Abstract
Particulate methane monooxygenase (pMMO) is a membrane-bound enzyme that catalyzes the oxidation of methane to methanol in methanotropic bacteria. Understanding how this enzyme hydroxylates methane at ambient temperature and pressure is of fundamental chemical and potential commercial importance. Difficulties in solubilizing and purifying active pMMO have led to conflicting reports regarding its biochemical and biophysical properties, however. We have purified pMMO from Methylococcus capsulatus (Bath) and detected activity. The purified enzyme has a molecular mass of ≈200 kDa, probably corresponding to an α2β2γ2 polypeptide arrangement. Each 200-kDa pMMO complex contains 4.8 ± 0.8 copper ions and 1.5 ± 0.7 iron ions. Electron paramagnetic resonance spectroscopic parameters corresponding to 40–60% of the total copper are consistent with the presence of a mononuclear type 2 copper site. X-ray absorption near edge spectra indicate that purified pMMO is a mixture of Cu(I) and Cu(II) oxidation states. Finally, extended x-ray absorption fine structure data are best fit with oxygen/nitrogen ligands and a 2.57-Å Cu-Cu interaction, providing direct evidence for a copper-containing cluster in pMMO.
One of the great challenges for the chemical and engineering communities is the selective oxidation of methane to methanol. Although world reserves of petroleum and natural gas are comparable, methane is used far less efficiently as an energy source because it has a low energy density and is hazardous and expensive to transport (1). Conversion of methane to a liquid such as methanol would solve this problem, but current industrial processes for this transformation are costly and inefficient, requiring high temperatures and pressures (2, 3). By contrast, methanotrophic bacteria (4) oxidize methane to methanol at ambient temperature and pressure by using methane monooxygenase (MMO) enzyme systems (5). Only one other enzyme, ammonia monooxygenase (6), can activate the C—H bond in methane (104 kcal/mol). Cytochromes P450 (7) and copper monooxygenases (8) oxidize larger, more reactive hydrocarbons, but cannot hydroxylate methane. Therefore, a detailed understanding of biological methane oxidation is of both fundamental chemical and potential commercial importance.
All methanotrophs produce a membrane-bound MMO called particulate MMO (pMMO) (5). Under conditions of low copper availability, several strains also express a soluble enzyme (sMMO) (9), which contains a catalytic diiron center (10). Whereas the structure, biochemistry, and mechanism of sMMO are well understood (11), studies of pMMO are less advanced because of difficulties in solubilizing and purifying active enzyme. The Methylococcus capsulatus (Bath) pMMO comprises three polypeptides, the α (≈47 kDa), β (≈24 kDa), and γ (≈22 kDa) subunits, encoded by the pmoB, pmoA, and pmoC genes, respectively (12, 13). It is not known how these three polypeptides are arranged in the pMMO holoenzyme. According to radiolabeling experiments with the suicide substrate acetylene, the active site is located on the β subunit (14, 15) and may also involve the α subunit (14).
The metal composition of the pMMO active site is the subject of much controversy. For purified samples, stoichiometries ranging from 2 (16) to 15 (14, 17, 18) copper ions per αβγ complex have been reported. Based on electron paramagnetic resonance (EPR) spectroscopic studies of membrane-bound (19, 20) and purified (17) M. capsulatus (Bath) pMMO, Chan and coworkers propose that the pMMO copper ions are organized in multiple trinuclear clusters composed of one Cu(II) and two Cu(I) ions. These sites are hypothesized to fall into two functional classes, catalytic and electron transfer (20). The primary evidence for these trinuclear clusters derives from the analysis of the hyperfine splitting pattern of an isotropic signal at g = 2.06 (19). A similar signal has been detected for Methylosinus trichosporium OB3b pMMO (18, 21).
By contrast, other researchers observe only a typical type 2 Cu(II) EPR signal for purified M. capsulatus (Bath) pMMO (14, 16) and for membrane-bound and whole-cell suspensions of M. capsulatus (Bath) and Methylomicrobium album BG8 pMMO (22–24). Antholine and coworkers (22) suggest that the isotropic signal attributed to a trinuclear cluster can instead be explained by the presence of a radical and a mononuclear Cu(II) ion coordinated by three or four nitrogen donors in a square planar geometry. In addition to copper, 1–2.5 iron ions have been detected in some preparations of purified pMMO from both M. capsulatus (Bath) (14, 16) and M. trichosporium OB3b (18). To address some of the many unresolved issues regarding pMMO, we have purified the enzyme from M. capsulatus (Bath), determined its molecular mass and metal ion content, and investigated the metal centers by x-ray absorption (XAS) and EPR spectroscopies.
Materials and Methods
Bacterial Growth.
