Abstract
Ionizing radiation damages DNA in several ways, including through formation of a single-nucleoside gap in one DNA strand. We have developed a two-dimensional gel electrophoresis method to investigate the effect of a strand gap on DNA structure. We generate a library of gapped DNA molecules by treating a DNA restriction fragment with the hydroxyl radical, generated by the reaction of Fe(II) EDTA with hydrogen peroxide. The DNA molecule studied contains a fixed bend produced by a set of phased adenine tracts. The A-tract bend serves as a reference bend for investigating the conformational nature of a strand gap. In the first electrophoretic dimension, a bent DNA molecule that has been treated with the hydroxyl radical is electrophoresed on a native gel. Smearing of the band on the native gel indicates that the library of gapped DNA molecules contains a variety of DNA conformations. In the second electrophoretic dimension, gapped DNA molecules having different native gel mobilities are electrophoresed on separate lanes of a denaturing gel to reveal how each strand gap affects the native gel mobility (and thus shape) of the DNA. Our results demonstrate that a single-nucleoside gap in a DNA duplex leads to an anisotropic, directional bend in the DNA helix axis. The implications of our findings for recognition of this lesion by DNA repair proteins are discussed.
One of the lesions inflicted by ionizing radiation on DNA is a single-nucleoside gap in an otherwise-intact DNA duplex (1). Because these strand gaps are not sequencespecific, there is no DNA sequence clue that a repair enzyme could use to recognize this sort of damage. An alternative means for recognition of such a lesion could be its effect on DNA structure and dynamics. Little is known, though, about the structural effect of a single-nucleoside gap in DNA.
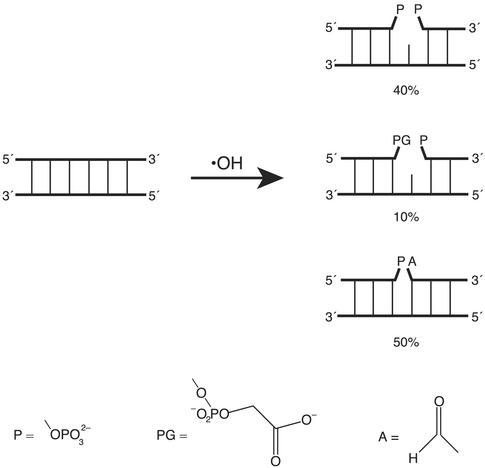
Ionizing radiation produces the hydroxyl radical (·OH) via the homolysis of a water molecule. Attack of the hydroxyl radical on a deoxyribose residue in the DNA backbone leads to various types of damage, including a single-nucleoside gap (ref. 2; Fig. 1). For our experiments, we use the Fenton reaction of Fe(II) EDTA with hydrogen peroxide (3) as a convenient source of the hydroxyl radical.
 |
1 |
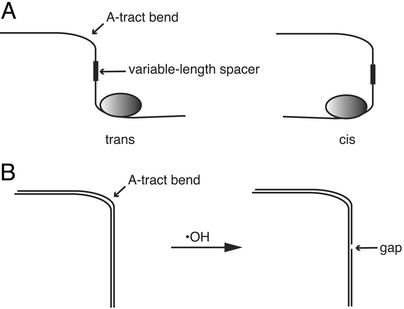
To study the effect of a strand gap on DNA structure we have developed a variant of the phasing experiment that was introduced by Zinkel and Crothers (4) to study protein-induced DNA bending (Fig. 2A). We start with a DNA molecule having a fixed bend (Fig. 2B) that is induced by a set of phased adenine tracts (5). Instead of moving a protein binding site relative to an adenine-tract bend (4), we construct a library of DNA molecules, each of which has one single-nucleoside gap somewhere in the backbone of the A-tract-containing DNA (Fig. 2B). We prepare the library of gapped duplex DNA molecules by treating the sample of bent DNA with the hydroxyl radical. We then use a two-dimensional gel electrophoresis system to monitor the effect of a strand gap on the overall shape of the DNA molecule. As a bonus, these experiments reveal which nucleotides in an adenine⋅thymine tract are most important for static DNA curvature, providing additional information to the ongoing debate about the origin of DNA bending (5).
Figure 1.
Structures of some of the lesions produced by reaction of the hydroxyl radical with duplex DNA. Three types of single-strand break have been detected by gel electrophoretic analysis (13, 14): a single-nucleoside gap, flanked either by two phosphates (Top) or by a 3′ phosphoglycolate and a 5′ phosphate (Middle); and a nick flanked by a 3′ phosphate and a 5′ aldehyde (Bottom). The relative yields of these three lesions, as estimated by analysis of deuterium kinetic isotope effect experiments (13, 14), are indicated.
