Abstract
The protein CopC from Pseudomonas syringae has been found capable of binding copper(I) and copper(II) at two different sites, occupied either one at a time or simultaneously. The protein, consisting of 102 amino acids, is known to bind copper(II) in a position that is now found consistent with a coordination arrangement including His-1, Glu-27, Asp-89, and His-91. A full solution structure analysis is reported here for Cu(I)-CopC. The copper(I) site is constituted by His-48 and three of the four Met residues (40, 43, 46, 51), which are clustered in a Met-rich region. Both copper binding sites have been characterized through extended x-ray absorption fine structure studies. They represent novel coordination environments for copper in proteins. The two sites are ≈30 Å far apart and have little affinity for the ion in the other oxidation state. Oxidation of Cu(I)-CopC or reduction of Cu(II)-CopC causes migration of copper from one site to the other. This behavior is observed both in NMR and EXAFS studies and indicates that CopC can exchange copper between two sites activated by a redox switch. CopC resides in the periplasm of Gram-negative bacteria where there is a multicopper oxidase, CopA, which may modulate the redox state of copper. CopC and CopA are coded in the same operon, responsible for copper resistance. These peculiar and novel properties of CopC are discussed with respect to their relevance for copper homeostasis.
Copper is an essential element for all living organisms, where it constitutes the prosthetic, active group of many proteins and enzymes (1). Copper is also very toxic in the free form because of its ability to produce radicals by cycling between oxidized Cu(II) and reduced Cu(I). Therefore, intracellular copper should be always kept complexed, within a tightly controlled copper homeostatic system (2, 3). An excess of copper is toxic particularly for lower organisms, such as bacteria and fungi, and indeed copper compounds are extensively used in agriculture to control plant pathogens. Some organisms, however, develop resistance on exposure to high copper levels (4–6).
Copper-resistant strains of Pseudomonas syringae pathovar (pv.) tomato are characterized by a 35-kb plasmid pPT23D (7). The plasmid was first isolated in Californian tomato fields exposed to high levels of copper compounds (7) and contains an operon (cop) encoding four proteins, CopABCD, acting under the control of a copper inducible promoter requiring the regulatory genes CopR and CopS, coded immediately downstream of CopD (8). This plasmid is able to confer copper resistance to the host strains of Gram-negative bacteria and protects the cells by sequestering the excess of copper in the periplasm and in the outer membrane (4); however, the molecular mechanisms of this process are not known.
The sequences of 18 proteins highly similar to CopC are known (9). In Gram-negative bacteria, such as P. syringae, CopC is a soluble protein of ≈100 amino acids and is located in the periplasm. Homologous proteins have been also found in Gram-positive bacteria. The structure of apoCopC from P. syringae has been solved by NMR (9). It shows a Greek key β-barrel similar to that of blue-copper proteins. The protein binds one Cu(II) ion (9) at the N-terminal end of the barrel, in a region encompassing some loops connecting various β-strands. The position of this Cu(II) site is similar to that of the redox site in cupredoxins although copper is of type II in this Cu(II)-CopC (9) and in a homologous protein (10, 11). CopC also shows a region containing four Met residues arranged as [M(X)nM]m motif, which is found in other proteins involved in copper homeostasis, like the copper transport permeases responsible for copper transport inside eukaryotic cells (12). Recently, the Met-rich motifs of these high-affinity copper transporters have been shown to be essential for copper uptake in yeast and humans (13).
To understand the copper binding properties of CopC, we have carried out NMR and extended x-ray absorption fine structure (EXAFS) studies on both the oxidized and reduced Cu-loaded proteins. Our results indicate a redox-induced reversible metal transfer between two regions of the protein far apart and with very different coordination environments. The relevance of this peculiar process is discussed within the frame of a genome-wide analysis of possible pathways for copper trafficking.
Materials and Methods
Protein and NMR Sample Preparation.
CopC was over-expressed and purified from the construct pAT2 in the Escherichia coli strain BL21 (DE3) pLysS as described (9). Briefly, overnight induction was performed in rich Luria-Bertani (LB) broth for unlabeled samples or M9 medium (14) supplemented with trace metals, vitamins and 2 g/liter 15NH4(SO4)2 and 3 g/liter 13C-labeled glucose as 15N and 13C sources. The protein was extracted by using a freeze–thaw procedure and isolated by ion exchange chromatography. NMR samples were prepared in 100 mM sodium phosphate buffer, pH 7, containing 10% D2O, with a protein concentration of ≈2–3 mM.
NMR Spectroscopy and Structure Calculations.
Binding of Cu(II) to apoCopC was followed through electronic and NMR spectroscopy as previously described (9). Binding of Cu(I) to apoCopC was followed through NMR spectra. To a 3 mM 13C/15N apoCopC sample a stepwise addition of up to 2 equivalents of Cu(I), obtained by reducing a CuSO4 solution with sodium ascorbate, was done under inert atmosphere and 2D 1H-15N heteronuclear single quantum coherence (HSQC) spectra were acquired for each addition.
Reoxidation of the Cu(I)-CopC sample was carried out by adding H2O2. The concentration of H2O2 was determined spectrophotometrically at 240 nm by using an extinction coefficient of 39.4 M−1⋅cm−1 (15). Reduction of the Cu(II)-CopC sample was carried out by addition of ascorbate in stoichiometric amounts under inert atmosphere.
