Abstract
Iron concentrations in the ocean are low enough to limit the growth of marine microorganisms, which raises questions about the molecular mechanisms these organisms use to acquire iron. Marine bacteria have been shown to produce siderophores to facilitate iron(III) uptake. We describe the structures of a suite of amphiphilic siderophores, named the amphibactins, which are produced by a nearshore isolate, γ Proteobacterium, Vibrio sp. R-10. Each amphibactin has the same Tris-hydroxamate-containing peptidic headgroup composed of three ornithine residues and one serine residue but differs in the acyl appendage, which ranges from C-14 to C-18 and varies in the degree of saturation and hydroxylation. Although amphiphilic siderophores are relatively rare, cell-associated amphiphilic siderophores are even less common. We find that the amphibactins are cell-associated siderophores. As a result of the variation in the nature of the fatty acid appendage and the cellular location of the amphibactins, the membrane partitioning of these siderophores was investigated. The physiological mixture of amphibactins had a range of membrane affinities (3.8 × 103 to 8.3 × 102 M−1) that are larger overall than other amphiphilic siderophores, likely accounting for their cell association. This cell association is likely an important defense against siderophore diffusion in the oceanic environment. The phylogenetic affiliation of Vibrio sp. R-10 is discussed, as well as the observed predominance of amphiphilic siderophores produced by marine bacteria in contrast to those produced by terrestrial bacteria.
A defining characteristic of vast regions of the world's oceans is the surprisingly low level of iron in surface waters, in contrast to the abundance of iron in most terrestrial environments. Iron is an essential nutrient for the growth of microorganisms (1) and an important bioactive metal in seawater. The low level of iron is now known to limit primary production by autotrophic microorganisms in ocean regions that are replete in major nutrients such as nitrate, phosphate, and silicate (2–8). Results from four separate mesoscale additions of iron (1–2 nM, FeSO4⋅7H2O) to large patches of ocean water (e.g., ≈70–100 km2) in the equatorial Pacific (2, 7, 8) and the Southern Ocean (5, 6) show that primary production by photosynthetic microorganisms increased markedly in response to iron. Bacterial production can also be either directly or indirectly stimulated by the addition of iron (9–11), although in some situations this occurs only after carbon limitation has been relieved (12). In any case, bacteria have relatively high Fe:C ratios and clearly play a role in biogeochemical cycling of iron (9).
To acquire iron, many aerobic bacteria, including marine species (13–20), produce siderophores. These low molecular weight compounds chelate iron(III) with high affinity and are synthesized in response to low iron concentrations (21, 22). Siderophores function in the receptor-mediated transport of iron into microbial cells, as well as in the extracellular chelation of iron(III).
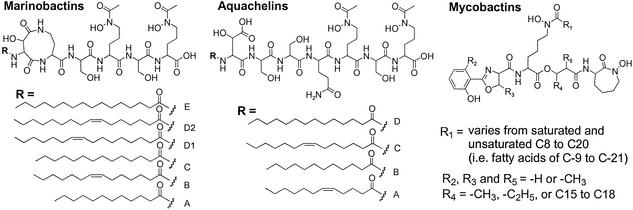
The relative scarcity of iron in the oceanic environment, where the diffusion of siderophores may be of concern, raises questions about the types of siderophores that marine bacteria produce. Recently, we reported two suites of amphiphilic peptide siderophores, the marinobactins and aquachelins, which are produced by distinct genera of marine bacteria (Fig. 1) (16). The marinobactins, produced by the Marinobacter sp. strain DS40M6, are characterized by a 6-aa peptide headgroup that coordinates iron through two hydroxamate groups and a β-hydroxy-aspartic acid residue. The N terminus contains an appendage of one of a series of fatty acid residues, ranging from C-12 to C-16. The aquachelins, produced by Halomonas aquamarina, are characterized by a 7-aa peptide with N-terminal appendages ranging from C-12 to C-14 fatty acids. The suite of fatty acids found in marinobactins and aquachelins comprises both saturated and unsaturated alkyl chains containing one double bond in the cis configuration. The marinobactins have been shown to partition into semisynthetic vesicles (23), indicating that these amphiphilic peptides could potentially associate with bacterial membranes as a means of preventing siderophore diffusion. However, under our growth and isolation conditions, both the marinobactins and aquachelins are primarily observed in the growth medium. Thus, although the marinobactins and aquachelins can associate with biological membranes, they are also quite water-soluble.
Figure 1.
Structures of the amphiphilic marinobactins and aquachelins produced by Marinobacter sp. strain DS40M6 and H. aquamarina strain DS40M3, respectively, and the lipophilic mycobactins produced by mycobacteria (16, 25).