M. capsulatus (Bath) was obtained from the American Type Culture Collection and Stephen J. Lippard (Massachusetts Institute of Technology, Cambridge). Cultures were maintained on agar plates containing potassium nitrate mineral salts (NMS) growth medium (25), 10 μM CuSO4 (26), and 0.25 mg/liter Na2MoO4. Plates were stored in a BBL GasPak vented environmental chamber that was partially filled with methane gas (BOC Gases, Murray Hill, NJ) and placed in a 42°C incubator. Methane was replenished daily and plates were restreaked every month. Inoculum for large-scale fermentations was prepared by transferring a single colony to 1 ml of NMS supplemented with 50 μM CuSO4 and 0.25 mg/liter Na2MoO4 in a 25-ml culture tube. The tube was sealed with a septum, and 5 ml of the headspace was evacuated and replaced with methane. After several days in a 42°C shaking incubator at 250 rpm, the solution was subcultured into 5 ml and then into 50 ml of the above medium. The 50-ml flasks were recharged daily with methane and subcultured every third week. For 10-liter fermentations, 50-ml cultures were added to 10 liters of NMS with 50 μM CuSO4 and 0.25 mg/liter Na2MoO4 until an OD600 of at least 0.2–0.4 was reached. Cells were grown at 42°C by using a 1:4 methane/air mixture at 1 liter/min, agitating at 300 rpm. When the OD600 reached 12–14, after ≈48 h, cells were harvested by centrifugation at 3,000 × g for 20 min, washed three times with 10 mM Pipes buffer (pH 7.1–7.3), frozen in liquid nitrogen, and stored at −80°C. Typically, 1 liter of cells was retained in the fermenter as inoculum, and 9 liters of new growth medium was added.
Membrane Isolation and Protein Purification.
Cells were lysed by sonication, and cell debris was removed by centrifugation at 24,000 × g for 1 h. The membrane fraction was isolated by ultracentrifugation for 1 h at 160,000 × g. Using a Dounce homogenizer, membranes were resuspended three times with 25 mM Pipes, 0.25 M NaCl (pH 7.2). After the final wash, the ≈10–20 mg/ml membrane suspension was frozen in 1-ml aliquots in liquid nitrogen and stored at −80°C. The detergent dodecyl-β-d-maltoside (12DM, Anatrace, Maumee, OH) was used to solubilize pMMO for purification (27). Membranes were thawed on ice and added to an equal volume of 50 mg/ml of 12DM in double-distilled H2O (ddH2O). Before loading onto a Source 30Q (Amersham Biosciences) anion exchange column, solubilized membranes were diluted 4-fold with the column equilibration buffer, 50 mM Hepes (pH 7.5)/0.05% 12DM. pMMO was eluted at 0.3–0.4 M NaCl by using a 0–1 M NaCl gradient, concentrated in a Centriprep 50 (Millipore), and loaded onto a Sephacryl S200 gel filtration column (Amersham Biosciences) equilibrated with 50 mM Hepes, 0.25 M NaCl (pH 7.5), and 0.05% 12DM. Purified pMMO was concentrated in a Centriprep 50 to 5–10 mg/ml and stored at 4°C for immediate use or at −80°C for future use. The concentration of solubilized pMMO was determined by the Detergent-Compatible Lowry assay (Bio-Rad). Purified pMMO concentrations were determined by using the extinction coefficient (585,016 M−1⋅cm−1 per 200-kDa enzyme) determined by amino acid analysis (Protein Chemistry Laboratory, Texas A&M University, College Station).
Molecular Mass Determination.
SDS/PAGE (28), pentadeca fluorooctanoic acid/PAGE (PFO/PAGE) (29), and blue-native/PAGE (BN/PAGE) (30) were performed as described with the following modifications. All samples were loaded onto gels immediately after adding sample buffers without boiling. Gradient gels (4–20%) for SDS/PAGE were obtained from ISC BioExpress, Kaysville, UT. Tris⋅HCl gradient gels (4–15%, Bio-Rad) were run on ice at 140 V for PFO/PAGE and on ice for 5 h at 100 V for BN/PAGE. Coomassie brilliant blue G-250 for BN-PAGE was obtained from Sigma, and gels were destained for 2 days. Molecular mass markers typically used for analytical gel filtration (Amersham Biosciences) were used for PFO/PAGE and BN/PAGE as 1 mg/ml solutions in ddH2O.
Activity Assays.
Enzyme activity was measured by the epoxidation of propylene (31). In a typical experiment, 49 μl of membrane-bound pMMO was mixed with 1 μl of 1 mg/ml NADH in a 2-ml septum-sealed serum vial (Wheaton Science Products). The reaction was initiated by evacuating 2 ml of air and adding 2 ml of propylene (BOC Gases). After shaking in a 45°C water bath for 3 min, propylene oxide concentration was analyzed on a Hewlett–Packard 5890 Gas Chromatograph with a Porapak Q column (Supelco). The amount of propylene oxide produced was determined by a standard curve with 1–10 mM propylene oxide (Aldrich) standards diluted in ddH2O. The specific activity was calculated as nmol of propylene oxide produced per min per mg of pMMO. To investigate the dependence of pMMO activity on metal ions, stock solutions of CuSO4 and Na2H2EDTA were added to final concentrations of 2–200 μM Cu(II) and 0.2–72 μM EDTA. For solubilized and purified pMMO, duroquinol was substituted for NADH as a reductant (32). Duroquinone was reduced as described (14). All enzyme assays were performed in at least duplicate.
Metal Ion Content.
All glassware for metal ion analyses was acid-washed for a minimum of 24 h, rinsed thoroughly with chelexed ddH2O, and dried. Metal ion content was measured by inductively coupled plasma-atomic emission spectroscopy using a Thermo Jarrell (Franklin, MA) Ash Atomscan model 25 Sequential ICP Spectrometer. Copper and iron atomic absorption standards (Aldrich) were prepared immediately before use at 2 ppm in chelexed ddH2O for instrument calibration. In parallel with all protein samples, 1-ppm copper and iron standards were measured. Before measurement, solubilized pMMO was diluted into chelexed ddH2O.