The NMR spectra for resonance assignment and structure determination were acquired on 13C/15N-enriched CopC, loaded with one equivalent of Cu(I) (Cu(I)-CopC hereafter) and are summarized in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. All of these experiments were recorded on a Bruker DRX 500 spectrometer operating at a proton nominal frequency of 500.13 MHz and equipped with a cryoprobe, with the exception of 2D and 3D 13C- and 15N-edited NOESY maps, which were recorded on a Ultra Shield 700 spectrometer operating at 700.13 MHz, by using a triple resonance probe equipped with pulsed field gradients along the xyz-axes. All 3D and 2D spectra were collected at 298 K, processed by using the standard Bruker software (xwinnmr) and analyzed with xeasy (16). NOESY maps were also collected at 313 K.
Distance constraints for structure determination were obtained by converting nuclear Overhauser effect (NOE) cross-peak intensities from 3D and 2D NOESY maps into upper distance limits of interproton distances with the program caliba (17). Stereospecific assignments of diastereotopic protons have been obtained by the analysis of the HNHB experiment (18) and through the program glomsa (17). 3JHNHα coupling constants were determined through the HNHA experiment and transformed into backbone dihedral φ angles through the Karplus equation (19). Backbone dihedral ψ angles for residue (i-1) were also determined from the ratio of the intensities of the dHαHN(i-1,i) and dHαHN(i,i) NOEs. χ1 torsion angle constraints were determined from the HNHB experiment as reported (18). Hydrogen bond constraints were obtained by using a TROSY version of the long-range HNCO experiment (20) and used as structural constraints.
The secondary structure elements were determined on the basis of the chemical shift index (21), of the 3JHNHα coupling constants and of the backbone NOEs.
Structure calculations were performed through iterative cycles of dyana (22). The program corma (23) was used to check that all of the cross-peaks expected on the basis of the structure were assigned and that no observed NOE (above the noise level) was left unassigned.
The 35 conformers with the lowest target function constitute the final family. REM was then applied to each member of the family by using the AMBER 6 package (24). The NOE and torsion angle constraints were applied with force constants of 50 kcal⋅mol−1⋅Å−2 and 32 kcal⋅mol−1⋅rad−2, respectively.
The quality of the structures was evaluated by using the program procheck-nmr (25). Experimental restraints were analyzed by using the program aqua (25, 26). Structure calculations were run on a Linux processor cluster.
Relaxation experiments were performed on 15N labeled Cu(I)-CopC. Data were collected on Bruker Avance 600 spectrometer, operating at proton nominal frequency of 600.13 MHz. A triple resonance probe equipped with pulsed field gradients along the z axis was used. The 15N backbone relaxation rates, R1 and R2, were measured as described (27). R2 were also measured with longer refocusing times (τCPMG) to determine the occurrence of exchange processes (27, 28). The overall rotational correlation time τm value and the rotational diffusion tensor were estimated from the R2/R1 ratio with the program quadric_diffusion (29, 30), by using the 3D structure of Cu(I)-CopC.
X-Ray Absorption Spectroscopy (XAS) Data Collection and Analysis.
For XAS measurements, two samples were prepared under nitrogen atmosphere immediately before recording the spectra. To two 3.0 mM apoCopC solutions in 100 mM phosphate buffer at pH 7.0, one equivalent each of Cu(II) (as CuSO4) was added. In one of the samples, Cu(II) was reduced by addition of one equivalent of sodium ascorbate. The two protein samples were prepared, always under inert atmosphere, in plastic cells covered with Kapton windows. The sample cells were then mounted in a two-stage Displex cryostat (modified Oxford instruments) and kept at 20 K during data collection.
XAS data were collected at Deutsches Elektronen Synchrotron (DESY, Hamburg, Germany) at the European Molecular Biology Laboratory bending magnet beam line D2 by using a Si (111) double monochromator for the measurement at the copper edge, with the DESY storage ring operating under normal conditions (4.5 GeV, 90–140 mA). Ionization chambers in front and behind the sample were used to monitor the beam intensity.
The XAS data were recorded by measuring the Cu-Kα fluorescence by using a Canberra 13-element solid-state detector over the energy range from 8,735 to 9,875 eV by using variable energy step widths. In the x-ray absorption near-edge structure and EXAFS regions steps of 0.3 and 0.5–1.2 eV were used, respectively. An absolute energy calibration of the spectra was obtained by following a reported procedure (31). Ten and six scans were recorded for Cu(II)-CopC and Cu(I)-CopC, respectively, for a total of more than 1.0 and 0.6 million counts per experimental point. The scans were then averaged to obtain good signal-to-noise statistics. Data reduction to extract the EXAFS spectrum was based on standard procedures and performed with a local set of programs (32). The full, k3 weighted, EXAFS spectra (17–750 eV above E0) and their Fourier transforms (FT) calculated over the range 3.0–14.0 Å−1 have been compared with theoretical simulations obtained by using the rapid curved multiple scattering theory implemented in the set of programs EXCURVE9.20 (33). The data analysis was carried out by using published protocols (34, 35).
The quality of the fit was assessed by the fit function through the parameter ɛ2, which accounts for the degree of over determinacy (36) and by the R factor as defined within EXCURVE9.20 (33).
Results
XAS Spectra Analysis.
To characterize the potential metal binding sites in CopC, x-ray absorption near-end structure and EXAFS spectra were recorded on protein samples with one equivalent of Cu(II) and after its reduction to Cu(I).