Hundreds of siderophores have been structurally characterized, yet few are amphiphilic (24). Other amphiphilic siderophores include the ornibactins, produced by Burkholderia (Pseudomonas) cepacia; corrugatin, produced by Pseudomonas corrugata; acinetoferrin, produced by Acinetobacter haemolyticus; rhizobactin 1021, produced by Sinorhizobium meliloti; and the carboxymycobactins, produced by mycobacteria, all of which are produced and secreted into the growth medium by terrestrial bacteria. With the exception of the ornibactins and carboxymycobactins, these are all single siderophores as opposed to a suite of siderophores that vary in the nature of the fatty acid tail (25–30). Mycobacteria, as well as other high G + C Gram-positive bacteria, also produce the only structurally characterized family of siderophores to date that are associated with the bacterial membrane and are not secreted into the growth medium. The mycobactins (Fig. 1) produced by mycobacteria are a suite of lipidic siderophores that differ primarily in fatty acid chain length (C-9 to C-21) or minor modifications in the head group (25, 28). Mycobactins coordinate iron(III) through both oxygen atoms of the hydroxamate groups and the nitrogen and oxygen of the 2-hydroxyphenyloxazoline moiety. The formobactins, nocobactins, and amamistatins, isolated from Nocardia asteroides, Rhodococcus bronchialis, Rhodococcus rubropertinctus, and Rhodococcus terrae, are nearly identical to the mycobactin siderophores in both head group and hydrocarbon tail and are also found within the bacterial cellular membrane (31–35).
We describe a suite of amphiphilic siderophores produced by a marine Gram-negative bacterium (Vibrio sp. R-10). These siderophores, named the amphibactins, are defined by a short peptide headgroup and one of a series of saturated, unsaturated, or hydroxylated fatty acid appendages ranging from C-14 to C-18. The amphibactins differ from the marinobactins and aquachelins (Figs. 1 and 2) by a smaller head group (four vs. six and seven amino acids, respectively) and generally longer fatty acid tails. Like the mycobactins, this suite of amphiphilic siderophores is cell-associated. Because siderophores are virulence factors for many pathogenic bacteria, including mycobacteria (36, 37), an understanding of the evolutionary advantages of amphiphilic siderophores in different environments may have implications for human health as well as for understanding iron uptake in the ocean. As a first step, we discuss the structural characterization and amphiphilic properties of the amphibactins, as well as the phylogenetic relationship of Vibrio sp. R-10 to other marine and terrestrial bacteria known to produce amphiphilic siderophores.
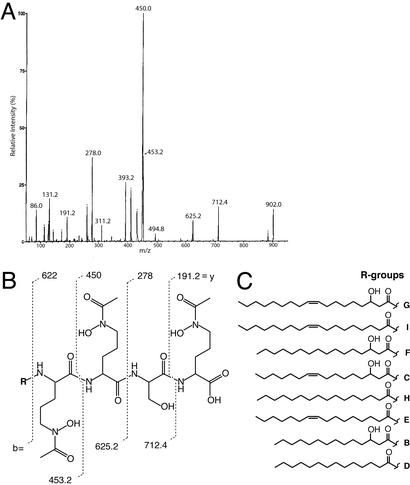
Figure 2.
(A) Tandem mass spectrum for amphibactin G from Vibrio sp. strain R-10. (B) Structure of the amphibactin headgroup. The vertical lines through the structure show the mass-to-charge ratio values for the y and b fragments obtained for amphibactin G by tandem MS. The “y” and “b” nomenclature refers to the charge when retained by the COOH-terminal fragment or the NH2-terminal fragment of the peptide, respectively (42). (C) Structures of the fatty acid tails for eight of the 10 amphibactins. The letters next to the structures refer to the amphibactin and correlate with their retention by HPLC (see Fig. 3).
Materials and Methods
Isolation of Siderophores.
Vibrio sp. R-10 was obtained from near-shore seawater at Roatan, Honduras by M.G.H. in June 1990 and isolated on seawater complete agar plates (13). For siderophore isolation, R-10 was grown in natural seawater medium (17) for 20 h on a rotary shaker (150 rpm; New Brunswick Scientific) at ambient temperature. The cells were harvested by centrifugation (10,800 × g), and the supernatant and pellet were tested for siderophore production with the chrome azurol S–shuttle solution assay (38). The bacterial pellet, from each 150-ml culture, was washed with doubly deionized water (ddH2O, Nanopure, Barnstead), transferred to 50-ml conical tubes and shaken with 45 ml of ethanol for 24 h at 4°C. The ethanol extract was filtered through Whatman filter paper, concentrated in vacuo to ≈20% of the original volume, and purified by HPLC as described below. When the cultures were in late stationary phase, the same set of siderophores normally associated with the cell pellets could, on rare occasions, be found in the supernatant as well. These secreted siderophores were isolated from the supernatant by batch adsorption onto Amberlite XAD-2 resin (Supelco) and elution with methanol as previously described (17). Siderophore-containing fractions, corresponding to elution with 100% methanol, were pooled, dried in vacuo, and applied to a C4 reversed-phase HPLC column (Vydac, Hesperia, CA). The siderophores were purified by using a gradient of 100/0 (% A/B) to 0/100 (% A/B) over 37 min, and the program was held an additional 10 min at 0/100 (% A/B), after which the gradient was reversed, returning to 100/0 (% A/B) over 10 min [A = 99.95% ddH2O (Nanopure) and 0.05% trifluoroacetic acid (TFA); B = 19.95% ddH2O, 0.05% TFA, and 80% methanol]. The absorbance of the eluent was monitored at 215 nm.
Structure Determination.