CD and EPR Spectroscopies.
CD spectra were measured in a 0.1-cm path length quartz cuvette by using a Jasco J-715 spectrometer. To remove Hepes, which interferes with CD signals at short wavelengths, pMMO was exchanged twice into a 0.1% 12DM solution in ddH2O by using a Microcon 100 (Millipore). X-band EPR spectra were acquired at 120 K by using a modified Varian E-4 spectrometer; Q-band EPR spectra were acquired at 2 K by using an instrument as described (33). EPR samples of membrane-bound and purified pMMO were frozen in liquid nitrogen. The concentration of EPR-active copper in pMMO was obtained by double integration using [CuEDTA]2− as a standard. A 0.1 M [CuEDTA]2− stock solution was prepared by adding equimolar CuCl2 and Na2H2EDTA to a 1% NaOH solution. This solution was then serially diluted to 500 and 250 μM with ddH2O.
XAS Spectroscopy.
Purified samples for XAS were diluted in 50 mM Hepes (pH 7.5), 0.25 M NaCl, 0.05% 12DM, and 50% glycerol and concentrated to 1–10 mM copper. Samples for reduction were vacuum-degassed for 1.5 h, transferred to a Coy anaerobic chamber, and chemically reduced by adding dithionite to a final concentration of 20 mM. Samples were loaded into Lucite cells wrapped with Kapton tape in air for as-isolated pMMO or anaerobically for reduced pMMO. Samples were flash-frozen in liquid nitrogen and stored at −80°C until data collection. XAS measurements were performed on reproducible independent samples.
Data were collected at beamline 7–3 at the Stanford Synchrotron Radiation Laboratory (SSRL) (Menlo Park, CA) and at the Biophysics Collaborative Access Team (BioCAT) beamline at the Advanced Photon Source (APS) (Argonne, IL) by using Si(220) double crystal monochromators detuned 50% for harmonic rejection. Fluorescence excitation spectra at SSRL were recorded by using a 30-element Ge solid-state array detector whereas APS data were collected by using dual photo-multiplier detectors. Samples were maintained at 10 K by using an Oxford Instruments (Scotts Valley, CA) continuous-flow liquid helium cryostat at SSRL and at 30 K by using a He Displex Cryostat at APS. XAS spectra were measured at SSRL by using 5-eV steps in the pre-edge, 0.25-eV steps in the edge (9,636–9,710 eV), and 0.05-Å−1 increments in the extended XAS fine structure (EXAFS) region, integrating from 1 to 30 s in a k3-weighted manner for a total scan length of 40 min. XAS near edge structure spectra were collected at APS by using 0.25-eV point spacing at a constant 37 ms per point over an eV range of 8,900–9,100 for a total collection time of 30 s per scan. X-ray energies were calibrated by simultaneous measurement of a Cu foil absorption spectrum, assigning the first inflection point as 8980.3 eV. Each channel of every scan was examined for glitches before averaging, and spectra were closely monitored for photoreduction. SSRL data represent the average of 10–12 scans whereas APS data represent the average of 30 scans.
XAS data were analyzed by using the Macintosh os x version of the exafspak program suite integrated with FEFF 7.0 for theoretical model generation (ref. 34 and www-ssrl.slac.stanford.edu/exafspak.html). Data reduction used a second-order polynomial in the pre-edge and a three-region cubic spline through the EXAFS region. Data were converted to k-space by using E0 = 9,000 eV. The k3-weighted EXAFS was truncated at 1.0 and 13.0 Å−1 for filtering purposes and Fourier-transformed. Data were then Fourier-filtered to isolate the EXAFS for each peak in the Fourier transform. Fitting analysis was performed on both Fourier-filtered and raw/unfiltered data. The best-fit bond lengths, coordination numbers, and Debye-Waller factors (see Table 3) were determined from fitting Fourier-filtered (R = 1.0–3.8 Å) data. Fits to both Fourier-filtered and unfiltered data gave equivalent structural parameters, however. EXAFS data were fit by using amplitude and phase functions calculated with FEFF 7.0 for a Cu-O at 2.00 Å, a Cu-C at 2.9 Å, a Cu-S at 2.25 and 2.40 Å, and a Cu-Cu at 2.60 and 2.90 Å. A scale factor of 0.86 and an E0 of −12.2 were calibrated by fitting EXAFS data from crystallographically characterized complexes. Coordination numbers were fixed at half-integer values whereas R and σ2 were freely variable parameters. The mean square deviation between data and fit, corrected for degrees of freedom, was used to determine the best simulation and justification for adding interactions to the simulation (35).
Table 3.
Summary of pMMO EXAFS fitting analysis
| Fit no.
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
|||||||||
| C.N. | R, Å | σ2, Å2* | C.N. | R, Å | σ2, Å2* | C.N. | R, Å | σ2, Å2* | C.N. | R, Å | σ2, Å2* | |
| Cu–N/O | 2.5 | 1.97 | 3.17 | 2.5 | 1.97 | 3.02 | 2.5 | 1.97 | 2.82 | 2.5 | 1.97 | 3.70 |
| Cu–C | 1.0 | 2.60 | 1.70 | |||||||||
| Cu–S | 1 | 2.71 | 3.13 | |||||||||
| Cu–Cu | 0.5 | 2.57 | 3.73 | |||||||||
| F† | 271 | 69 | 59.6 | 40.7 | ||||||||
R, best-fit bond lengths; C.N., coordination numbers. The best fit is shown in bold.