Figure 2.
(A) A classic phasing experiment for investigating DNA bending by protein (4). A fixed bend in the DNA helix, imposed by a set of phased adenine tracts, is placed at various distances from a protein binding site, by changing the length of the spacer region between the two sites. The fixed adenine-tract bend serves as a reference bend for testing the ability of the protein to bend DNA. If the protein bends DNA on binding, then the electrophoretic mobility of the complex will vary in a sinusoidal fashion as the length of the spacer is changed. The two limiting forms are trans, in which the DNA and protein bends cancel and the electrophoretic mobility of the complex is greatest, and cis, in which the two bends add and electrophoretic mobility is lowest. (B) Modified phasing experiment for investigating the effect of a strand gap on DNA shape. A DNA molecule having a fixed adenine tract bend is treated with the hydroxyl radical to produce a library of gapped DNA molecules. Each DNA molecule has a single gap located somewhere in the duplex. If a strand gap introduces a fixed bend, the electrophoretic mobility of a gapped DNA molecule will depend on the phasing of the gap relative to the fixed bend (as in the classic phasing experiment shown in A).
Experimental Methods
DNA Preparation.
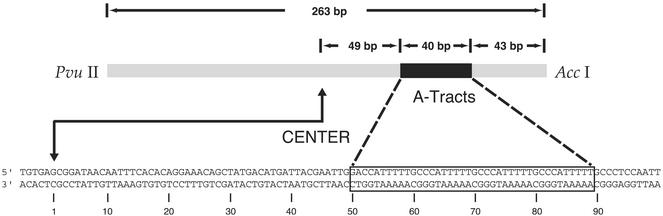
We previously described (6) the construction of a pUC18-based plasmid containing the insert (A5N5)4 (Fig. 3). The plasmid was purified by ultracentrifugation through a cesium chloride gradient and linearized by treatment with the restriction endonuclease AccI. The linear plasmid was radiolabeled at the 3′ end by reaction with [α-32P]dCTP and the Klenow fragment of DNA polymerase (pol) I or radiolabeled at the 5′ end by reaction with [γ-32P]ATP and polynucleotide kinase. A second restriction cut using PvuII gave the desired singly end-labeled 263-bp fragment.
Figure 3.
Schematic representation of the test DNA molecule, which contains four phased A-tracts. Only the sequence around the A-tract insert is shown in detail. The base pair at the center of the fragment is numbered 1, as indicated in the numbering scheme below the sequence.
Gel Electrophoresis.
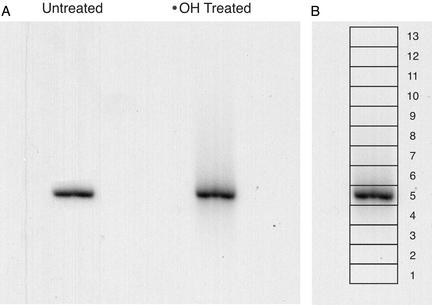
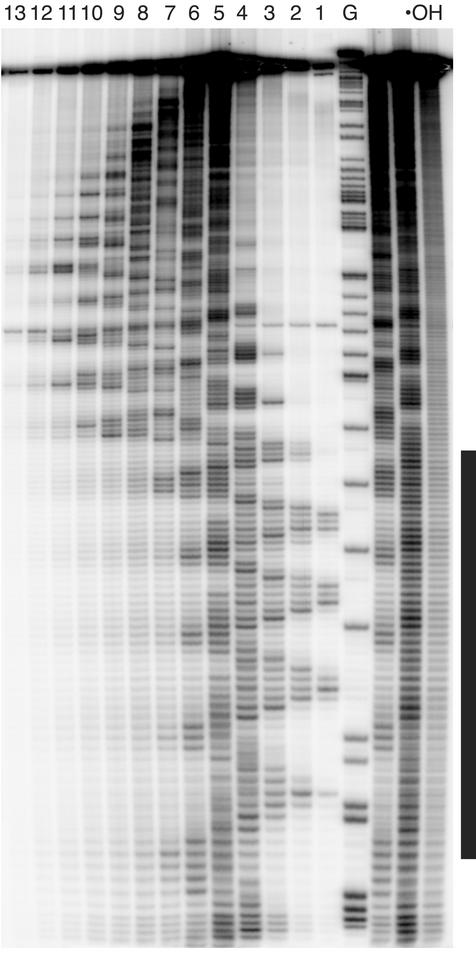
After reaction with the hydroxyl radical (7, 8), the DNA was electrophoresed in the first dimension for 18 h at 250 V at room temperature on a 31 cm × 38.5 cm × 0.4 mm nondenaturing polyacrylamide gel [6%; 29:1 acrylamide:bis; 1× TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3); Fig. 4A]. Radiolabeled 1-kb ladder (New England Biolabs) was run in an adjacent lane as a reference for gel mobility determination (not shown). The gel was cut into 13 slices of ≈6 × 15 mm (height × width) around the main band (Fig. 4B). DNA was isolated from the gel slices by the crush-and-soak procedure (9). DNA recovered from the slices was electrophoresed in a second dimension on an 8% denaturing gel (Fig. 5). Nucleotides were assigned by reference to the products of a Maxam–Gilbert guanine-specific sequencing reaction (10) that was run in an adjacent lane.