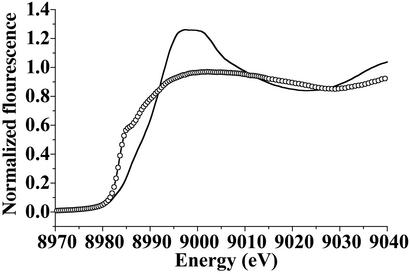
Fig. 1 shows the edge region of Cu(II)-CopC and Cu(I)-CopC where the changes in the edge energy and shape clearly identify the two different copper oxidation states. The preedge and edge absorptions of Cu(II) and Cu(I) compounds are known to be related to the coordination geometry of the metal environment (37–39). The preedge region at ≈8,980 eV in Cu(II) complexes shows a characteristic absorption caused by electronic transitions between the 1s and empty 3d atomic orbitals whose intensity is proportional to the deviation from centrosimmetry of the Cu(II) coordination geometry (37, 40). In Cu(II)-CopC (Fig. 1, solid line), the 1s-3d preedge is essentially absent, indicating a nearly centrosymmetrical coordination environment for Cu(II) bound to CopC. Cu(I)-CopC shows the characteristic edge of Cu(I) compounds (Fig. 1, open circles), with the typical absorption at 8,984.5 eV, assigned to a 1s → 4p absorption (38). A number of studies have shown that the Cu(I) coordination number and/or geometry are correlated with the intensity of this transition (38, 39). The height of the 1s-4p transition present in the edge of the Cu(I)-CopC spectrum suggests 3- or 4-coordination for Cu(I) (39, 41).
Figure 1.
Comparison of the Cu K-edge regions of Cu(II)-CopC (solid line) and Cu(I)-CopC (open circles).
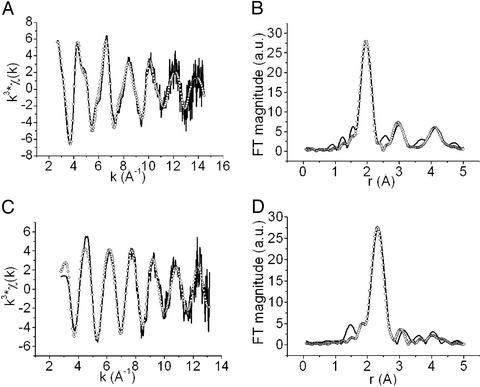
The k3 weighted EXAFS spectra and the respective Fourier transforms for the two samples superimposed with the best fits (open circles) from theoretical simulations are shown in Fig. 2; the parameters obtained from these fits are reported in Table 1.
Figure 2.
Experimental (solid line) and simulated (open circles) EXAFS spectra (A and C) and their FTs (B and D) of Cu(II)-CopC (A and B) and Cu(I)-CopC (C and D).
Table 1.
Fitting results of the EXAFS spectra on copper-bound CopC
| Ligand | Distance, Å* | DW-factor 2σ2, Å2* | ɛ2 × 101 | R-factor | |
|---|---|---|---|---|---|
| Cu(II)-CopC† | |||||
| Fit 1 | 2 His | 1.99 (1) | 0.016 (6) | 8.9 | 0.40 |
| 2 N/O | 1.97 (1) | 0.004 (4) | |||
| Fit 2 | 2 His | 1.99 (1) | 0.016 (7) | 7.2 | 0.34 |
| 2 N/O | 1.97 (1) | 0.004 (4) | |||
| 2 N/O | 2.83 (2) | 0.020 (6) | |||
| Cu(I)-CopC† | |||||
| Fit 1 | 1 His | 1.95 (1) | 0.014 (6) | 9.4 | 0.37 |
| 2 S | 2.31 (2) | 0.008 (4) | |||
| Fit 2 | 1 His | 1.95 (1) | 0.009 (4) | 7.7 | 0.33 |
| 3 S | 2.30 (2) | 0.012 (6) |
The numbers in parentheses represent the estimated standard deviations, obtained from the least-squares covariance matrix.
The samples are in 100 mM phosphate buffer at pH 7.0.
The EXAFS spectrum of Cu(II)-CopC (Fig. 2A) shows the typical camelback features of the oscillations, revealing histidine binding to the metal ion. This is reflected by the outer shell peaks at ≈3.0 and 4.2 Å present in the FT because of the histidine carbon backscattering. The fitting of the spectrum reveals that the Cu(II) ion is bound to two His at 1.99 Å and two other O/N ligands at 1.97 Å. Addition of two further O/N ligands at 2.83 Å improves the fit of the spectrum by 6% bringing the R factor to 0.34. The Cu(II) location within the frame of the apoprotein structure was already determined through NMR titrations (9). The present EXAFS data indicates that the two His (1 and 91) occupy two equatorial positions. Two other equatorial ligands are probably oxygen atoms from the carboxylate groups of Asp or Glu residues, in view of the quite short coordination distance found. An apical position is taken by one water molecule, which was already observed through water relaxation measurements (9). Finally, we may note that Lys-3 is close to the Cu(II) binding site, but no further info is available for it. A tetragonal arrangement is consistent with the EPR data (9). This copper binding site is called from now on the A site.