Debye-Waller factor × 103.
Degrees of freedom weighted mean square deviation between data and fit ×102.
Results and Discussion
Isolation and Purification of pMMO.
Large quantities of M. capsulatus (Bath) cells were produced in 10-liter fermentations by using nitrate mineral salts medium supplemented with 50 μM CuSO4. In contrast to previous reports, high CuSO4 concentrations did not inhibit cell growth (14, 17). It was therefore not necessary to gradually increase the copper levels (14) or supplement the medium with CuEDTA (17, 20). Cells harvested at OD600 = 12 yield ≈10 g of cell paste per liter of medium. Membranes were washed thoroughly to remove adventitious metal ions and loosely bound proteins and remained active after many months of storage at −80°C. Reducing agents, catalase, and high ionic strength buffers (17) did not enhance stability.
Our purification protocol differs from those reported (14, 16, 17). The Source 30Q anion exchange column removes contaminating proteins as well as lipids. Low molecular mass proteins not resolved on the Source 30Q column are then separated by gel filtration on Sephacryl S200. The final pMMO sample is purified to homogeneity and comprises three polypeptides of molecular masses ≈47, ≈24, and ≈22 kDa (Fig. 1A). A 35-kDa polypeptide, initially assigned as a pMMO subunit (26) and subsequently recognized to be a proteolytic fragment of the β subunit (17), is not observed even at high protein concentrations (Fig. 1A, lane 3). A 63-kDa putative formaldehyde or methanol dehydrogenase suggested to form a functional complex with pMMO (16) is typically removed before purification by washing the membranes. Samples of pMMO purified by this method are stable to proteolysis for many months at −80°C or for 1–2 weeks at 4°C.
Figure 1.
Gel electrophoresis analyses of purified pMMO. (A) SDS/PAGE. Lane 1, 1.5 mg/ml purified pMMO; lane 2, Invitrogen molecular mass standards: 116.3, 97.4, 66.3, 55.4, 36.5, 31, 21.5, 14.4, and 6 kDa; lane 3, 6 mg/ml purified pMMO. (B) PFO/PAGE. Lane 1, ovalbumin (43 kDa); lane 2, albumin (67 kDa); lane 3, adenylate cyclase (70 kDa), lane 4, catalase (232 kDa); lane 5, urease monomer (91 kDa), lane 6, ferritin (440 kDa); lane 7, 2 mg/ml purified pMMO. (C) BN/PAGE. Lane 1, ovalbumin (43 kDa); lane 2, albumin (67 and 134 kDa); lane 3, catalase (232 kDa); lane 4, ferritin (440 kDa); lane 5, 0.8 mg/ml purified pMMO.
Molecular Mass Determination.
Although pMMO from M. capsulatus (Bath) is composed of three polypeptides, neither the subunit stoichiometry nor the total molecular mass has been determined. The ratio of subunits has always been assumed to be 1:1:1, corresponding to an αβγ heterotrimer of ≈100 kDa. For M. trichosporium OB3b pMMO, comprising two subunits of molecular masses 25 and 41 kDa, the overall mass is estimated to be 326 kDa by analytical gel filtration (21). The molecular mass of a multisubunit membrane protein cannot be measured accurately by analytical gel filtration chromatography, however (36). For this reason, other techniques, such as PFO/PAGE (29) and BN/PAGE (30), have been developed. These techniques are similar to SDS/PAGE except that the dodecylsulfate anion is replaced with a nondenaturing alternative that preserves the quarternary structure of the membrane protein.
PFO/PAGE analysis of purified pMMO reveals a band immediately below that of catalase (232 kDa) as well as one just above ovalbumin (43 kDa) (Fig. 1B). The ≈200-kDa band suggests that purified pMMO has an α2β2γ2 subunit structure, although other compositions cannot be excluded. The lower molecular mass band likely corresponds to the α subunit (47 kDa), but could also be a complex between the β (24 kDa) and γ (22 kDa) subunits. PFO is thus partly denaturing under the experimental conditions. Analytical ultracentrifugation sedimentation velocity experiments using PFO running buffer also indicate the presence of two species corresponding to analogous molecular masses (R.L.L., B. Demeler, and A.C.R., unpublished results). A molecular mass of ≈200 kDa is further corroborated by BN/PAGE (Fig. 1C). Lower molecular mass species are not observed in this experiment. Instead, an ≈440-kDa molecular mass band is observed, perhaps corresponding to a tetramer. It is possible that pMMO can form higher-order species considering the high levels of pMMO found in M. capsulatus (Bath) intracytoplasmic membranes (26). Most importantly, an ≈100-kDa band is not apparent in either of these gel experiments. Because both techniques yield an ≈200-kDa pMMO species, the amount of PFO or brilliant blue G-250 bound to the enzyme does not appreciably alter its mass.
Enzyme Activity.