Figure 4.
Native gel electrophoretic analysis of gapped DNA (first electrophoretic dimension). (A) Hydroxyl radical-treated DNA was electrophoresed on a 6% nondenaturing polyacrylamide gel. The DNA was radiolabeled at the 3′ end with 32P. Whereas untreated DNA runs in a tight band (left lane), the band for hydroxyl radical-treated DNA (right lane) is smeared above and below the main band. (B) Boxes show the slices that were excised from the lane of the native gel containing hydroxyl radical-treated DNA.
Figure 5.
Phosphorimage of an 8% denaturing polyacrylamide gel (second electrophoretic dimension). Lanes 1–13 contain DNA eluted from slices 1–13 of the nondenaturing gel (see Fig. 4B). Lane G contains the products of a Maxam–Gilbert guanine-specific sequencing reaction for a DNA molecule 17 bp longer than the test DNA, to provide size markers. Lane ·OH contains DNA treated with the hydroxyl radical that was not run on a native gel (control). The vertical bar to the right of the gel image marks the A-tract region. The sinusoidal pattern of gel mobility discussed in the text may be seen more easily if the gel image is viewed from a distance of a few feet, and perhaps rotated 90°.
Gel Analysis.
An imaging plate was exposed to the denaturing gel, and the plate was scanned by using a Molecular Dynamics PhosphorImager (Model 400E). To determine the average mobility on the native gel of a particular gapped DNA, the peaks in a one-dimensional scan of the lanes of the denaturing gel were Fourier-sharpened and then integrated (11).
Results and Discussion
We studied a DNA restriction fragment of 263 bp having four tandem copies of the sequence A5⋅T5 phased with the DNA helical repeat. The center of the four A-tracts is 71 bp from the center of the DNA molecule (Fig. 3). The sequence A5⋅T5 is highly curved, as judged by its anomalous mobility in gel electrophoresis (12). We use the curved A-tract segment as a reference bend for phasing analysis (4) of the bending of gapped DNA.
To prepare a library of singly gapped DNA duplexes, we utilized the reaction of the hydroxyl radical with DNA (7, 8). The hydroxyl radical cleaves DNA by abstracting a hydrogen atom from a deoxyribose in the DNA backbone (13). The structure of the lesion depends on which deoxyribose hydrogen is removed by ·OH (2, 13, 14). Roughly half the reaction products are DNA molecules having a single gap, flanked either by two phosphates or by a 5′ phosphate and a 3′ phosphoglycolate (refs. 2, 13, and 14; Fig. 1).
We treated the DNA molecule in Fig. 3 with the hydroxyl radical to produce a library of gapped DNA molecules. Under our reaction conditions, each DNA molecule has no more than a single gap; ≈70% of the DNA molecules are left intact. Because of the nonspecific nature of the hydroxyl radical cleavage reaction (7), gaps are fairly equally distributed among all nucleotides in the molecule (see Fig. 5, lane ·OH).
When the library of singly gapped duplex DNAs was subjected to native gel electrophoresis, we observed that the DNA migrated mainly as a single band (Fig. 4A, right lane). However, a noticeable smear of radioactivity was seen both above and below the main band. We presume that the smear is the consequence of the effect of a gap on the electrophoretic mobility of the DNA duplex (15).
