Abstract
DNA polymerases replicate DNA by adding nucleotides to a growing primer strand while avoiding frameshift and point mutations. Here we present a series of up to six successive replication events that were obtained by extension of a primed template directly in a crystal of the thermostable Bacillus DNA polymerase I. The 6-bp extension involves a 20-Å translocation of the DNA duplex, representing the largest molecular movement observed in a protein crystal. In addition, we obtained the structure of a “closed” conformation of the enzyme with a bound triphosphate juxtaposed to a template and a dideoxy-terminated primer by constructing a point mutant that destroys a crystal lattice contact stabilizing the wild-type polymerase in an “open” conformation. Together, these observations allow many of the steps involved in DNA replication to be observed in the same enzyme at near atomic detail. The successive replication events observed directly by catalysis in the crystal confirm the general reaction sequence deduced from observations obtained by using several other polymerases and further refine critical aspects of the known reaction mechanism, and also allow us to propose new features that concern the regulated transfer of the template strand between a preinsertion site and an insertion site. We propose that such regulated transfer is an important element in the prevention of frameshift mutations in high-fidelity DNA polymerases. The ability to observe processive, high-fidelity replication directly in a crystal establishes this polymerase as a powerful model system for mechanistic studies in which the structural consequences of mismatches and DNA adducts are observed.
Accurate DNA replication is a fundamental requirement for genomic stability. DNA polymerases achieve remarkably high replication fidelity, often allowing only a single insertion error for every 105 incorporated nucleotides (1, 2). Selection of the correct base for insertion requires discrimination against both nucleotide misinsertion (mispair) and strand misalignment (frameshift) errors. Here, we report studies on a DNA polymerase I (pol I) large fragment from a thermostable strain of Bacillus stearothermophilus (Bacillus fragment, BF) that catalyzes template-directed DNA replication in the crystal (3). These catalytically active crystals allowed us to determine the structures of the product complexes representing six successive replication events.
The molecular structures of a wide variety of DNA polymerases and their complexes have become available during the past few years (4), including several members of the A-family (pol I): E. coli Klenow fragment (KF) (5); BF (3, 6); Thermus aquaticus (Taq) (7–10); and bacteriophage T7 (T7) (11). The structures of all polymerases determined to date show that these enzymes include three subdomains that are associated with binding of the DNA primer⋅template and an incoming dNTP, termed “palm,” “fingers,” and “thumb” subdomains because of their resemblance to a right hand (5). The palm is the most conserved of these structural regions, and contains two universally observed carboxylate residues (12) that play a direct role in the phosphoryl transfer reaction (4). Significant structural and sequence divergence is observed in finger and thumb subdomains.
Structural studies of A-family polymerases (pol I) identified two distinct conformational states of the fingers subdomain. In polymerase⋅DNA⋅dNTP ternary complexes of Taq (9, 10) and T7 (11), the fingers are “closed” around the dNTP and DNA template, forming a binding site that is sterically complementary to a Watson–Crick base pair. Polymerase⋅DNA binary complexes form an “open” conformation (3, 9). Similar open and closed conformations have been observed in X-family polymerases (13, 14), B-family polymerases (15–19), and HIV reverse transcriptase (20–22).
BF polymerase, a member of the A-family, is uniquely suited for a detailed structural analysis of the polymerase activity because the BF⋅DNA cocrystals retain the ability to synthesize DNA in the crystal. A previous study (3) demonstrated that formation of a new base pair and translocation of the DNA is observed in the crystal structure when BF crystals containing adenosine in the DNA template are placed in a stabilization solution containing dTTP. It remained to be shown whether the BF crystals could select the correct base from a pool of dNTPs and whether the activity was processive. We have now introduced each of the four deoxynucleotides into the crystal soak solution and observed both accurate and processive replication. These observations allowed us to identify open conformation-associated structural features that play a role in polymerase fidelity. We also captured a closed ternary complex of BF by mutagenic disruption of crystal contacts that exist in the open crystal form. Comparison of the open binary complexes with the closed ternary complex suggests a mechanism in which the polymerase maintains control over the template position throughout each catalytic cycle, thereby suppressing frameshift mutations.
Methods
Protein Preparation.
Mutagenesis of the Bacillus fragment was performed by using the QuikChange Site-Directed Mutagenesis Kit from Stratagene to construct single alanine mutants at positions Asp-329 (D329A) and Asp-598 (D598A), as well as the double mutant (D329A–D598A). Proteins were purified as described (6).
Cocrystallization of BF with DNA Primer Template and dNTP Substrates.