The average specific activity of the membrane-bound pMMO is 19.4 nmol/mg⋅min, comparable to other preparations (14, 16, 17, 19, 20) (Table 1). For membrane-bound pMMO, both NADH and duroquinol (32) were effective reducing agents. According to early reports, the addition of CuSO4 either directly to membrane fractions (19) or to the cells before lysis and membrane isolation (37) resulted in increased pMMO activity. The activity of our membrane-bound pMMO was unchanged with the addition of up to 200 μM CuSO4, however. The cells for the previous experiments were grown in the presence of only 0–20 μM CuSO4, which could lead to incomplete metallation of the pMMO active site. In the present experiments, the cells were grown with 50 μM CuSO4 and may already contain fully loaded active sites. The effect of EDTA on pMMO activity was also investigated. The addition of 0.2–72 mM EDTA decreased activity, but approximately half of the original specific activity remained even with a 40-fold molar excess of EDTA. A similar effect has been observed for purified pMMO from M. trichosporium OB3b (18) and for partially purified pMMO from M. capsulatus (Bath) (16). These observations suggest that the metal ions in pMMO are necessary for activity, but may differ in accessibility. The specific activity decreased on solubilization with 12DM (Table 1). No activity was observed when NADH was used as the reductant, but was detected with duroquinol. Attempts to correlate solubilized pMMO activity with detergent levels did not yield an optimal detergent-to-protein ratio. After purification, most samples were not active. The 17.7-nmol/mg⋅min activity measured for the few active preparations (Table 1) is higher than the values reported for other purified samples (14, 17). The ability to detect activity could not be correlated with metal ion content or particular protein impurities.
Table 1.
Average specific activity of pMMO
| pMMO sample | Reductant | Specific activity, nmol/min⋅mg |
|---|---|---|
| Membrane-bound | NADH | 21 |
| Duroquinol | 16 | |
| Solubilized | Duroquinol | 3.9 |
| Purified | Duroquinol | 17.7 |
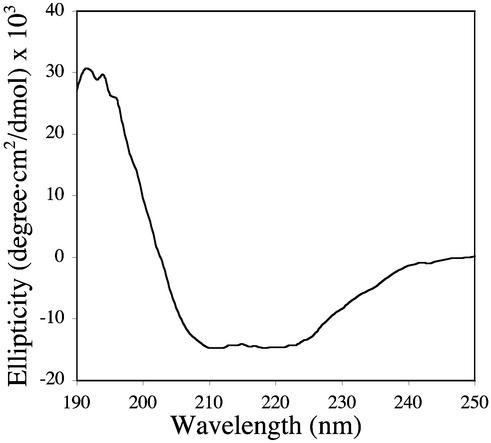
For inactive samples, numerous approaches for restoring activity were pursued. Attempts to remove bound detergent by dialysis, add back other proteins separated on the anion exchange column, and supplement the assay mixture with reducing agents or CuSO4 and CuCl were not successful. A lipid reconstitution procedure reported to yield recovered activities of 2–5 nmol/mg⋅min (17) did not restore activity either. There are several possible explanations for the lack of activity. First, the detergent could interfere with activity. Second, a cofactor or protein essential to the electron transport chain might be removed during purification. Potential candidates include cytochromes associated with pMMO (14, 17), an NADH:quinone oxidoreductase (38), or a putative dehydrogenase suggested to form an active complex with pMMO (16). Finally, solubilized, purified pMMO might not be folded properly. The CD spectrum shows secondary structure with a significant α-helical component (Fig. 2), but it is still possible that pMMO is present in an inactive conformation. Until these issues are fully investigated, the variability in pMMO activity levels observed in our and other preparations (17) will likely remain a problem.
Figure 2.
CD spectrum of 0.77 μM purified pMMO in 12DM. Data were collected at room temperature from 300 to 190 nm at 1-nm resolution.
Metal Ion Analyses.
According to inductively coupled plasma-atomic emission spectroscopy measurements, membrane-bound pMMO contains 20.8 ± 4.5 copper ions per 200 kDa, and purified pMMO contains 4.8 ± 0.8 copper ions per 200 kDa (Table 2). By contrast, values of 8–118 copper ions per 200 kDa for membrane-bound pMMO (14, 19, 20, 37) and 24–30 copper ions per 200 kDa for purified pMMO (14, 17) have been reported. There are several likely explanations for the presence of significantly fewer copper ions in our pMMO samples. First, the wide variation in membrane-bound pMMO copper content reported by others suggests that cells and membranes may not have been washed thoroughly. Large amounts of copper present in the growth medium could remain adventitiously bound to the membranes, and could also account for the high levels of copper found in purified pMMO. For our preparations, both the harvested cells and membrane fractions were washed carefully three times. Second, the protein concentration may not have been determined accurately, leading to incorrect stoichiometry. For example, we found that the Bradford assay is inaccurate in the presence of 12DM. Thus, for our inductively coupled plasma-atomic emission spectroscopy analysis, the concentration of solubilized pMMO was determined by using a detergent-compatible version of the Lowry assay, and the concentration of purified pMMO was calculated by using an extinction coefficient determined from amino acid hydrolysis. In support of our results, Dalton and coworkers (16) also measured four copper ions per 200 kDa by using a detergent-compatible protein assay. Although it is possible that our purification procedure removes some essential copper ions, the activity is in fact similar to that observed for preparations containing significantly more copper (14, 19, 20). Thus, it seems likely that the catalytic metal center(s) remain(s) intact.
Table 2.
Metal ion analysis of pMMO by inductively coupled plasma-atomic emission spectroscopy
| pMMO sample | Mol Cu per 200 kDa | Mol Fe per 200 kDa |
|---|---|---|
| Solubilized* | 20.8 ± 4.5 | 2.2 ± 0.7 |
| Purified† | 4.8 ± 0.8 | 1.5 ± 0.7 |
Average of seven samples.