The EXAFS spectrum of Cu(I)-CopC is indicative of a completely different coordination sphere (Fig. 2C). The EXAFS and the FT of Cu(I)-CopC are well reproduced by a coordination polyhedron constituted by two or three S atoms at 2.30 Å and one His nitrogen at 1.95 Å. The fit of these data shows unequivocally the presence of sulfur in the first coordination shell of the Cu(I) ion. The quality of the fit is slightly better if three instead of two sulfur atoms are at a distance of 2.30 Å, typical of 3–4-coordinated Cu(I) complexes (38, 39). By inspection of the apoCopC structure, a region located in the loop between strands β3 and β4a and containing four Met residues (40, 43, 46, 51) and one His (His-48) is present at the C-terminal end of the β-barrel. Three of these four Met residues might be the ligands of Cu(I) together with His-48. This site is called B site.
Structural and Dynamical Characterization of Cu(I)-CopC in Solution.
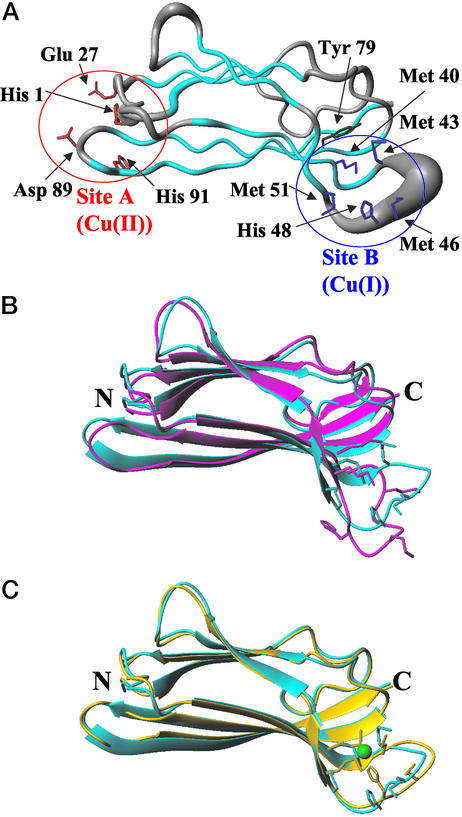
Addition of one equivalent of Cu(I) to apoCopC produces some spectral changes which are essentially localized in the B site and on the residues around it. Resonance assignments of Cu(I)-CopC are reported in Table 3, which is published as supporting information on the PNAS web site. Scalar correlation spectra and NOESY maps of apo- and Cu(I)-CopC show very similar patterns with a few changes in shifts and an increased number of NOEs around the B site in the latter system (Fig. 5 A and B, which is published as supporting information on the PNAS web site). From chemical shift index analysis and 1H-1H connectivities, it can be seen that the secondary structure elements, i.e., β-strands arranged in a β-barrel fold, are completely maintained. The 2D and 3D NOESY spectra provide 1,464 meaningful 1H-1H distances (listed in Table 4, which is published as supporting information on the PNAS web site) and 51 stereospecific assignments (Table 5, which is published as supporting information on the PNAS web site) which, together with 129 dihedral angle (Table 6, which is published as supporting information on the PNAS web site) and 34 hydrogen bond constraints (Table 7, which is published as supporting information on the PNAS web site), allowed us to calculate the solution of the Cu(I)-CopC protein. A statistical analysis of the structure is reported in Table 8, which is published as supporting information on the PNAS web site. In Fig. 3A, a 35 conformer family of Cu(I)-CopC is represented as a tube, whose radius is proportional to the backbone rms deviation (rmsd) of each residue (Fig. 6, which is published as supporting information on the PNAS web site). It can be seen that the structure is very well defined, with a slight increase in disorder in two loops connecting β-strands. In Fig. 3B, the mean energy minimized structures of Cu(I)- and apo-CopC are superimposed. The global backbone rmsd between Cu(I)- and apo-CopC structures is 2.1 Å. The region where the two structures differ more than the structural resolution (i.e., the rmsd between the two mean minimized structures is larger than the sum of rmsd of each family, as shown in Fig. 5C) involves the loop between strands β3 (residues 35–43) and β4a (residues 51–53) (see Fig. 3B). This loop is protruded toward the solvent to a different extent for the two systems. Four Met residues (i.e., 40, 43, 46, and 51), which are potential Cu(I) ligands, are located in this region. Met-40 and -51 are on two different antiparallel β-strands and are facing each other. Met-43 is found at the end of strand β4a, where residues 41 and 42 form a β-bulge with Thr-78 on strand β6, which is in turn stabilized by a hydrogen bond between the OH group of the conserved Tyr-79 and the backbone of Thr-75. Met-46 is located at the extreme end of the loop which is very exposed to the solvent. Finally, His-48 is found in front of Met-43. Thus, three of five potential Cu(I) binding residues of site B, i.e., Met-40, -43, and -51, are constrained to some extent by the secondary structure in this region. The ring of His-48 is very close to this cluster of three Met residues, as supported by several NOEs of its ring proton Hδ2. The EXAFS and NMR data point at His-48 as a metal ligand together with three of four Met residues.
Figure 3.
(A) Solution structure of Cu(I)-CopC represented as a tube whose radius is proportional to the backbone rmsd of each residue. β-Strands are colored in cyan. Conserved residues in the Cu(I) binding site (Site B) are shown in blue. Conserved residues in the Cu(II) binding site (Site A) are shown in red. Tyr 79 is shown in green. The figure was generated with MOLMOL (56). (B) Overlay of the apoCopC structure (in magenta) (9) and the structure of Cu(I)-CopC calculated without any link to the copper atom (in cyan). (C) Overlay of the structures of Cu(I)-CopC calculated without (cyan) and with (yellow) links between Cu(I) and the sulfur atoms of Met-40, -43, and -51, and the ring of His-48.
