leads to direct strand cleavage. Upon photoactivation of [Rh(bpy)2phzi]3+ in the presence of a mismatch-containing DNA duplex, site-specific photocleavage is observed at the base 3′- to the mismatch site. Both the position of strand cleavage and the level of specificity are in accordance with that seen with the chrysi complex. Significantly, however, specific cleavage of the mismatch site by [Rh(bpy)2phzi]3+ can be seen at metal concentrations as low as 10 nM.
Figure 3.
(a) Phosphoimagery of 20% polyacrylamide gels shows mismatch-specific cleavage by the rhodium(III) complex after DNA photocleavage by 1 μM [Rh(bpy)2chrysi]Cl3 (BC) and 0.1–100 μM [Rh(bpy)2phzi]Cl3 (BZ) on 5′-32P-labeled (indicated by *) CC mismatch-containing 31/41-mer DNA duplex (1 μM). (b) All samples were prepared in 10 mM Tris, pH 8.0/50 mM NaCl. All irradiations were at 313 nm for 15 min after DNA photocleavage by varying the concentration of [Rh(bpy)2chrysi]Cl3 (BC) and [Rh(bpy)2phzi]Cl3 (BZ), as well as at different wavelengths for [Rh(bpy)2phzi]Cl3. It is notable that BC does not cleave the DNA at concentrations up to 100 nM, whereas BZ already shows a cleavage at 10 nM at the mismatch site. AG and CT, Maxam-Gilbert sequencing reactions; DC, dark control DNA sample without irradiation; LC, the light control sample irradiated in the absence of metal complex.
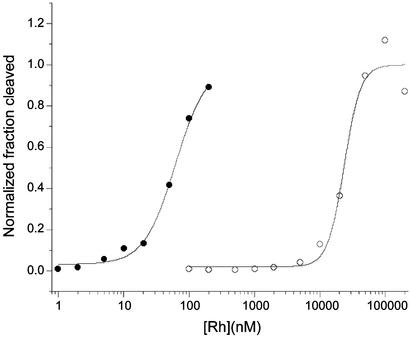
Titrations as a function of concentration, where the ratio of metal/DNA is kept constant, can be used to establish the specific binding constant to an individual site. Fig. 4 shows the quantitation for such a photocleavage titration. The specific binding constant for [Rh(bpy)2phzi]3+ at the CC mismatch in this oligonucleotide is 1 × 107 M−1. This compares to a value of 3 × 105 M−1 for [Rh(bpy)2chrysi]3+ under comparable conditions.
Figure 4.
Binding isotherms for [Rh(bpy)2phzi]3+ targeted to a CC mismatch-containing oligonucleotide (●) and the highest affinity well matched (○) site in the same duplex. Photocleavage reactions were performed at 313 nm for 15 min. The concentration of the oligomer varied from 1 × 10−10 to 4 × 10−4 M with the rhodium complex at a 10-fold lower concentration. Samples were eluted through 20% denaturing polyacrylamide gels, and the data were analyzed by using PHOSPHORIMAGER and IMAGEQUANT software (Molecular Dynamics). Cleavage is observed at the base 3′ to the CC mismatch on the shorter strand. For comparison, the specific binding affinity of the corresponding chrysi complex to this site is 3.1 × 10−5 M−1.
Not only does [Rh(bpy)2phzi]3+ bind more tightly to mismatched sites, photoactivation experiments also reveal that the complex has approximately a fivefold higher photoefficiency than the chrysi analogue in cleaving the CC mismatch. The last three lanes in Fig. 3 also show the photoefficiency for the complex at different wavelengths. Remarkably, the cleavage can be seen even at visible wavelengths of 525 nm.
Site-specific binding by [Rh(bpy)2phzi]3+ can also be compared quantitatively to nonspecific binding to well matched sites on the duplex (Fig. 4). For this oligonucleotide, the highest cleavage intensity other than that for the CC site is evident within the duplex region 5′-GTCGG-3′. The determination of the binding constant for this site yielded a value of 4 × 105 M−1, almost two orders of magnitude weaker than that for the mismatch. The basis for reaction at this site is unclear. This site represents the highest well matched site affinity, so that the average nonspecific affinity is weaker. This level of specificity is comparable to that seen with the chrysi complex (4).