The masses and amino acid connectivity of the amphibactins were determined by electrospray MS and tandem MS with instrumentation similar to that described previously (16, 17). The amino acid composition of the amphibactins, including the enantiomeric configuration, was determined with Marfey's reagent (1-fluoro-2,4-dinitrophenyl-5-l-alanine amide) (39). The position of the d- and l-Orn residues could not be determined by partial peptide fragmentation and subsequent amino acid analysis, because none of the peptide fragments would have a unique d- or l-Orn composition. The UV-visible absorption spectra of the ferric-bound amphibactins were compared with spectra from other Tris-hydroxamates (24). Acid-hydrolyzed amphibactins were treated with methanolic HCl to produce methyl esters of the fatty acids for identification of the fatty acid moieties. Gas chromatography of the fatty acid methyl esters were compared with bacterial fatty acid methyl ester standards (Supelco). The position of the double bond in the unsaturated fatty acids was determined by reductive work-up of ozonolysis products.
Partition Experiments with Amphibactins.
The partition coefficients for the suite of amphibactins between membrane and aqueous phases were measured by HPLC. Unilamellar vesicles were prepared by extrusion of l-α-dimyristoylphosphatidylcholine (DMPC) thin films through polycarbonate filters (200-nm average pore size) in 10 mM Tris⋅HCl/0.1 M KCl, pH 8.0 (23). DMPC vesicles (concentration range of 0–15 mM) were incubated with a 70 μM amphibactin mixture in acid-washed centrifuge tubes in the dark at 27°C with gentle shaking. After 2.5 h of equilibration, the siderophore/vesicle mixture was centrifuged for 3 h at 200,000 × g. The supernatant, containing nonpartitioned siderophore, was removed and assayed by HPLC for siderophore concentration by using an analytical C4 reversed-phase column (Vydac) with a linear gradient of 0–100% B over 37 min [A = 99.9% ddH2O and 0.1% trifluoroacetic acid (TFA); B = 19.9% ddH2O, 0.1% TFA, and 80% methanol]. The absorbance of the eluent was monitored at 215 nm. The concentration of amphibactin siderophores remaining in the supernatant (water phase) was determined by comparing the area of the chromatographic peak to a standard curve for the physiological mixture of amphibactins. Controls were completed to monitor and correct for any adsorption of the siderophore onto the centrifuge tubes. Comparison of membrane partition coefficients between the amphibactins and marinobactins (23) was completed.
SSU rRNA Gene Amplification and Phylogenetic Analysis.
Nucleic acids from colonies of bacteria on plates were extracted, and the small subunit (SSU) rRNA genes were amplified by using standard methods (17) and the oligonucleotide primers 27F and 1492R (40). Both strands of the amplified DNA were sequenced by using conserved bacterial primers (40) and ABI Prism BigDye (Ver. 2.0, Perkin–Elmer) dye-terminator chemistry. SSU rRNA gene sequences from the isolates, their close relatives, and other known siderophore-producing bacteria [these sequences were obtained from the Ribosomal Database Project (RDP) (41) or GenBank] were aligned by using clustalw and then corrected by hand by using sequencher (Gene Codes, Ann Arbor, MI) before importing the alignment into paup 4.0b10 (Sinauer, Sunderland, MA) for phylogenetic analysis. Phylogenetic analysis was completed as in ref. 16 with 344 phylogenetically informative characters, except the bootstrap analysis used 500 rounds of resampling. Distance and maximum parsimony analyses gave similar results, except that distance methods placed the Halomonas group in the Marinobacter rather than the Shewanella branch of the γ Proteobacteria.
Results and Interpretation
Amphibactin Structure Determination.
The Vibrio sp. R-10 produces a suite of at least 10 siderophores, named the amphibactins, ranging in mass from 816 to 902 protonated molecular ion (M+H)+. This mixture of siderophores was isolated from the cellular pellet of R-10 by extraction with ethanol, making this one of the few bacteria shown to produce cell-associated siderophores.
The structure of the amphibactin peptidic head group was determined by using a combination of amino acid analysis and MS. The peptide headgroup of each amphibactin contained the same amino acids (one l-serine, two d-ornithines, and one l-ornithine). The connectivity and derivatization of the amino acids was established by tandem MS. Similar to the marinobactins and aquachelins (16), the ornithine residues of the amphibactins are N-hydroxylated and acetylated, forming the hydroxamate group that coordinates iron(III). Fragment analysis established the connectivity of the amino acids (Fig. 2 A and B). The “y” and “b” nomenclature refers to the charge when retained by the COOH-terminal fragment or the NH2-terminal fragment of the peptide, respectively (42). The y4 fragment, corresponding to 622 (m/z), was seen in low abundance in all mass spectra, because it is directly attached to the R group. A similar phenomenon was observed when analyzing spectra for the marinobactins and aquachelins (16). The structural assignment was further supported by observation of internal fragmentation corresponding to a unit with two N-OH, N-Ac-ornithine residues and also a unit with one N-OH, N-Ac-ornithine, and one serine residue. Each amphibactin siderophore was found to have identical y fragments but different b fragments. Thus the tandem MS and amino acid analysis data suggested that the difference in the amphibactins was a result of different fatty acid appendages.