In the attempt to identify the Met residue not involved in Cu(I) coordination we have performed four different structure calculations by linking the copper atom to the ring of His-48 (without selecting Nɛ or Nδ coordination) and to three of four sulfur atoms of Met residues, excluding one Met at a time. Slightly better results in terms of target function are obtained when Met-46 is excluded (0.7 Å2 with respect to an average value of 1.0 Å2 in the other cases); however, the rmsd values between the mean structures obtained in each of these calculations and the mean structure calculated without any links to the copper are lower than the sum of rmsd of the families. This indicates that, with the present structure resolution, it is not possible to rule out one of the four possible combinations of ligands. In Fig. 3C the structure obtained when Met-46 is excluded is compared with the structure of Cu(I)-CopC obtained without any links to the metal. It is evident that the two structures are essentially identical and that the metal binding site is formed by experimental structural constraints without the need of the links to copper. In all cases the four donor atoms are found in a roughly tetrahedral geometry around the metal ion. Moreover, a very low solvent accessibility of copper in all cases indicates that the three Met ligands and His-48 provide a protective environment for the Cu(I) ion.
From the ratio between the transverse and longitudinal 15N relaxation rates, the correlation time for molecule reorientation (τm) can be determined, which is related to the actual molecular mass of the protein (42). For Cu(I)-CopC, τm is estimated to be 7.3 ± 0.3 ns, which compares with values of 6.2 ± 0.2 ns and 6.4 ± 0.3 ns for apoCopC and Cu(II)-CopC, respectively (9). The latter values are consistent with a monomeric state for a protein of this size. The ≈15% increase in τm observed for Cu(I)-CopC indicates that some aggregation occurs on Cu(I) binding. The partial aggregation of the protein may be the result of interactions of the Cu(I)-bound B site of one molecule with the ligands of the A site of another molecule in a head-tail type of interaction, either as discrete dimers or in a polymer fashion. The presence of aggregation is also supported by the behavior with temperature: at 313 K, the τm values of apo and Cu(I)-CopC are the same, indicating a monomeric state.
Reversibility of the Oxidation-Dependent Copper Binding.
From all these data it appears that two distinct copper-binding sites are present in the protein and that they are highly specific for the oxidation state. We have confirmed this result by characterizing the reduction of the Cu(II)-CopC sample and the oxidation of the Cu(I)-CopC sample, through 1H-15N HSQC spectra.
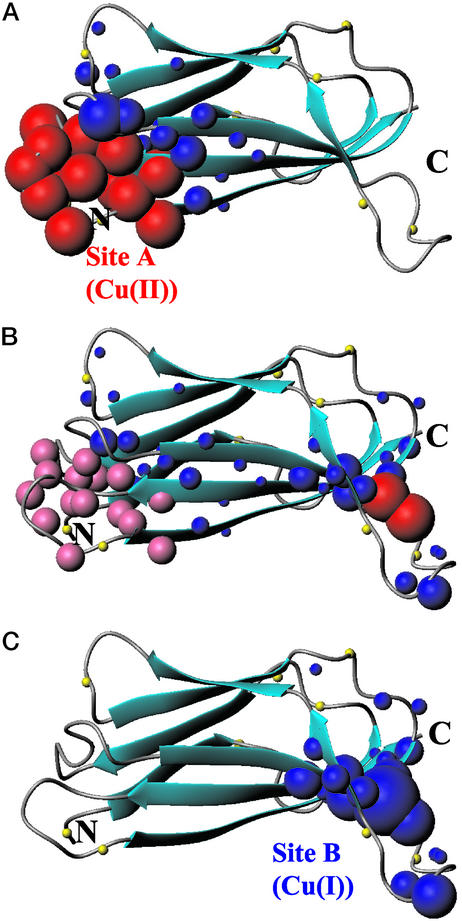
If the reducing agent is progressively added stepwise to a Cu(II)-CopC sample, we observe in the NMR spectra: (i) the reappearance of signals which were undetectable in the Cu(II)-CopC sample; (ii) the recovery of intensity for those cross-peaks that experience different shifts in the Cu(II)-CopC sample; (iii) a progressive variation in shift for residues at the B site giving rise, at the end of the titration, to a 1H-15N HSQC spectrum superimposable to that of the Cu(I)-CopC sample. These spectral changes are mapped in a schematic representation on the structure of apoCopC (Fig. 4). The reoxidation of the sample was carried out by adding H2O2 to the Cu(I)-CopC sample. The spectral changes involve the disappearance of the signals of residues in a sphere of ≈10 Å around A site and chemical shift changes of a second set of signals. The behavior of the different sets of signals indicates the reversibility of the process as all of the spectral changes occur in the reverse way with respect to the reduction and produce, at the end, a 1H-15N HSQC spectrum matching with that already reported for Cu(II)-CopC (9).
Figure 4.
Spectral changes occurring in 1H-15N HSQC spectra of apoCopC on addition of one equivalent of Cu(II) (A) and, on the latter adduct, of half (B) and one (C) equivalent of ascorbate are mapped on the apoCopC structure (9). Residues whose amide 1H-15N cross-peaks are broadened beyond detection are represented as red spheres. Pink spheres indicate residues whose signals experience slow exchange between the Cu(II) and the Cu(I) forms, giving rise to two sets with half intensity at 0.5 equivalent of ascorbate (B). Residues experiencing shift changes are shown as blue spheres whose radius is proportional to the shift change. The N atoms of proline residues are represented as yellow spheres.