[Rh(bpy)2phzi]3+ targets not only the CC mismatch but other mismatches as well. The full family of mismatches was examined through photocleavage experiments on a 31-bp oligomer by varying the base sequence at the central mismatch site. The pyrimidine mismatches in particular, including CT, TT, and TC, are also readily cleaved. By quantitative photocleavage titration, the binding affinity for the CA mismatch is found to be 3 × 106 M−1, whereas that for CT is 6 × 107 M−1. Overall, targeting seems to be mismatch-specific, not base-specific. As with the parent chrysi complex, then, site-specificity depends on the local helix destabilization associated with mispairing.
Also of interest, whereas [Rh(bpy)2phzi]3+ has the ability to target a variety of different mismatches with high affinity, it is not able to cleave all mismatches with equal photoefficiency. This variation may be determined through quantitative photocleavage titration; at maximum binding, different intensities of photocleavage are observed. The different photoefficiencies can be understood in terms of variations in bound geometries within the different mismatch site, because photocleavage is expected to depend on access of the photoexcited ligand radical to hydrogen atoms on the sugar (7). Several mismatches, such as the CC and TT, are cleaved with much better efficiency than with [Rh(bpy)2(chrysi)]3+; others, such as CA, are bound tightly but do not exhibit efficient cleavage.
Photocleavage of DNA from Cell Lines Proficient or Deficient in Mismatch Repair.
As a test of the specificity and applicability of these complexes, photocleavage was also carried out on genomic DNA isolated from human cancer cell lines with or without compromised mismatch repair. Specifically, the HCT15, SW620, SKOV3, and DU145 cell lines were used. SW620 cells are fully competent in mismatch repair. HCT15 cells have an MSH6 mutation. SKOV3 and DU145 cells, containing mutations in MLH1 and both MLH1 and PMS2, respectively, are deficient in mismatch repair (2, 3, 16–19).
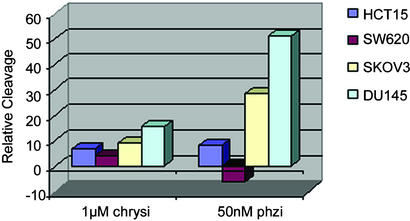
The genomic DNA was first digested into ≈5-kb lengths by restriction, radioactively end-labeled, treated with either [Rh(bpy)2chrysi]3+ or [Rh(bpy)2phzi]3+, and irradiated. Analysis involved integration of lower molecular weight bands found after denaturing agarose gel electrophoresis of the fragments. Fig. 5 shows relative levels of photocleavage obtained. Significant cleavage is evident with [Rh(bpy)2phzi]3+ at 50 nM concentration for DNA from SKOV3 and DU145, the two cell lines that lack MLH1. A low level of photocleavage is apparent with HCT15, lacking MSH6, and no significant cleavage over the controls is apparent with SW620, a cell line competent in mismatch repair. The photocleavage results correlate directly with independent analyses of these cell lines for microsatellite instabilities (16–19).
Figure 5.
Comparisons of photocleavage of genomic DNA obtained from various cancer cell lines. SW620 is a mismatch-repair competent cell line; HCT15 contains a mutation in MSH6, a redundant repair protein, whereas SKOV3 and DU145 are mismatch-repair compromised owing to mutations in essential repair proteins. Photocleavage of genomic DNA was monitored by using denaturing gel electrophoresis, as described in Materials and Methods. Photocleavage by the mismatch-specific probes correlates with previous studies of mismatch-repair activity in these cell lines.
Similar trends are evident for the chrysi complex, although higher concentrations of the metal complex must be used and only a small differential effect is apparent. Interestingly, the highest level of discrimination in cleavage among the different cell lines was found at 0.1 nM [Rh(bpy)2phzi]3+.
Discussion
Design of a High-Affinity Reagent Targeted to Mismatches.
A mismatch recognition agent has been synthesized with a heterocyclic aromatic ligand system based on the condensation technique. As with the parent rhodium intercalators, [Rh(bpy)2phzi]3+ promotes direct strand cleavage at its target site with photoactivation. Importantly, photocleavage experiments using [Rh(bpy)2phzi]3+ show that this intercalator can recognize DNA mismatches with very high specificity at concentrations in the nanomolar range.