The fatty acid moieties were structurally characterized by comparison of mass and fragmentation of amphibactin methyl esters to those of bacterial standards by GC-MS. Positions of unsaturation were determined by GC-MS after ozonolysis of the fatty acid. The side chains of the amphibactins range in length from C-14 to C-18 and differ by the degree of unsaturation and hydroxylation (Fig. 2C). Microorganisms are known to make a variety of saturated and unsaturated oxyfunctionalized fatty acids (43). Interestingly, the amphibactins are similar in fatty acid length to the mycobactins, which are also easily extracted with ethanol from the cellular membranes of mycobacteria.
The UV-visible spectrum of the Fe(III)–amphibactin siderophore complexes is consistent with Tris-hydroxamate ligation. The absorption maximum at 423 nm is a result of the hydroxamate to Fe(III) charge–transfer band, which is similar to other Tris–hydroxamate ferric iron complexes (21, 22, 24).
Membrane Partitioning of the Amphibactins.
A distinctive feature of the amphibactins is their fatty acid appendage. Variations in the nature of the hydrophilic head group and the hydrophobic fatty acid tail will likely affect the partitioning of these amphiphilic siderophores into the cell membranes and thus affect functional features of the iron transport mechanism. Structurally, the amphibactins have smaller head groups and longer fatty acid tails than the marinobactins. The chain length and degree of unsaturation in the fatty acid greatly affect partitioning of marinobactins in phosphatidylcholine (PC) vesicles (23). To compare the partition propensity of the amphibactins to the marinobactins, we have investigated the partitioning of the amphibactins into PC vesicles.
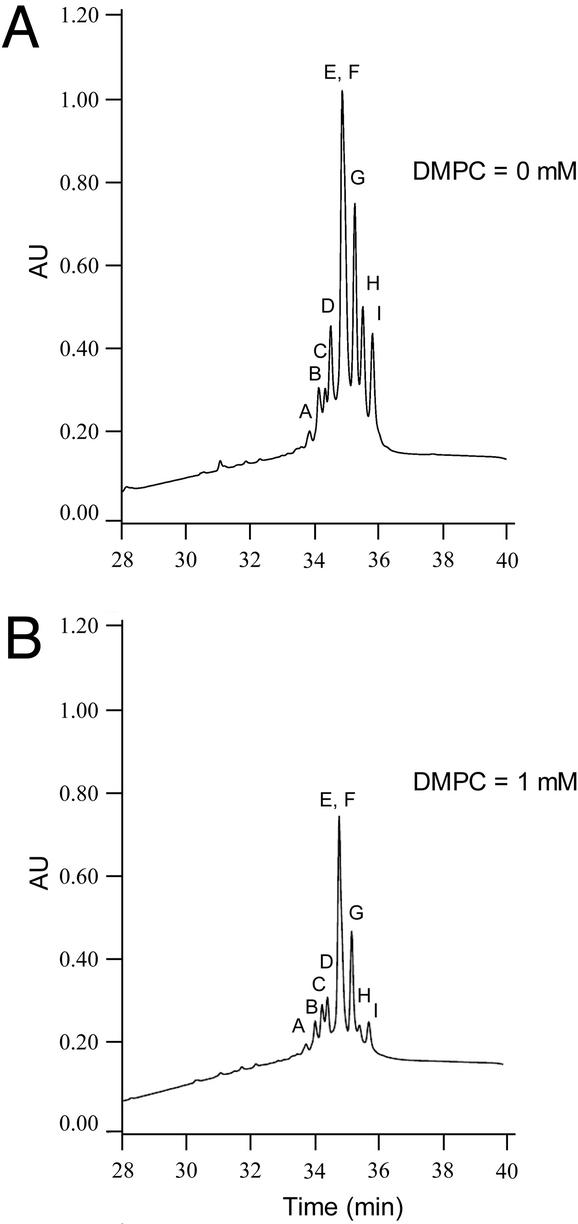
The HPLC chromatogram of the suite of amphibactin siderophores before and after addition of 1 mM DMPC vesicles shows that all of the amphibactins partition substantially into PC vesicles (Fig. 3). Thus the partition coefficients can be determined by equilibrating a fixed concentration of the physiological suite of amphibactins with increasing concentrations of DMPC vesicles and plotting the ratio of siderophore remaining in solution (Dw) to the total siderophore concentration (Dt) (i.e., Dw/Dt) vs. lipid vesicle concentration (Table 1), as previously described (23). Table 1 summarizes the partition coefficients for each of the predominant amphibactin siderophores (apo-AB, D, G-I) as compared with the previously determined partition coefficients for the physiological mixture of marinobactins (apo-MA-E). The partition coefficient experiments show that the physiological mixture of amphibactins has larger partition coefficients overall than the marinobactins. Whereas the partition coefficients of the marinobactins dropped by an order of magnitude on introduction of one double bond in the fatty acid or on chain shortening by two methylene carbons, the difference in partition coefficients is less significant for similar changes in the amphibactins (Table 1).
Figure 3.
HPLC chromatograms of the amphibactin physiological mixture equilibrated with DMPC model membranes. (A) The amphibactin physiological mixture (70 μM) before equilibration with 1 mM DMPC. (B) The remaining mixture of amphibactins in the water phase after equilibration with 1 mM DMPC. The lettering above each peak identifies the amphibactin species as shown in Fig. 2. Only the nine most predominant amphibactins are shown.
Table 1.