To determine how a particular strand gap affects the gel mobility of DNA, we subjected the smeared band to a second dimension of electrophoresis. The native gel was sliced into 13 strips around the main band (Fig. 4B). DNA was eluted from each strip and electrophoresed in a separate lane of a denaturing gel (Fig. 5). Remarkably, a highly periodic two-dimensional pattern was obtained that extends throughout the length of the DNA molecule. In a control experiment using a restriction fragment without an A-tract insert, we observed no modulation in the two-dimensional gel pattern (11). This result suggests that the strikingly periodic pattern seen in Fig. 5 is caused by the variation in the positions of strand gaps relative to the fixed A-tract bend. This effect is analogous to the sinusoidal variation in gel mobility of a DNA–protein complex in the phasing assay (ref. 4; see Fig. 2A) that is the hallmark of protein-induced DNA bending.
We analyzed the periodic pattern by determining its Fourier power spectrum (11) and by fitting of a cosine curve (16). Both methods demonstrated that the two-dimensional cleavage pattern has a periodicity of ≈10.5 nucleotides, the same period as the DNA helix repeat (7). The pattern is clearly made up of two distinct domains, one that is within the A-tract region and one that is outside of the A-tracts.
Two-Dimensional Gel Pattern Outside the A-Tract Region.
We attribute the highly modulated pattern outside of the A-tract region to the phasing of a bend at a strand gap with the bend at the A-tract. In the classic phasing experiment of Zinkel and Crothers (ref. 4; Fig. 2A), a standard (A-tract) bend is placed some distance away from the bend to be tested. When the two bends are disposed in a cis arrangement relative to each other, the overall curvature of the DNA molecule reaches a maximum. When the bends are trans, overall curvature is minimized (Fig. 2A).
In this experiment, we observe the effect on native gel mobility of moving a gap around the DNA helix, relative to an A-tract bend (Fig. 2B). The oscillatory behavior we see in the denaturing gel pattern (Fig. 5) shows that a gap in the DNA backbone causes a directional (anisotropic) bend. If instead a gap acted as a swivel or universal joint (a nondirectional bend), there would be no phase relationship between the gap-induced bend and the fixed A-tract bend, and so no modulated pattern would be observed. We note that our results also are consistent with a gap being a site of anisotropic flexibility rather than a static, fixed bend. Nevertheless, we emphasize that our observation of a strongly modulated pattern that repeats with the DNA helical period shows that a strand gap induces a directional, and not isotropic, curvature to the DNA helix.
To further explore the nature of the gap-associated bend, we compared the patterns for the two strands of the DNA duplex (Fig. 6). The two patterns are precisely opposite in phase, consistent with the expectation that a gap in one strand of the DNA duplex creates a bend in the opposite direction from a bend induced by gap at the same base pair in the complementary strand.
Figure 6.
Mean native gel electrophoretic mobilities of DNA molecules containing a single-nucleoside gap at one position in the sequence are shown. The data for this plot were derived from the phosphorimage shown in Fig. 5. (One could think of this plot as depicting the gel mobility data in Fig. 5 turned 90°.) A gray horizontal bar indicates the A-tract region. The intact DNA molecule runs in slice 5 of the native gel (Main Band). Filled triangles and thick line, DNA radiolabeled at the 5′ end; open rectangles and thin line, DNA radiolabeled at the 3′ end. The positions of adenines and thymines in the A-tracts are marked. The numbering system corresponds to that of Fig. 3. The 5-nt phase shift between the patterns for the two strands shows that a gap in one strand results in a bend in the opposite direction from a gap at the same base pair in the complementary strand.
A dramatic feature of the pattern outside of the A-tracts is that the cosine wave oscillates about a position well above the mobility of the main band on the native gel, the position where the intact bent DNA molecule runs (see Fig. 5). When the native gel (first electrophoretic dimension) is run at higher temperature, we observed that the elevated retardation of gel mobility is even more pronounced (11). Whereas the intact A-tract-containing DNA molecule has an RL value (ratio of apparent length to actual length; ref. 12) of 1.34, introduction of a single gap outside of the A-tract causes the average RL value to increase to 1.52. In the most extreme case, when a single gap is perfectly in phase with the A-tract bend, the RL value is 1.78. We applied the algorithm of Kerppola and Curran (17) to our data and estimated the bending angle of a single gap to be ≈12°. Recently, in experiments on a nick-sensing DNA repair protein from Arabidopsis (18), atomic force microscopy was used to study the shape of a gapped DNA molecule. A bend angle of ≈17° was measured, in good agreement with our value.