Average of 28 samples.
Although copper is always detected in pMMO, the presence of iron has been disputed (17). For membrane-bound pMMO, iron stoichiometries ranging from 2 to 20 iron ions per 200 kDa have been reported (14, 19, 20). For purified pMMO, values of zero (17), two (16), or five (14) iron ions per 200 kDa have been reported. Our membrane-bound pMMO contains 2.2 ± 0.7 iron ions per 200 kDa, and our purified pMMO contains 1.5 ± 0.7 iron ions per 200 kDa (Table 2). Chan and coworkers (17, 20) did not detect iron in their purified pMMO and suggested that high iron concentrations in the growth medium lead to pMMO contamination by soluble MMO (sMMO) and other iron-containing proteins. Although our cell growth medium includes 80 μM FeEDTA, sMMO is not associated with our membrane-bound or purified samples. The small amount of iron in our purified samples may instead be caused by cytochrome impurities. Nevertheless, we cannot exclude the possibility that iron plays a functional role in pMMO.
EPR Spectroscopy.
The Q-band (35 GHz) EPR spectra of both membrane-bound and purified pMMO exhibit signals characteristic of type 2 Cu(II) with g∥ = 2.24, g⊥ = 2.05, and A∥ = 530 MHz (Fig. 3). These parameters are similar to those reported for both whole-cell suspensions (24) and purified samples (14, 16) from M. capsulatus (Bath) and are consistent with a mononuclear Cu(II) site coordinated by three or four histidine imidazoles (22–24, 39). By contrast, the signal attributed by Chan and coworkers (17, 19, 20) to a mixed valence trinuclear copper cluster is not observed in either spectrum. A radical at g = 2.00 and a signal from trace amounts of rhombic iron are observed in the membrane-bound sample, but are not observed on purification. Integration of the X-band EPR spectrum indicates that 40–60% of the total copper (four to six copper ions per 200 kDa) in purified pMMO are EPR-active. An increase in signal intensity is not obtained by adding ferricyanide, in contrast to previous reports (20). On addition of 20 mM dithionite, the Cu(II) EPR signal disappears completely.
Figure 3.
Q-band EPR spectra of purified (Upper) and membrane-bound (Lower) pMMO. The same signal is observed for both samples with g∥ = 2.24, g⊥ = 2.05, and A∥ (Cu) = 530 MHz. Spectrometer conditions: microwave frequency, 35.72 GHz; microwave power, 50 μW; modulation amplitude, 1.3 G; scan time, 4 min; time constant, 128 msec; temperature, 2 K.
XAS Spectroscopy.
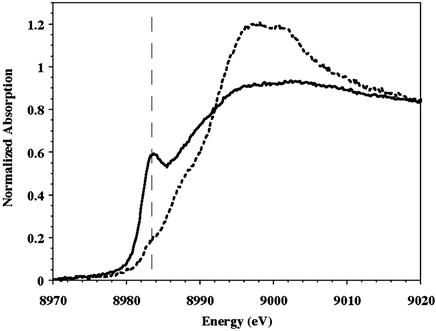
XAS near edge structure spectra for purified and purified dithionite-reduced pMMO are compared in Fig. 4. A substantial (≈3 eV) edge energy shift observed on chemical reduction indicates that the average copper oxidation state of as-isolated pMMO is higher than that of the dithionite-reduced sample. An absorption feature at ≈8,984 eV, attributed to a Cu(I) 1s → 4p transition (40), is present in both spectra, however. Therefore, some fraction of the copper in both pMMO samples is in the Cu(I) oxidation state, and this fraction is clearly greater for the reduced protein. The presence of Cu(I) in the as-isolated purified protein is consistent with the EPR quantitation. Previous XAS near edge structure spectra of pMMO membrane preparations also suggested a mixture of copper oxidation states, although the ≈8,984-eV feature was less pronounced on reduction (20).
Figure 4.
Cu K-edge XAS near edge spectra of purified pMMO. Dashed line, as isolated purified pMMO; solid line, dithionite-reduced pMMO. The 1s → 4p feature at ≈8,984 eV is marked by the dashed vertical line.
The Fourier transform of the Cu EXAFS data (Fig. 5) exhibits two peaks, indicating that two shells of backscatterers are present. The EXAFS generating the more intense peak at 1.5 Å are best fit with oxygen/nitrogen (O/N) ligands at average bond lengths of 1.97 Å (Table 3, fit 1). The corresponding coordination number is 2.5 O/N ligands per copper ion, but this value may be underestimated. Cu-O/N bond lengths of ≈2 Å are typically observed for four-coordinate Cu(I) and Cu(II) species, whereas two- and three-coordinate Cu-O/N bond lengths are on average 0.15–0.06 Å shorter, respectively [Cambridge Structural Database (CSD), associated software systems (including CONQUEST), and documentation are proprietary products in which the rights are held by the Cambridge Crystallographic Data Centre, Cambridge, U.K.]. Two scenarios would result in an underestimate of the Cu-O/N coordination number. First, the low coordination number may reflect the presence of two sets of Cu-O/N ligand environments at close but distinct bond lengths. Two such environments cannot be fit independently given the data resolution. Alternatively, a low percentage of Cu-S scattering might destructively interfere with the Cu-low Z signal and dampen the EXAFS amplitude for the Cu-O/N interaction. There is no direct XAS evidence for a Cu-S interaction, however.