Partition coefficients for the physiological mixtures of amphibactin and marinobactin siderophores into DMPC vesicles
| Siderophore* | Fatty acid tail† | Partition coefficient, M−1 |
|---|---|---|
| apo-AG | C18:1; 3-OH | 833 ± 108 |
| apo-AI | C18:1 | 1,018 ± 194 |
| apo-AB | C14:0; 3-OH | 1,338 ± 161 |
| apo-AD | C14:0 | 1,915 ± 140 |
| apo-AH | C16:0 | 3,784 ± 225 |
| apo-MA | C12:0 | 36 ± 7 |
| apo-MB | C14:1 | 25 ± 4 |
| apo-MC | C14:0 | 195 ± 21 |
| apo-MD | C16:1 | 209 ± 28 |
| apo-ME | C16:0 | 5,818 ± 694 |
Phylogenetic Analysis.
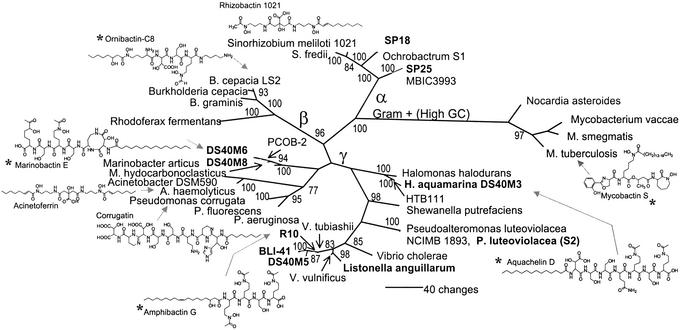
The phylogenetic affiliation of Vibrio sp. R–10, based on SSU rRNA gene sequences, is shown in Fig. 4. Nine other marine bacteria whose siderophore structure has been partially or completely determined are also shown. Several additional marine bacteria with characterized siderophores were not included in the tree, including a number of aerobactin-producing Vibrio strains (44), a Pseudomonas sp. isolated as a contaminant in a cyanobacteria culture (45), and the bisucabarin-producing deep-sea mud bacterium Alteromonas haloplanktis (46). The SSU rRNA gene sequences of the isolates Listonella anguillarum (47), DS40M5, 6, 8 and BLI-41 have been previously described (13, 16, 17). All of the DS40M strains, which were isolated from a depth of 40 m off the continental slope of the eastern equatorial Atlantic, are free-living γ Proteobacteria. DS40M3 has been identified as a H. aquamarina strain from the Oceanospirillum group; the Marinobacter group contains the isolates DS40M6 and 8; and DS40M5 is a member of the Vibrio group. Other marine siderophore producers in the γ Proteobacteria include the Pseudoalteromonas (Alteromonas) luteoviolacea strain S2 isolated off Chub Cay in the Bahamas (15). The SSU rRNA gene sequence of this strain is most closely related to Pseudoalteromonas (Alteromonas) luteoviolacea National Collections of Industrial and Marine Bacteria 1893 (RDP 0.863), confirming the identification that was done previously with more classical methods (15). The R-10 isolate, which was isolated from nearshore seawater off Roatan, Honduras, is also a γ Proteobacterium. Although this bacterium is clearly from the Vibrio group, it is not possible to identify the strain to species with certainty from the sequence data available. Close relatives include Vibrio tubiashii (RDP 0.939); Vibrio Lu1 (RDP 0.962), a marine luminous bacterium; and Vibrio rumoiensis (RDP 0.962). The remaining marine isolates whose siderophore structures have been partially characterized are the α Proteobacteria SP18 (which produces a C-10:1 amphiphilic siderophore with a short peptide headgroup; structure determination in progress, J.D.M. and A.B.) and SP25. Both of these strains are from the Rhizobium–Agrobacterium group. SP18 was isolated from seawater taken off Scripps Pier in San Diego, CA, during a phytoplankton bloom and is closely related (RDP 0.982) to Ochrobactrum strain S1. SP25 was isolated from the sediment of an estuary in San Diego and is closely related (RDP 0.988) to two environmental isolates, MBIC3993 and MBIC1535, from the Stappia stellulata subgroup.
Figure 4.
Phylogenetic tree of siderophore-producing bacteria based on maximum parsimony analysis of SSU rRNA DNA sequences. The α, β, and γ Proteobacteria as well as the high G + C Gram-positive bacteria are marked. This latter clade, containing Mycobacterium and Nocardia, was used as the outgroup. Marine isolates are in bold type. The tree shown here is one of two equally parsimonious trees, which differ only in a minor rearrangement within the Vibrio group. Bootstrap values >75% are shown. For clarity, the bootstrap value (87) between the γ Proteobacteria DS40M6, 8 and PCOB-2 has been omitted. Amphibactin G, marinobactin E, and aquachelin D (16) are amphiphilic siderophores produced by marine bacteria. Also included are most of the terrestrially produced amphiphilic siderophores, corrugatin (29), ornibactin (26), rhizobactin 1021 (30), acinetoferrin (27), and mycobactin S (25). Some bacteria, designated with a star (*) next to their name, make suites of siderophores like the amphibactins. P. luteoviolacea refers to the previously characterized marine isolate, Alteromonas luteoviolacea strain S2 (15).