Ligation ladder experiments (19) demonstrated that a single-nucleotide gap (a gap flanked by hydroxyls on the ends of the broken strand) causes the DNA molecule to run with retarded mobility on a native gel. The authors of this study invoked flexibility rather than bending as the source of the retardation in gel mobility because of the lack of dependence of gel mobility on the spacing of multiple single gaps. In contrast, our experiments show that for a gap flanked by phosphates there is a pronounced dependence on gel mobility of the position of the gap relative to the fixed A-tract bend. This is a clear indication that there is directional bending at the site of the gap that we have studied. Because we prepared our gapped DNA by treatment with the hydroxyl radical, the primary DNA damaging agent produced by ionizing radiation, we presume that this bent structure is representative of the sort of DNA structure that must be recognized by repair enzymes that deal with radiation-induced damage.
DNA having an internal loop as the result of a three base pair mismatch shows very little retardation in electrophoretic mobility, yet cyclizes with remarkable efficiency when treated with DNA ligase (20). Furthermore, there is very little dependence of the cyclization probability (J factor) on the length of the DNA. These results led to the suggestion that this kind of abnormal DNA structure has substantial torsional, as well as bending, flexibility (20). It is possible that the gapped DNA we study here is both directionally bent and torsionally flexible. Bending is apparent in the retardation in gel electrophoretic mobility of gapped DNA that we have observed. The issue of torsional flexibility in gapped DNA could be resolved by a cyclization experiment (20).
Two-Dimensional Gel Pattern Within the A-Tract Region.
A transition in the oscillatory pattern occurs at the junction of the A-tract segment and the flanking mixed-sequence DNA (Figs. 5 and 6). Whereas for the flanking DNA the cosine wave is centered at a position above the main band, the cosine wave for gaps introduced into the A-tracts oscillates around a position slightly below that of the main band. Therefore, introducing a single gap in the A-tract region usually results in increased native gel mobility (less bending), whereas a gap outside the A-tract leads to decreased mobility (more bending).
Our library of gapped DNAs allows us to systematically study the effect of the loss of each nucleotide in the A-tract region on A-tract-induced curvature. Our results show that adenine nucleotides at the 3′ end of the A-tract are most responsible for maintenance of DNA bending, because a gap at these positions causes the greatest increase in gel mobility compared with the intact bent duplex (Fig. 6). This is consistent with the junction model for DNA bending (12, 21), which asserts that the transition between the unusual structure adopted by an A-tract and the normal structure of mixed-sequence DNA is responsible for DNA curvature. A⋅T base pairs at the 3′ end of an A-tract adopt a conformation most distinct from that of B-form DNA, and so removing an adenine base and sugar at this site affects the bending most. Our results are less consistent with another model for DNA bending (22), which asserts that G/C-rich sequences between A-tracts are the actual site of DNA bending.
Implications for Recognition of Gapped DNA by Repair Proteins.
Recently a remarkable set of x-ray cocrystal structures was published (23) that reveals intimate details of DNA repair. Three structures involving human DNA pol β were solved: one in which the polymerase was bound to gapped DNA, another with polymerase in a ternary complex with gapped DNA and a dideoxynucleoside triphosphate, and a third in which polymerase was bound to nicked DNA. These structures represent three intermediates in a DNA repair reaction. A striking feature of all three structures is that the DNA is bent ≈90° at the site of the gap or nick. The authors suggest in the conclusion of the paper that “. . . kinking . . . is probably an identifying feature of pol β's gap substrate. Pol β's ability to recognize this feature may facilitate recognition of damaged DNA.” We note that the anisotropic bending that we have observed by our two-dimensional gel experiment offers support to this proposal, because our work shows that a gap is more than just a site of enhanced isotropic flexibility, but indeed induces a directional bend in the DNA duplex.
Additional structural insight into repair of DNA strand gaps comes from the solution NMR structure of the mammalian x-ray cross-complimenting group protein, XRCC1 (24). XRCC1 has been implicated in the process of assembling the proteins involved in repair of oxidative lesions in DNA (25). XRCC1 was shown to bind to nicked DNA and to a complex of pol β with gapped DNA (24). The ternary XRCC1/pol β/nicked DNA complex was modeled based on the pol β/gapped DNA cocrystal structure (23) and the NMR structure of XRCC1 (24). This modeling exercise showed that XRCC1 could bind to the concave surface of bent DNA found in the pol β cocrystal structure, with the polymerase bound to the convex side of the bend.