Complementary oligonucleotides (complex I, 5′- GACGTACGTGATCGCA-3′ and 5′-GCGATCACG-3′; complex II, 5′-ACGTCGCTGATCCG-3′ and 5′-GGATCAGCG-3′) were annealed as described (3). The resulting DNA substrates contain a 9-bp duplex region, a 5′ single-stranded template overhang (complex I, 5′-GACGTA; complex II, 5′-ACGT), and a single nucleotide 3′ template overhang (complex I, 3′-A; complex II, 3′-G). The 3′ template overhang favors binding of the DNA with the 5′ overhang at the active site. The binary BF⋅DNA complex I was obtained by incubating 12 mg/ml wild-type protein with DNA at a 3:1 molar ratio. A BF⋅DNA⋅dCTP ternary complex was similarly obtained by incubating the D329A mutant with DNA complex II, 3.75 mM ddATP, 3.75 mM dCTP, and 10 mM MnSO4. Crystals were grown by hanging drop vapor diffusion from 49% saturated ammonium sulfate, 2.5% (vol/vol) 2-methyl-2,4-pentanediol (MPD), 100 mM Mes buffer (pH 5.8), and 10 mM MgSO4.
Data Collection, Structure Determination, and Analysis.
Diffraction data were collected at 98 K by using an Raxis IV detector (Molecular Structure, The Woodlands, TX) on a Rigaku rotating anode x-ray generator, and on beamlines X12B and X25 at the National Synchrotron Light Source, Brookhaven National Laboratory. Data were processed, phases were calculated, and models were refined and built as described in ref. 3 (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). The BF⋅DNA binary complex crystals are space group P212121 with one molecule per asymmetric unit (3); the BF⋅DNA⋅dCTP crystals are P31 with two molecules per asymmetric unit. The structure of the ternary complex was solved by molecular replacement using the binary complex. The P31 crystal data were treated as perfectly twinned (48.5% twinning). Molecule A of the ternary complex was used for analysis because it was the best ordered in the P31 asymmetric unit. The BF binary and ternary complexes were superimposed by using the Cα atoms of the palm subdomain (residues 646–655, 823–838, and 863–869).
Catalysis in the Crystal.
Catalysis was initiated by transfer of crystals into stabilization solutions containing 30 mM dNTP (10-bp structure, 30 mM dTTP; 11-bp structure, 15 mM dTTP and dATP; 12-bp structure, 10 mM dTTP, dATP, and dCTP; 15-bp structure, 7.5 mM dTTP, dATP, dCTP, and dGTP; ref. 3). Nucleotide soaks were allowed to proceed for 24 h at room temperature to ensure completeness of the reaction (partial incorporation can be observed after a 1-h soak).
Results and Discussion
DNA Synthesis in the Crystals.
DNA synthesis was observed in the BF polymerase crystals by cocrystallization with an annealed DNA primer⋅template containing a 9-bp primer⋅template duplex and a 6-nt 5′ single-stranded template overhang (Fig. 1A). These crystals were then transferred into stabilization solutions containing varying compositions of nucleotides. The template was designed to control the number of incorporation events in an accurately replicating polymerase⋅DNA complex by altering the nucleotide composition of the crystal stabilization solution. Four product complexes were obtained in separate experiments by addition of dTTP, dTTP+dATP, dTTP+dATP+dCTP, and dTTP+dATP+dCTP+dGTP to the starting primer⋅template, resulting in the formation of 10-, 11-, 12-, and 15-bp structures, respectively, as intended (Fig. 1A; and see Table 1). The maximally observed increase in DNA length observed in the 15-bp product corresponds to a 20-Å extension of the DNA duplex. This large increase in length is accommodated by the presence of large solvent channels in the crystal lattice, located at the site on the polymerase where the nascent DNA duplex emerges from the protein (Fig. 1B).
Figure 1.
DNA synthesis in the BF polymerase crystal. The BF polymerase domain is yellow and the putative 3′–5′ exonuclease domain is cyan. The corresponding DNA sequences of the template (red) and primer (green) strands for each complex are shown. (A) The structure of the initial 9-bp primer⋅template complex (Upper). Arrows indicate the product complexes resulting from four separate nucleotide soaking experiments. Nucleotides incorporated into the primer strand by the polymerase during crystal soaks are blue (underlined). (B) The 9-bp (Left) and 15-bp (Right) complexes are displayed with neighboring molecules in the crystal lattice. This view highlights a large solvent channel, ≈20 Å wide, that accommodates the nascent DNA.
The quality of the electron density maps allowed for unambiguous identification of the nucleotide sequence along the entire length of the DNA in the crystal (Fig. 4, which is published as supporting information on the PNAS web site), enabling us to deduce that all newly synthesized DNA forms the expected, cognate Watson–Crick base pairs. Furthermore, each round of nucleotide incorporation is accompanied by translocation of the DNA relative to the protein. Our choice of template sequence and extension mix, therefore, permitted all of the four possible Watson-Crick base pairs to be observed at the 3′ end of the primer terminus in the active site.
No base pair mismatches are observed in these experiments, within the limits of detection by crystallography, further confirming high-fidelity copying of the template in the crystal. Rather than inserting a mispair, the polymerase stalls, even under conditions where only noncognate dNTP was present in the extension mix. Furthermore, in the full-length product obtained in the presence of all four dNTPs, there is no density suggesting any additional, non-template-directed primer extension of the blunt-ended DNA duplex.