Discussion
Hundreds of siderophores have been structurally characterized to date, but few siderophores produced by terrestrial bacteria are amphiphilic (e.g., ornibactin, corrugatin, acinetoferrin, rhizobactin 1021, carboxymycobactins, and the mycobactins, Fig. 4) (25–30). The discovery of the amphibactins extends the number of relatively rare amphiphilic siderophores to the Vibrionaceae family of the γ Proteobacteria. Furthermore, the amphibactins are the first fully characterized class of cellular bound siderophores outside of the high G + C Gram-positive bacterial species.
It is intriguing that nearly half of the known marine siderophore structures are amphiphilic, a far higher percentage than for known terrestrial siderophores. We have found amphiphilic siderophores in diverse marine bacteria spanning the α and γ subgroups of Gram-negative bacteria (Fig. 4), and we have found them in high percentages relative to nonamphiphilic siderophores. There is also some evidence for a cell-associated catecholate siderophore in the freshwater cyanobacteria Synechococcus PCC 6301 (19). Why do aquatic bacteria seem to produce more amphiphilic siderophores than their terrestrial counterparts? Certainly it could be argued that our selection of marine siderophores is biased by the ability of certain species to survive in standard laboratory culture conditions. For instance, very little is known about the iron requirements or uptake mechanisms of bacteria with SSU rRNA gene sequences that are abundant in environmental clone libraries, such as the SAR11 cluster of α Proteobacteria (48, 49). However, one could argue that similar biases exist for terrestrial bacteria whose siderophores have been structurally characterized and where amphiphilic siderophores are relatively rare.
These results suggest that marine (and freshwater) bacteria have evolved Fe(III) scavenging strategies to suit aquatic environments, where diffusion of siderophores away from the cell is potentially a serious problem (50, 51). The addition of fatty acid tails to siderophore scaffolds broadens the iron(III) uptake strategies available to marine bacteria. The continuum of marine bacterial strategies is perhaps bracketed on one side by use of traditional freely diffusible siderophores (i.e., alterobactins, ref. 15) and on the other side by the use of tethered and cell-associated siderophores (i.e., amphibactins, this study). Within this continuum, siderophores with large variations in membrane partitioning, such as the marinobactins and the aquachelins, could produce a gradient of siderophores with differing hydrophobicities extending away from the cell (Table 1, ref. 23). Each strategy may impact how bound iron(III) is imported into bacterial cells, and each may represent an adaptation to a different niche in the oceanic environment.
Iron uptake by cells that secrete freely diffusible siderophores may be efficient in aquatic environments only if the concentration of iron chelators in the bulk medium is high enough that the bacteria can use siderophores produced by other cells (of their own species or others) (50). In contrast, cells that secrete suites of siderophores with different membrane affinities (marinobactins and aquachelins) may counter diffusion by creating gradients of siderophores, with cell association of the more hydrophobic siderophores (i.e., saturated and longer-chain fatty acids) and release of the more hydrophilic siderophores (i.e., extending beyond the bacterium). The transport of iron, then, could be accomplished with a “bucket-brigade” approach analogous to that proposed for iron exchange between the mycobacteria siderophores, carboxymycobactin and mycobactin. These compounds primarily differ by the length and terminal modification of fatty acids, as well as cellular location. The mycobactins have significantly longer fatty acids and are cell-associated, whereas the carboxymycobactins are secreted extracellularly. The iron accumulation strategy of mycobacteria has similarly been suggested to proceed through a shuttle mechanism whereby the freely diffusible carboxymycobactins transfer iron to the cell-associated mycobactins (28). In contrast, the membrane partition coefficients of the amphibactins span a smaller range than the marinobactins (Table 1, ref. 23) and may be more specifically cell-associated. In this case, Vibrio sp. R-10 may counter diffusion by simply limiting the release of siderophores from the cell surface.
Siderophores have been studied for many years. The pioneering studies that elucidated the many facets of siderophore-mediated iron acquisition, including ferric stability constants, iron(III) exchange kinetics, the specificity and stereoselectivity of iron(III)-siderophore uptake into bacteria, and recently the structures of siderophore outer membrane receptors (21, 22, 24, 52–58), have paved the way for studies of iron acquisition by marine and other aquatic bacteria. The majority of these studies have been with water-soluble and secreted siderophores as a result of the predominance of water-soluble siderophores discovered to date in the terrestrial environment. Our discovery of the amphibactins, another suite of amphiphilic peptidic siderophores, demonstrates the increasing recognition of the prevalence of amphiphilic siderophores produced by marine bacteria, and extends the traditional continuum of iron(III)-chelation strategies.
Acknowledgments
We thank Deeanne Edwards for technical assistance. We are grateful to the National Science Foundation and the U.S. Department of Energy for funding through the Environmental Molecular Science Institute (The Center for Environmental Bioinorganic Chemistry, Princeton), National Science Foundation Grant CHE9810248 (to A.B. and M.G.H.), National Institutes of Health Grant GM38130 (to A.B.), and the W. M. Keck Foundation (to A.B.). Partial support was provided by California Sea Grant NA66RG0447, Project R/MP-76 (to A.B.), and Sea Grant Fellowship support (to J.N.C.-F. and J.S.M.) funded by a grant from the National Sea Grant College Program, National Oceanic and Atmospheric Administration, and the U.S. Department of Commerce.