Figure 5.
Cu EXAFS fitting analysis for pMMO. (A) Fourier transform comparison of raw/unfiltered pMMO EXAFS (solid line) with two-environment Cu-O/N and Cu-Cu theoretical fits (dashed line). (B) Comparison of unfiltered k3-weighted pMMO EXAFS (solid line) with a theoretical two-environment Cu-O/N and Cu-Cu k3-weighted EXAFS (dashed line).
The second peak in the Fourier transform, observed reproducibly at a phase-shifted bond length of 2.2 Å, corresponds to a second backscattering shell at ≈2.6 Å (Fig. 5A). Attempts to fit this feature used three models: a Cu–C interaction, possibly from a nearby histidine imidazole ring (Table 3, fit 2), a long Cu–S interaction (Table 3, fit 3), and a direct Cu–Cu interaction (Table 3, fit 4). In each case, addition of a second ligand shell produced a mean squared deviation value (F′) better than a single environment fit (Table 3, fit 1), but the interaction was best simulated by a second Cu atom at 2.57 Å (Fig. 2B). Notably, the 2.57 Å Cu–Cu distance is strikingly similar to Cu–Cu distances observed in the nitrous oxide reductase tetranuclear CuZ cluster (41). Short Cu–Cu distances are also present in the cytochrome c oxidase CuA site (42), but optical and EPR spectroscopic features characteristic of CuA (43) or CuZ (44) are not observed for pMMO. Thus, pMMO may contain a new type of copper center. Although it is likely that the second metal ion in pMMO is copper, further analysis is required to rule out the possibility that another metal ion, such as iron, is involved. Nevertheless, this result provides direct evidence that some component of the pMMO metal ions exist in a multinuclear, copper-containing cluster.
Acknowledgments
This work was supported by American Chemical Society–Petroleum Research Fund Grant 32798-G4 (to A.C.R.), funds from the David and Lucile Packard Foundation (to A.C.R.), National Institutes of Health Grant HL13531 (to B.M.H.), and American Heart Association Grant 0130527Z (to T.L.S.). R.L.L. is supported in part by National Institutes of Health Training Grant GM08382. SSRL is funded by the U.S. Department of Energy (Basic Energy Sciences, Brookhaven Executive Round Table) and the National Institutes of Health (National Center for Research Resources; National Institute of General Medical Sciences). Use of the APS was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-ENG-38. The Biophysics Collaborative Access Team is National Institutes of Health-supported Research Center RR-08630.
Abbreviations
- MMO methane monooxygenase
pMMO, particulate MMO
- 12DM
dodecyl-β-d-maltoside
- XAS
x-ray absorption
- EXAFS
extended XAS fine structure
- SSRL
Stanford Synchrotron Radiation Laboratory
- ddH2O
double-distilled H2O
- PFO/PAGE
pentadeca fluorooctanoic acid/PAGE
- BN/PAGE
blue-native/PAGE
- APS
Advanced Photon Source
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Voss D. Technol Rev. 2002;105:69–72. [Google Scholar]
- 2.Periana R A, Taube D J, Gamble S, Taube H, Satoh T, Fujii H. Science. 1998;280:560–564. doi: 10.1126/science.280.5363.560. [DOI] [PubMed] [Google Scholar]
- 3.Periana R A, Taube D J, Evitt E R, Löffler D G, Wentrcek P R, Voss G, Masuda T. Science. 1993;259:340–343. doi: 10.1126/science.259.5093.340. [DOI] [PubMed] [Google Scholar]
- 4.Hanson R S, Hanson T E. Microbiol Rev. 1996;60:439–471. doi: 10.1128/mr.60.2.439-471.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murrell J C, McDonald I R, Gilbert B. Trends Microbiol. 2000;8:221–225. doi: 10.1016/s0966-842x(00)01739-x. [DOI] [PubMed] [Google Scholar]
- 6.Hyman M R, Murton I B, Arp D J. Appl Environ Microbiol. 1988;54:3187–3190. doi: 10.1128/aem.54.12.3187-3190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ortiz de Montellano P R. Cytochrome P450: Structure, Mechanism, and Biochemistry. New York: Plenum; 1986. [Google Scholar]
- 8.Prigge S T, Mains R E, Eipper B A, Amzel L M. Cell Mol Life Sci. 2000;57:1236–1259. doi: 10.1007/PL00000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stanley S H, Prior S D, Leak D J, Dalton H. Biotechnol Lett. 1983;5:487–492. [Google Scholar]
- 10.Woodland M P, Patil D S, Cammack R, Dalton H. Biochim Biophys Acta. 1986;873:237–242. [Google Scholar]
- 11.Merkx M, Kopp D A, Sazinsky M H, Blazyk J L, Müller J, Lippard S J. Angew Chem Int Ed. 2001;40:2782–2807. [PubMed] [Google Scholar]