The fact that this bulky intercalator, like the parent chrysi complex, is able to specifically target mismatches suggests the generality of this targeting strategy. By increasing the expanse of the intercalating ligand beyond that of a base pair, binding to well matched DNA sites is disfavored owing to shape-selection. Given the inherent destabilization associated with mismatched sites, however, binding within such sites is energetically more favorable. The specificity for a mismatch, therefore, does not depend on base sequence directly nor the presence or absence of specific functionalities in the major or minor groove. Instead, recognition is associated with the local helix instability of a mispair. Any sequence dependence observed, therefore, should be related to the sequence dependence of mismatch stability (1).
The specificity of [Rh(bpy)2phzi]3+ resembles that for [Rh(bpy)2chrysi]3+ (11). This observation also supports the general targeting strategy. Because the expanse of the recognition ligand is comparable for the two complexes, one expects a comparable level of discrimination vs. well matched B-DNA sites. It is interesting, nonetheless, that the disposition of the phzi ligand somewhat further from the octahedral center than for chrysi does not lead to a relative loss of specificity.
The primary source of increased sensitivity of targeting with [Rh(bpy)2phzi]3+ is derived from the increase in binding affinity, both for specific and nonspecific sites. This significant increase in affinity can be attributed to the presence of the endocyclic nitrogen atoms within the phzi ligand. The increased polarizability of the phzi ligand as compared with the chrysi ligand leads to increased stabilization through stacking. Intercalative stacking in DNA, in contrast to groove binding, is favored not through hydrophobic interactions but through dipole-induced dipole interactions associated with heterocyclic aromatic ligands (20). This increased affinity of the phzi complex containing the endocyclic nitrogens serves as an illustration of the affinity to be gained through polarized stacking. Indeed, the increase in affinity suggests that the phzi is intercalatively stacked within the mismatched site; the positioning of the mismatched bases is unclear, however. Thus, in addition to shape considerations, a tuning of the electronic structure by using nitrogen substitution in the aromatic ring results in mismatch targeting with increased affinity.
Differential Targeting of DNA from Mismatch Repair Deficient vs. Proficient Cell Lines.
Mismatch repair increases the level of fidelity of replication, but if the mismatch repair system does not function properly, mispairs can accumulate (21). Failure of mismatch repair results in microsatellite instability, and this result has provided the common assay for mismatch repair within cells. Here, we describe discrimination among genomic DNAs based on small molecule recognition of mismatches.
Our results show agreement between earlier studies of microsatellite instability and expression of mismatch repair proteins in various cell lines (16–19) vs. photocleavage by the mismatch probes. A significant level of photocleavage is apparent with DU145 cells, which contain mutations in MLH1 and PMS2; these mutations are expected to severely compromise the mismatch repair system. Indeed, particularly low mismatch repair activity has been seen with this cell line (19). For SKOV3 cells, mutations are present in MLH1 but not in PMS2. Consistent with mismatch accumulation in this cell line, significant photocleavage of genomic DNA from SKOV3 cells is found. It is noteworthy that photocleavage here occurs to a lesser extent than for DU145 DNA. Importantly, no significant photocleavage is apparent on genomic DNA from SW620 cells, a cell line proficient in mismatch repair. Interestingly, for HCT15 DNA, little photocleavage above controls is evident. This cell line is deficient in MSH6, but owing to redundancy in the mismatch repair system, such a mutation is not expected to lead to mismatch accumulation and, consistent with our results, increases in microsatellite instability are not observed (20). Hence, our photocleavage results provide a means to distinguish among these different cell lines based on their mismatch repair activity.
Targeting and discrimination among different genomic DNAs, therefore, can be considered based on the increased frequency of mispairing in genomic DNA from mismatch repair deficient cells. Because the metal complexes are specific in targeting mismatches, their levels of photocleavage should reflect the levels of mismatches accumulated. The heterogeneity in fragment length both before and after photocleavage makes absolute quantitation of the frequency of mismatches in these cell lines difficult. Nonetheless, differential DNA cleavage is evident among these cell lines with nanomolar concentrations of the metal complex. It is also noteworthy that the results described thus far reflect overall mismatch accumulation in the genome. By probing individual genes through photocleavage, gene-specific analyses could also be envisioned.
Implications for Therapeutic Design.