Other recent work has shown that additional DNA repair proteins associate with XRCC1 that is bound to damaged DNA (25). Initial sensing of single-strand breaks in DNA is thought to fall to poly(ADP-ribose) polymerase-1 (PARP) (26), which interacts with XRCC1 and may nucleate the formation of a repair complex. We suggest that the structural and dynamic properties of gapped DNA itself may be important to the recognition and repair of the lesion.
Conclusions
Our experiments show that a single-nucleoside gap in one strand of duplex DNA is not merely a site of isotropic flexibility, but instead permits bending of the helix axis in only one direction. These results provide new information on the structure and dynamics of DNA that has been damaged by ionizing radiation, and may give insight into how such damage is recognized by cellular repair proteins.
Acknowledgments
This work was supported by U.S. Public Health Service Grant GM40894.
Abbreviation
- pol
polymerase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ward J E. In: DNA Repair in Higher Eukaryotes, DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Vol. 2. Totowa, NJ: Humana; 1988. pp. 65–84. [Google Scholar]
- 2.Pogozelski W K, Tullius T D. Chem Rev. 1998;98:1089–1107. doi: 10.1021/cr960437i. [DOI] [PubMed] [Google Scholar]
- 3.Udenfriend S, Clark C T, Axelrod J, Brodie B B. J Biol Chem. 1954;208:731–739. [PubMed] [Google Scholar]
- 4.Zinkel S S, Crothers D M. Nature. 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 5.Hagerman P J. Annu Rev Biochem. 1990;59:755–781. doi: 10.1146/annurev.bi.59.070190.003543. [DOI] [PubMed] [Google Scholar]
- 6.Price M A, Tullius T D. Biochemistry. 1993;32:127–136. doi: 10.1021/bi00052a018. [DOI] [PubMed] [Google Scholar]
- 7.Tullius T D, Dombroski B A. Science. 1985;230:679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- 8.Price M A, Tullius T D. Methods Enzymol. 1992;212:194–219. doi: 10.1016/0076-6879(92)12013-g. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 10.Maxam A M, Gilbert W. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 11.Guo H. Dissertation. Baltimore: The Johns Hopkins Univ.; 1997. [Google Scholar]
- 12.Koo H S, Wu H M, Crothers D M. Nature. 1986;320:501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- 13.Balasubramanian B, Pogozelski W K, Tullius T D. Proc Natl Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tullius, T. D. (2000) Radiat. Res.2, Conf. Proc., 333–335.
- 15.Werel W, Schickor P, Heumann H. EMBO J. 1991;10:2589–2594. doi: 10.1002/j.1460-2075.1991.tb07800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayes J J, Tullius T D, Wolffe A P. Proc Natl Acad Sci USA. 1990;87:7405–7409. doi: 10.1073/pnas.87.19.7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kerppola T K, Curran T. Science. 1991;254:1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 18.Petrucco S, Volpi G, Bolchi A, Rivetti C, Ottonello S. J Biol Chem. 2002;277:23675–23683. doi: 10.1074/jbc.M201411200. [DOI] [PubMed] [Google Scholar]
- 19.Mills J B, Cooper J P, Hagerman P J. Biochemistry. 1994;33:1797–1803. doi: 10.1021/bi00173a024. [DOI] [PubMed] [Google Scholar]
- 20.Kahn J D, Yun E, Crothers D M. Nature. 1994;368:163–166. doi: 10.1038/368163a0. [DOI] [PubMed] [Google Scholar]
- 21.Crothers D M, Haran T E, Nadeau J G. J Biol Chem. 1990;265:7093–7096. [PubMed] [Google Scholar]
- 22.Goodsell D S, Kopka M L, Cascio D, Dickerson R E. Proc Natl Acad Sci USA. 1993;90:2930–2934. doi: 10.1073/pnas.90.7.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawaya M R, Prasad R, Wilson S H, Kraut J, Pelletier H. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 24.Marintchev A, Mullen M A, Maciejewski M W, Pan B, Gryk M R, Mullen G P. Nat Struct Biol. 1999;6:884–893. doi: 10.1038/12347. [DOI] [PubMed] [Google Scholar]
- 25.Whitehouse C J, Taylor R M, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko D D, Weinfeld M, Caldecott K W. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 26.de Murcia G, Menissier de Murcia J. Trends Biochem Sci. 1994;19:172–176. doi: 10.1016/0968-0004(94)90280-1. [DOI] [PubMed] [Google Scholar]