These results indicate that the copper ion migrates from site A to B and vice versa on change in its redox state, and that the A and B metal binding sites have very high specificity for Cu(II) and Cu(I), respectively, being their affinity constants strongly dependent on the copper oxidation state.
Analysis of the NMR spectra of a solution containing CopC and both one equivalent of Cu(I) and one of Cu(II) shows that the protein is able to bind simultaneously Cu(II) at the A site and Cu(I) at the B site. Indeed, the final HSQC spectrum displays the sum of spectral changes observed in the spectra of Cu(I)- and Cu(II)-CopC. The same sample can be obtained by adding one equivalent of ascorbate to Cu(II)2-CopC or one equivalent of H2O2 to Cu(I)2-CopC.
Discussion
The present characterization points to a very peculiar and unprecedented behavior of the interaction of copper with the protein CopC. This protein has two completely distinct copper binding sites ≈30 Å apart. They seem specifically designed to selectively bind two copper ions, one in the reduced state and the other in the oxidized one. Cu(I) is bound to three sulfur atoms from Met residues and a histidyl nitrogen in a tetrahedral geometry (site B). The Cu(II) site can be classified as type II, with a tetragonal ligand environment including residues His-1, Glu-27, Asp-89, and His-91, and an apical position occupied by a water molecule (site A). Both sites represent novel copper binding sites in proteins, as confirmed by a search in the Scripps Metalloprotein Database (www.scripps.edu/research/metallo/).
Although redox-dependent changes of copper coordination in proteins have been reported so far (43–45), this is the first case, to our knowledge, that two distinct binding sites are found for the two copper oxidation states in the same protein. The evidence that CopC can be either simultaneously loaded with Cu(I) and Cu(II) or with one copper ion in either site suggests that no cooperativity is present between the two sites. Besides differences in copper coordination, the two binding sites in CopC provide two different types of potential molecular recognition and interaction patterns: site A is highly charged and would prefer an electrostatic type of interaction, at variance with site B, at the other end of the β-barrel, which is essentially neutral and could interact with other Met-rich domains.
CopC is encoded in the cop operon; the complete operon CopABCD, with the regulatory genes CopRS, provides resistance to increased extra cellular copper through an accumulation mechanism (4). It has been shown that the function of all these proteins is tightly connected and all are involved in copper homeostasis (4). CopA and CopB encode, respectively, a 72-kDa periplasmic protein binding 11 copper atoms and a 39-kDa outer membrane protein of unknown copper binding capacity. CopA is homologous to the multicopper oxidase CueO from E. coli, whose crystal structure (PDB ID 1KV7) was recently solved (46), and contains a large number of motifs rich in Met, His, and Asp. In particular, CopC and CopA share the MX2MXHX2M motif, which is now found to bind preferentially Cu(I). A few proposals have been suggested for the specific role of CopC in partnership with CopA, which might imply a metal-transfer function, an electron-transfer function or both (10). In the latter hypothesis, CopC and CopA may function together in copper resistance with CopA oxidizing Cu(I) bound to CopC to the less toxic Cu(II) form, as supported by genetic studies on the two homologues of these proteins from E. coli (47, 48). Alternatively, copper delivered by CopC to CopA could catalyze the oxidase activity of CopA, as suggested by the copper-inducible oxidation of siderophores by CueO in E. coli (49).
The outer membrane protein CopB is likely involved in copper efflux from the cell and has 5 repeats of the sequence DHSXMX2M as potential copper binding motifs. Either CopA or CopC can deliver Cu(I) to CopB by docking of the Met-rich regions, shared by all these proteins.
CopD is a 33-kDa and inner membrane protein containing several predicted transmembrane regions and some conserved His residues, but no Met-rich motifs.
In proteins homologous to CopC from Gram-positive bacteria, which lack a periplasmic space, and in some chromosomal copies of Gram-negative bacteria the Met-rich region is absent, as already pointed out (9), and the neighbor CopA and CopB genes are also missing. On the contrary, CopC is always found next to CopD, and Cu(II) binding residues are highly conserved in CopC-like sequences (9). Expression of CopC and CopD together increased sensitivity to copper and increased copper uptake, which was not seen when only CopC or CopD were expressed (50). Interestingly, Gram-positive bacteria express a CopCD fusion protein (9), providing an intriguing clue that CopC may functionally and directly interact with CopD to facilitate Cu(II) transfer, perhaps ensuring that copper essential for cellular function reaches the cytoplasm while CopA and B sequester and/or export the excess (51). CopC may act as a molecular switch between these two different pathways depending on the redox state of copper.
Still, the process of copper translocation might occur in a different manner if trafficking pathways of Cu(I) and Cu(II) intersect. Within this frame, we may speculate that the water molecule interacting with the solvent accessible Cu(II) ion in site A can be displaced by Met residues, either from CopA, CopB or from site B of a second CopC molecule in a head–tail interaction, but that only copper reduction triggers metal transfer to the new Met-rich site, having higher affinity for Cu(I). If the two sites with different affinity for the two redox states of copper are nearby, the mechanism of copper release and uptake occurs in a protective local environment, reminiscent of that of metallochaperones (52, 53), ensuring a safer metal transfer in the periplasmic space.