Whereas chemotherapy in the past has focused largely on applications of cytotoxic agents lacking selectivity, current strategies are directed at developing means to discriminate and selectively target cancerous cells. Discrimination based on mismatch recognition offers the opportunity for such selective targeting. Defects in the mismatch repair system are commonly associated with different cancers (2, 16). By specifically targeting the genomic DNA of such cancerous cells with small molecules that recognize and generate lesions in mismatch-containing DNA, preferential damage to cancerous cells could be generated. The design and application of bulky metallointercalators that bind mismatches with high affinity, as described here, represent a foundation for such strategies. Complexes have been designed with high selectivity for mismatches and with high affinity. Moreover, the differential targeting of genomic DNA from cell lines based on their compromised mismatch repair systems has been demonstrated. Creating new generations of complexes with applicability in vivo remains a challenge, but the diversity in structure afforded by octahedral metallointercalators should make such design viable.
Acknowledgments
Financial support of this research was provided by National Institutes of Health Grant GM33309. H.J. thanks the Deutsche Akademie der Naturfoscher Leopoldina for a postdoctoral fellowship; J.R.H. and J.K. thank the National Institutes of Health for National Research Service Award predoctoral and postdoctoral fellowships, respectively.
Abbreviations
- phzi
benzo[a]phenazine-5,6-quinone diimine
- chrysi
chrysene-5,6-quinone diimine
- phi
phenanthrene quinone diimine
- bpy
2,2′-bipyridine
References
- 1.Peyret N, Seneviratne A, Allawi H T, SantaLucia J., Jr Biochemistry. 1999;28:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 2.Duval A, Hamelin R. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 3.Jacob S, Praz F. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 4.Jackson B A, Barton J K. J Am Chem Soc. 1997;119:12986–12987. [Google Scholar]
- 5.Boon E M, Kisko J L, Barton J K. Methods Enzymol. 2002;353:506–522. doi: 10.1016/s0076-6879(02)53073-1. [DOI] [PubMed] [Google Scholar]
- 6.Erkilla K E, Odom D T, Barton J K. Chem Rev. 1999;88:2777–2796. doi: 10.1021/cr9804341. [DOI] [PubMed] [Google Scholar]
- 7.Sitlani A, Long E C, Pyle A M, Barton J K. J Am Chem Soc. 1992;114:2303–2312. [Google Scholar]
- 8.Krotz A H, Hudson B P, Barton J K. J Am Chem Soc. 1993;115:12577–12578. [Google Scholar]
- 9.Kielkopf C L, Erkilla K E, Hudson B P, Barton J K, Rees D C. Nat Struct Biol. 2000;7:117–121. doi: 10.1038/72385. [DOI] [PubMed] [Google Scholar]
- 10.Jackson B A, Alekseyev V Y, Barton J K. Biochemistry. 1999;38:4655–4662. doi: 10.1021/bi990255t. [DOI] [PubMed] [Google Scholar]
- 11.Jackson B A, Barton J K. Biochemistry. 2000;39:6176–6182. doi: 10.1021/bi9927033. [DOI] [PubMed] [Google Scholar]
- 12.Zincke T, Noack H. Justus Liebigs Ann Chem. 1897;295:6–27. [Google Scholar]
- 13.Mürner H, Jackson B A, Barton J K. Inorg Chem. 1998;37:3007–3012. [Google Scholar]
- 14. Gidney, P. M., Gillard, R. D. & Heaton, B. T. (1972) J. Chem. Soc. Dalton Trans., 2621–2628.
- 15.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 16.Taverna P, Liu L, Hanson A J, Monks A, Gerson S L. Cancer Chemother Pharmacol. 2000;46:507–516. doi: 10.1007/s002800000186. [DOI] [PubMed] [Google Scholar]
- 17.Umar A, Boyer J C, Thomas D C, Nguyen D C, Risinger J I, Boyd J, Ionov Y, Perucho M, Kunkel T A. J Biol Chem. 1994;269:14367–14370. [PubMed] [Google Scholar]
- 18.Chen Y, Wang J, Fraig M M, Metcalf J, Turner W R, Bissada N K, Watson D K, Schweinfest C W. Cancer Res. 2001;61:4112–4121. [PubMed] [Google Scholar]
- 19.Yeh C, Lee C, Dahiya R. Biochem Biophys Res Commun. 2001;285:409–413. doi: 10.1006/bbrc.2001.5187. [DOI] [PubMed] [Google Scholar]
- 20.Muller W, Crothers D M. Eur J Biochem. 1975;54:267–277. doi: 10.1111/j.1432-1033.1975.tb04137.x. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T A. J Biol Chem. 1992;267:18251–18354. [PubMed] [Google Scholar]