Abbreviations
- DMPC
dimyristoylphosphatidylcholine
- SSU
small subunit
- RDP
Ribosomal Database Project
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been reported in the GenBank database (accession nos. AY217768 Ochrobactrum SP18; AY217769 Agrobacterium SP25; AY217770 Vibrio R-10, 5′ end of the SSU rRNA gene; AY217771 Vibrio R-10, 3′ end; AY217772 Vibrio BLI-41; and AY217773 Pseudoalteromonas luteoviolacea strain S2.
References
- 1.Bruland K W, Donat J R, Hutchins D A. Limnol Oceanogr. 1991;36:1555–1577. [Google Scholar]
- 2.Mann E L, Chisholm S W. Limnol Oceanogr. 2000;45:1067–1076. [Google Scholar]
- 3.Landry M R, Barber R T, Bidigare R R, Chai F, Coale K H, Dam H G, Lewis M R, Lindley S T, McCarthy J J, Roman M R, et al. Limnol Oceanogr. 1997;42:405–418. [Google Scholar]
- 4.Hutchins D A, DiTullio G R, Zhang Y, Bruland K W. Limnol Oceanogr. 1998;43:1037–1054. [Google Scholar]
- 5.Boyd P W, Watson A J, Law C S, Abraham E R, Trull T, Murdoch R, Bakker D C E, Bowie A R, Buesseler K O, Chang H, et al. Nature. 2000;407:695–702. doi: 10.1038/35037500. [DOI] [PubMed] [Google Scholar]
- 6.Gervais F, Riebesell U, Gorbunov M Y. Limnol Oceanogr. 2002;47:1324–1335. [Google Scholar]
- 7.Behrenfeld M J, Bale A J, Kolber Z S, Aiken J, Falkowski P G. Nature. 1996;383:508–511. [Google Scholar]
- 8.Coale K H, Johnson K S, Fitzwater S E, Gordon R M, Tanner S, Chavez F P, Ferioli L, Sakamoto C, Rogers P, Millero F, et al. Nature. 1996;383:495–501. doi: 10.1038/383495a0. [DOI] [PubMed] [Google Scholar]
- 9.Tortell P D, Maldonado M T, Granger J, Price N M. FEMS Microbiol Ecol. 1999;29:1–11. [Google Scholar]
- 10.Cochlan W P. Limnol Oceanogr. 2001;46:428–435. [Google Scholar]
- 11.Pakulski J D, Coffin R B, Kelley C A, Holder S L, Downer R, Aas P, Lyons M M, Jeffery W H. Nature. 1996;383:133–134. [Google Scholar]
- 12.Kirchman D L, Meon B, Cottrell M T, Hutchins D A, Weeks D, Bruland K W. Limnol Oceanogr. 2000;45:1681–1688. [Google Scholar]
- 13.Haygood M G, Holt P D, Butler A. Limnol Oceanogr. 1993;38:1091–1097. [Google Scholar]
- 14.Butler A. Science. 1998;281:207–210. doi: 10.1126/science.281.5374.207. [DOI] [PubMed] [Google Scholar]
- 15.Reid R T, Butler A. Limnol Oceanogr. 1991;36:1783–1792. [Google Scholar]
- 16.Martinez J S, Zhang G P, Holt P D, Jung H-T, Carrano C J, Haygood M G, Butler A. Science. 2000;287:1245–1247. doi: 10.1126/science.287.5456.1245. [DOI] [PubMed] [Google Scholar]
- 17.Martinez J S, Haygood M G, Butler A. Limnol Oceanogr. 2001;46:420–424. [Google Scholar]
- 18.Winkelmann G, Schmid D G, Nicholson G, Jung G, Colquhoun D J. BioMetals. 2002;15:153–160. doi: 10.1023/a:1015206419613. [DOI] [PubMed] [Google Scholar]
- 19.Wilhelm S W, Trick C G. Limnol Oceanogr. 1994;39:1979–1984. [Google Scholar]
- 20.Granger J, Price N M. Limnol Oceanogr. 1999;44:541–555. [Google Scholar]
- 21.Raymond K N, Miller G, Matzanke B F. In: Topics in Current Chemistry. Boschke F L, editor. Heidelberg: Springer; 1984. pp. 50–102. [Google Scholar]
- 22.Crumbliss A L. In: Handbook of Microbial Iron Chelates. Winkelmann G, editor. Boca Raton, FL: CRC; 1991. pp. 172–233. [Google Scholar]
- 23.Xu G, Martinez J S, Groves J T, Butler A. J Am Chem Soc. 2002;124:13408–13415. doi: 10.1021/ja026768w. [DOI] [PubMed] [Google Scholar]
- 24.Winkelmann G. Handbook of Microbial Iron Chelates. Boca Raton, FL: CRC; 1991. [Google Scholar]
- 25.Ratledge C. In: The Mycobacteria, a Sourcebook. Kubica G P, Wayne L G, editors. New York: Dekker; 1984. pp. 603–627. [Google Scholar]
- 26.Stephan H, Freund S, Beck W, Jung G, Meyer J-M, Winkelmann G. BioMetals. 1993;6:93–100. doi: 10.1007/BF00140109. [DOI] [PubMed] [Google Scholar]
- 27.Okujo N, Sakakibara Y, Yoshida T, Yamamoto S. BioMetals. 1994;7:170–176. doi: 10.1007/BF00140488. [DOI] [PubMed] [Google Scholar]