In eukaryotes, Cu(II) is reduced before entering the cell (54). The one-electron reduction of Cu(II) would strongly facilitate the displacement of the ligands to which the Cu(II) ion is bound, making the metal available for transport across the plasma membrane. Then, a widely conserved family of high affinity copper transport proteins (CTR proteins) mediates copper uptake at the plasma membrane (13). Interestingly, CTR proteins contain a series of clustered Met residues arranged as [M(X)nM]m motifs similar to those found in some members of the bacterial cop operon. Finally, from a genome-wide analysis of protein sequences it can be noted that [M(X)nM]m motifs are present in all eukaryotic genomes.
The present results on CopC may suggest a more general mechanism of copper trafficking triggered by redox-dependent coordination properties of the metal, already exploited in artificial molecular machines (55).
Supplementary Material
Acknowledgments
We thank Prof. D. A. Cooksey for kindly providing the plasmid pCop65. This work was financially supported by the European Commission (Contract QLG2-CT-2002-00988), by Italian Consiglio Nazionale delle Ricerche (99.00950.CT03) and by Ministero dell'Università e della Ricerca Scientifica e Tecnologica COFIN 2001, and carried out under the Large Scale Facility program (European Union Contract No. HPRI-CT-1999-00009). A.T. acknowledges Marie Curie Fellowship No. QLK4-CT-1999-51551.
Abbreviations
- EXAFS
extended x-ray absorption fine structure
- NOE
nuclear Overhauser effect
- HSQC
heteronuclear single quantum coherence
- rmsd
rms deviation
- XAS
x-ray absorption spectroscopy
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates of the average minimized structure of Cu(I)-CopC have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1NM4).
References
- 1.Linder M C. Biochemistry of Copper. New York: Plenum; 1991. pp. 1–13. [Google Scholar]
- 2.Rae T, Schmidt P J, Pufahl R A, Culotta V C, O'Halloran T V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 3.Puig S, Thiele D J. Curr Opin Chem Biol. 2002;6:171–180. doi: 10.1016/s1367-5931(02)00298-3. [DOI] [PubMed] [Google Scholar]
- 4.Cooksey D A. FEMS Microbiol Rev. 1994;14:381–386. doi: 10.1111/j.1574-6976.1994.tb00112.x. [DOI] [PubMed] [Google Scholar]
- 5.Odermatt A, Suter H, Krapf R, Solioz M. Ann NY Acad Sci. 1992;671:484–486. doi: 10.1111/j.1749-6632.1992.tb43836.x. [DOI] [PubMed] [Google Scholar]
- 6.Silver S. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 7.Cha J S, Cooksey D A. Proc Natl Acad Sci USA. 1991;88:8915–8919. doi: 10.1073/pnas.88.20.8915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mills S D, Jasalavich C A, Cooksey D A. J Bacteriol. 1993;175:1656–1664. doi: 10.1128/jb.175.6.1656-1664.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnesano F, Banci L, Bertini I, Thompsett A R. Structure (London) 2002;10:1337–1347. doi: 10.1016/s0969-2126(02)00858-4. [DOI] [PubMed] [Google Scholar]
- 10.Huffman D L, Huyett J, Outten F W, Doan P E, Finney L A, Hoffman B M, O'Halloran T V. Biochemistry. 2002;41:10046–10055. doi: 10.1021/bi0259960. [DOI] [PubMed] [Google Scholar]
- 11.Carl P J, Baccam S L, Larsen S C. J Phys Chem B. 2000;104:8848–8854. [Google Scholar]
- 12.Lee J, Pena M M, Nose Y, Thiele D J. J Biol Chem. 2002;277:4380–4387. doi: 10.1074/jbc.M104728200. [DOI] [PubMed] [Google Scholar]
- 13.Puig S, Lee J, Lau M, Thiele D J. J Biol Chem. 2002;277:26021–26030. doi: 10.1074/jbc.M202547200. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Nelson D P, Kiesow L A. Anal Biochem. 1972;49:474–478. doi: 10.1016/0003-2697(72)90451-4. [DOI] [PubMed] [Google Scholar]
- 16.Eccles C, Güntert P, Billeter M, Wüthrich K. J Biomol NMR. 1991;1:111–130. doi: 10.1007/BF01877224. [DOI] [PubMed] [Google Scholar]
- 17.Güntert P, Braun W, Wüthrich K. J Mol Biol. 1991;217:517–530. doi: 10.1016/0022-2836(91)90754-t. [DOI] [PubMed] [Google Scholar]
- 18.Archer S J, Ikura M, Torchia D A, Bax A. J Magn Reson. 1991;95:636–641. [Google Scholar]
- 19.Vuister G W, Bax A. J Am Chem Soc. 1993;115:7772–7777. [Google Scholar]
- 20.Banci L, Felli I C, Kummerle R. Biochemistry. 2002;41:2913–2920. doi: 10.1021/bi011617b. [DOI] [PubMed] [Google Scholar]
- 21.Wishart D S, Sykes B D. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 22.Güntert P, Mumenthaler C, Wüthrich K. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 23.Borgias B, Thomas P D, James T L. Complete Relaxation Matrix Analysis (CORMA) San Francisco: Univ. of California; 1989. , Version 5.0. [Google Scholar]