- 12.Semrau J D, Chistoserdov A, Lebron J, Costello A, Davagnino J, Kenna E, Holmes A J, Finch R, Murrell J C, Lidstrom M E. J Bacteriol. 1995;177:3071–3079. doi: 10.1128/jb.177.11.3071-3079.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stolyar S, Costello A M, Peeples T L, Lidstrom M E. Microbiology. 1999;145:1235–1244. doi: 10.1099/13500872-145-5-1235. [DOI] [PubMed] [Google Scholar]
- 14.Zahn J A, DiSpirito A A. J Bacteriol. 1996;178:1018–1029. doi: 10.1128/jb.178.4.1018-1029.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cook S A, Shiemke A K. J Inorg Biochem. 1996;63:273–284. [Google Scholar]
- 16.Basu P, Katterle B, Andersson K K, Dalton H. Biochem J. 2003;369:417–427. doi: 10.1042/BJ20020823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen H H, Elliott S J, Yip J H, Chan S I. J Biol Chem. 1998;273:7957–7966. doi: 10.1074/jbc.273.14.7957. [DOI] [PubMed] [Google Scholar]
- 18.Takeguchi M, Okura I. Catal Surv Jpn. 2000;4:51–63. [Google Scholar]
- 19.Nguyen H-H T, Shiemke A K, Jacobs S J, Hales B J, Lidstrom M E, Chan S I. J Biol Chem. 1994;269:14995–15005. [PubMed] [Google Scholar]
- 20.Nguyen H-H T, Nakagawa K H, Hedman B, Elliott S J, Lidstrom M E, Hodgson K O, Chan S I. J Am Chem Soc. 1996;118:12766–12776. [Google Scholar]
- 21.Takeguchi M, Miyakawa K, Okura I. J Mol Catal. 1998;132:145–153. [Google Scholar]
- 22.Yuan H, Collins M L P, Antholine W E. J Am Chem Soc. 1997;119:5073–5074. [Google Scholar]
- 23.Yuan H, Collins M L P, Antholine W E. Biophys J. 1999;76:2223–2229. doi: 10.1016/S0006-3495(99)77378-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemos S S, Collins M L P, Eaton S S, Eaton G R, Antholine W E. Biophys J. 2000;79:1085–1094. doi: 10.1016/s0006-3495(00)76362-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whittenbury R, Phillips K C, Wilkinson J F. J Gen Microbiol. 1970;61:205–218. doi: 10.1099/00221287-61-2-205. [DOI] [PubMed] [Google Scholar]
- 26.Prior S D, Dalton H. J Gen Microbiol. 1985;131:155–163. [Google Scholar]
- 27.Smith D D S, Dalton H. Eur J Biochem. 1989;182:667–671. doi: 10.1111/j.1432-1033.1989.tb14877.x. [DOI] [PubMed] [Google Scholar]
- 28.Coligan J E. Current Protocols in Protein Science. New York: Wiley; 2001. [DOI] [PubMed] [Google Scholar]
- 29.Ramjeesingh M, Huan L-J, Garami E, Bear C E. Biochem J. 1999;342:119–123. [PMC free article] [PubMed] [Google Scholar]
- 30.Schägger H, von Jagow G. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- 31.Colby J, Stirling D I, Dalton H. Biochem J. 1977;165:395–402. doi: 10.1042/bj1650395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiemke A K, Cook S A, Miley T, Singleton P. Arch Biochem Biophys. 1995;321:421–428. doi: 10.1006/abbi.1995.1413. [DOI] [PubMed] [Google Scholar]
- 33.Werst M M, Davoust C E, Hoffman B M. J Am Chem Soc. 1991;113:1533–1538. [Google Scholar]
- 34.Ankudinov A L, Rehr J J. Phys Rev B Condens Matter. 1997;56:R1712–R1715. [Google Scholar]
- 35.Riggs-Gelasco P J, Stemmler T L, Penner-Hahn J E. Coord Chem Rev. 1995;144:245–286. [Google Scholar]
- 36.le Maire M, Champeil P, Møller J V. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 37.Semrau J D, Zolandz D, Lidstrom M E, Chan S I. J Inorg Biochem. 1995;58:235–244. doi: 10.1016/0162-0134(94)00056-g. [DOI] [PubMed] [Google Scholar]
- 38.Cook S A, Shiemke A K. Arch Biochem Biophys. 2002;398:32–40. doi: 10.1006/abbi.2001.2628. [DOI] [PubMed] [Google Scholar]
- 39.Katterle B, Gvozdev R I, Abudu N, Ljones T, Andersson K K. Biochem J. 2002;363:677–686. doi: 10.1042/0264-6021:3630677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kau L-S, Spira-Solomon D J, Penner-Hahn J E, Hodgson K O, Solomon E I. J Am Chem Soc. 1987;109:6433–6442. [Google Scholar]
- 41.Brown K R, Djinovic-Carugo K, Haltia T, Cabrito I, Saraste M, Moura J J G, Moura I, Tegoni M, Cambillau C. J Biol Chem. 2000;275:41133–41136. doi: 10.1074/jbc.M008617200. [DOI] [PubMed] [Google Scholar]
- 42.Wilmanns M, Lapplainen P, Kelly M, Sauer-Eriksson E, Saraste M. Proc Natl Acad Sci USA. 1995;92:11955–11959. doi: 10.1073/pnas.92.26.11955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farrar J A, Neese F, Lapplainen P, Kroneck P M H, Saraste M, Zumft W G, Thomson A J. J Am Chem Soc. 1996;118:11501–11514. [Google Scholar]
- 44.Prudêncio M, Pereira A S, Tavares P, Besson S, Cabrito I, Brown K, Samyn B, Devreese B, Beeumen J V, Rusnak F, et al. Biochemistry. 2000;39:3899–3907. doi: 10.1021/bi9926328. [DOI] [PubMed] [Google Scholar]