- 28.Gobin J, Horowitz M A. J Exp Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Risse D, Beiderbeck H, Taraz K, Budzikiewicz H, Gustine D. Z Naturforsch. 1998;53c:295–304. [Google Scholar]
- 30.Lynch D, O'Brien J, Welch T, Clarke P, Cuiv P O, Crosa J H, O'Connell M. J Bacteriol. 2001;183:2576–2585. doi: 10.1128/JB.183.8.2576-2585.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brennan P J. In: Microbial Lipids. Ratledge C, Wilkinson S G, editors. London: Academic; 1988. pp. 203–298. [Google Scholar]
- 32.Murakami Y, Kato S, Nakajima M, Matsuoka M, Kawai H, Shin-Ya K, Seto H. J Antibiotics. 1996;49:839–845. doi: 10.7164/antibiotics.49.839. [DOI] [PubMed] [Google Scholar]
- 33.Suenaga K, Kokubo S, Shinohara C, Tsuji T, Uemura D. Tetrahedron Lett. 1999;40:1945–1948. [Google Scholar]
- 34.Kokubo S, Suenaga K, Shinohara C, Tsuji T, Uemura D. Tetrahedron. 2000;56:6435–6440. [Google Scholar]
- 35.Ratledge C, Patel P V. J Gen Microbiol. 1976;93:141–152. doi: 10.1099/00221287-93-1-141. [DOI] [PubMed] [Google Scholar]
- 36.de Voss J J, Rutter K, Schroeder B G, Su H, Zhu Y Q, Barry C E., III Proc Natl Acad Sci USA. 2000;97:1252–1257. doi: 10.1073/pnas.97.3.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sritharan M. World J Microbiol Biotechnol. 2000;16:769–780. [Google Scholar]
- 38.Schwyn B, Neilands J B. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 39.Marfey P. Carlsberg Res Commun. 1984;49:591–596. [Google Scholar]
- 40.Lane D J. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. New York: Wiley; 1991. pp. 115–175. [Google Scholar]
- 41.Maidak B L, Cole J R, Lilburn T G, Parker C T, Jr, Saxman P R, Stredwidck J M, Garrity G M, Li B, Olsen G J, Pramanik S, et al. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roepstroff P, Fohlman J. Biomed Mass Spectrom. 1984;11:601. doi: 10.1002/bms.1200111109. [DOI] [PubMed] [Google Scholar]
- 43.Ratledge C, Wilkinson S G. Microbial Lipids. London: Academic; 1988. pp. 23–53. [Google Scholar]
- 44.Murakami K, Fuse H, Takimura O, Kamimura K, Yamaoka Y. J Mar Biotechnol. 1998;6:76–79. [Google Scholar]
- 45.Buyer J S, de Lorenzo V, Neilands J B. Appl Environ Microbiol. 1991;57:2246–2250. doi: 10.1128/aem.57.8.2246-2250.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi A. J Antibiotics. 1987;40:1671–1676. doi: 10.7164/antibiotics.40.1671. [DOI] [PubMed] [Google Scholar]
- 47.Jalal M A F, Hossain M B, van der Helm D, Sanders-Loehr J, Actis L A, Crosa J H. J Am Chem Soc. 1989;111:292–296. [Google Scholar]
- 48.Rappe M S, Connan S A, Vergin K L, Giovannoni S J. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 49.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volker C, Wolf-Gladrow D A. Mar Chem. 1999;65:227–244. [Google Scholar]
- 51.Hutchins D A, Rueter J G, Fish W. Limnol Oceanogr. 1991;36:1–12. [Google Scholar]
- 52.Carrano C J, Raymond K N. J Bacteriol. 1978;136:69–74. doi: 10.1128/jb.136.1.69-74.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carrano C J, Jordon M, Drechsel H, Schmid D G, Winkelmann G. BioMetals. 2001;14:119–125. doi: 10.1023/a:1016633529461. [DOI] [PubMed] [Google Scholar]
- 54.Cohen S M, Raymond K N. Inorg Chem. 2000;39:3624–3631. doi: 10.1021/ic990608c. [DOI] [PubMed] [Google Scholar]
- 55.Clarke T E, Braun V, Winkelmann G, Tari L W, Vogel H J. J Biol Chem. 2002;277:13966–13972. doi: 10.1074/jbc.M109385200. [DOI] [PubMed] [Google Scholar]
- 56.Buchanan S K, Smith B S, Venkatramani L, Xia D, Esser L, Palnitkar M, Chakraborty R, van der Helm D, Deisenhofer J. Nat Struct Biol. 1999;6:56–63. doi: 10.1038/4931. [DOI] [PubMed] [Google Scholar]
- 57.Ferguson A D, Hofmann E, Coulton J W, Diederichs K, Welte W. Science. 1998;282:2215–2220. doi: 10.1126/science.282.5397.2215. [DOI] [PubMed] [Google Scholar]
- 58.Ferguson A D, Chakraborty R, Smith B S, Esser L, van der Helm D, Deisenhofer J. Science. 2002;295:1715–1719. doi: 10.1126/science.1067313. [DOI] [PubMed] [Google Scholar]