- 24.Case D A, Pearlman D A, Caldwell J W, Cheatham T E, Ross W S, Simmerling C L, Darden T A, Merz K M, Stanton R V, Cheng A L, et al. AMBER 6. San Francisco: Univ. of California; 1999. [Google Scholar]
- 25.Laskowski R A, Rullmann J A C, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski R A, MacArthur M W, Thornton J M. Curr Opin Struct Biol. 1998;8:631–639. doi: 10.1016/s0959-440x(98)80156-5. [DOI] [PubMed] [Google Scholar]
- 27.Kay L E, Nicholson L K, Delaglio F, Bax A, Torchia D A. J Magn Reson. 1992;97:359–375. [Google Scholar]
- 28.Peng J W, Wagner G. Methods Enzymol. 1994;239:563–596. doi: 10.1016/s0076-6879(94)39022-3. [DOI] [PubMed] [Google Scholar]
- 29.Brüschweiler R, Liao X, Wright P E. Science. 1995;268:886–889. doi: 10.1126/science.7754375. [DOI] [PubMed] [Google Scholar]
- 30.Lee L K, Rance M, Chazin W J, Palmer A G., III J Biomol NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 31.Pettifer R F, Hermes C. J Appl Crystallogr. 1985;18:404–412. [Google Scholar]
- 32.Notling H F, Hermes C. EXPROG: EMBL EXAFS Data Analysis and Evaluation Program Package for PC/AT. Hamburg, Germany: European Molecular Biology Laboratory; 1993. [Google Scholar]
- 33.Binsted N, Hasnain S S. J Synchrotron Radiat. 1996;3:185–196. doi: 10.1107/S0909049596005651. [DOI] [PubMed] [Google Scholar]
- 34.Briganti F, Mangani S, Pedocchi L, Scozzafava A, Golovleva L A, Jadan A P, Solyanikova I P. FEBS Lett. 1998;433:58–62. doi: 10.1016/s0014-5793(98)00884-9. [DOI] [PubMed] [Google Scholar]
- 35.Jaron S, Blackburn N J. Biochemistry. 2001;40:6867–6875. doi: 10.1021/bi002849y. [DOI] [PubMed] [Google Scholar]
- 36.Lytle F W, Sayers D E, Stern E A. Phys Rev B. 1989;11:2795–2801. [Google Scholar]
- 37.Kau L S, Spira-Solomon D J, Penner-Hahn J E, Hodgson K O, Solomon E I. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 38.Pickering I J, George G N, Dameron C T, Kurz B, Winge D R, Dance I G. J Am Chem Soc. 1993;115:9498–9505. [Google Scholar]
- 39.Blackburn N J, Strange R W, Reedijk J, Volbeda A, Farooq A, Karlin K D, Zubieta J. Inorg Chem. 1989;28:1349–1357. [Google Scholar]
- 40.Wirt M D, Bubacco L, Peisach J. Inorg Chem. 1995;34:2377–2381. [Google Scholar]
- 41.Eisses J F, Stasser J P, Ralle M, Kaplan J H, Blackburn N J. Biochemistry. 2000;39:7337–7342. doi: 10.1021/bi000690j. [DOI] [PubMed] [Google Scholar]
- 42.Clore G M, Szabo A, Bax A, Kay L E, Driscoll P C, Gronenborn A M. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- 43.Bertini I, Sigel A, Sigel H. Handbook on Metalloproteins. New York: Marcel Dekker; 2001. pp. 1–1800. [Google Scholar]
- 44.Messerschmidt A, Huber R, Poulos T, Wieghardt K. Handbook of Metalloproteins. Chichester, U.K.: Wiley; 2001. pp. 1–1248. [Google Scholar]
- 45.Richard A, Gralla E B, Valentine J S. Copper Containing Proteins, Advances in Protein Chemistry. Vol. 60. Amsterdam: Elsevier Science; 2002. pp. 1–550. [Google Scholar]
- 46.Roberts S A, Weichsel A, Grass G, Thakali K, Hazzard J T, Tollin G, Rensing C, Montfort W R. Proc Natl Acad Sci USA. 2002;99:2766–2771. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Outten F W, Huffman D L, O'Halloran T V. J Biol Chem. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 48.Grass G, Rensing C. J Bacteriol. 2001;183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim C, Lorenz W W, Hoopes J T, Dean J F. J Bacteriol. 2001;183:4866–4875. doi: 10.1128/JB.183.16.4866-4875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cha J S, Cooksey D A. Appl Environ Microbiol. 1993;59:1671–1674. doi: 10.1128/aem.59.5.1671-1674.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Puig S, Rees E, Thiele D J. Structure (London) 2002;10:1292–1295. doi: 10.1016/s0969-2126(02)00863-8. [DOI] [PubMed] [Google Scholar]
- 52.Arnesano F, Banci L, Bertini I, Cantini F, Ciofi-Baffoni S, Huffman D L, O'Halloran T V. J Biol Chem. 2001;276:41365–41376. doi: 10.1074/jbc.M104807200. [DOI] [PubMed] [Google Scholar]
- 53.Huffman D L, O'Halloran T V. Annu Rev Biochem. 2001;70:677–701. doi: 10.1146/annurev.biochem.70.1.677. [DOI] [PubMed] [Google Scholar]
- 54.Hassett R, Kosman D J. J Biol Chem. 1995;270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 55.Amendola V, Fabbrizzi L, Mangano C, Pallavicini P. Acc Chem Res. 2001;34:488–493. doi: 10.1021/ar010011c. [DOI] [PubMed] [Google Scholar]
- 56.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.