Capture of a Closed BF⋅DNA⋅dNTP Ternary Complex.
Crystals of a BF⋅DNA⋅dNTP ternary complex were obtained by incorporating a chain terminating dideoxynucleotide into the DNA primer strand to prevent continued synthesis, a strategy previously used to capture a closed conformation in other polymerase systems, including Taq (9) and T7 (11). Unfortunately, this approach still resulted in crystals that contain an open conformation in the BF polymerase, with the addition of a dNTP bound along the O helix (data not shown) in a position similar to that observed in polymerase⋅dNTP binary complexes of KF (23) and Taq (24). Analysis of the protein–protein contacts in the lattice of the BF crystals showed that the side chains of Asp-329 and -598 form a crystal contact at the tip of the fingers subdomain. We hypothesized that this contact is a dominant factor in shifting the conformational equilibrium to the open state in the crystal lattice. To test this hypothesis, we mutated these residues to alanine, singly and in combination. A new crystal form was obtained with a D392A mutant (Table 1) under the same crystallization conditions used to obtain the wild-type open ternary structure. This new crystal is also a ternary complex, but the polymerase now adopts a closed conformation, as predicted (Fig. 2). Similar to what has been observed in the Taq and T7 systems, the fingers subdomain of the D329A mutant closes around a newly formed dCTP⋅G base pair. Nucleotide incorporation is prevented because the primer strand contains a 3′ dideoxy terminus. Two metal binding sites are observed, coordinated by Asp-653, Asp-830, and the triphosphate of the incoming nucleotide.
Figure 2.
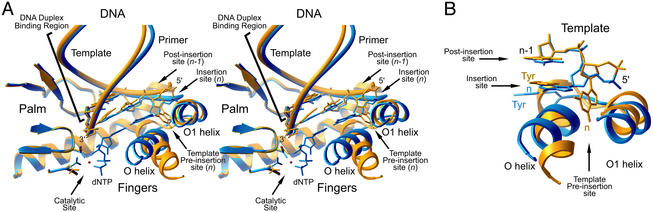
Active site superposition of open and closed BF structures. (A) The 11-bp open binary complex (yellow) and the closed ternary complex (blue) are shown in stereo view. The largest conformational differences occur in the fingers domain, including the O helix, O1 helix, and preinsertion site. The acceptor template base (n) occupies the preinsertion site in the open conformation and the insertion site in the closed conformation. (B) A close-up view of the preinsertion site. The locations of the conserved Tyr-714 are indicated.
Protein/DNA Interactions.
Four sites with important protein/DNA interactions have been described in the A-family polymerases (3, 9–11): the nucleotide “insertion site” adjacent the 3′ primer terminus, where the incoming nucleotide forms a Watson–Crick pair with the acceptor base on the template strand (template base n); the “catalytic site,” located adjacent to the insertion site, where formation of the new phosphodiester bond is catalyzed (the insertion and catalytic site together form the “replication site”); a “postinsertion site,” immediately adjacent to the insertion site, where the previously synthesized base pair forms the 3′ terminal base pair of the extending primer⋅template DNA duplex (base pair n-1); and a “DNA duplex binding region” where the newly synthesized duplex base pairs still interact with the polymerase (base pairs n-2 to n-11 in BF). The open BF product complexes described here allow us to further define an additional site, the “template preinsertion site,” where the acceptor template base (n) is located before moving into the replication site. Each of these sites is shown in structural detail in Fig. 2 and schematically in Fig. 3.
Figure 3.
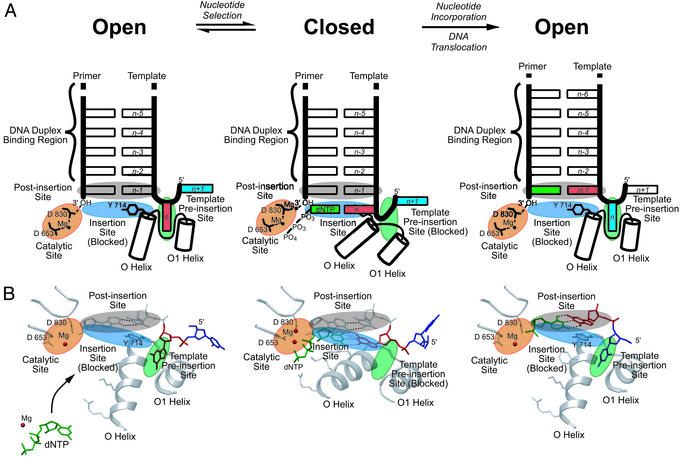
Conformational interlocks during DNA synthesis. A schematic overview of the polymerase active site (A) and atomic coordinates (B) derived from the open and closed BF structures represent a complete round of DNA synthesis. The conformational changes described here are presented in animated form in Movie 1, which is published as supporting information on the PNAS web site. The reaction cycle starts with the acceptor template base (n, red) bound at the template preinsertion site (between the O and O1 helices; green shading); Tyr-714 blocks access to the insertion site (blue shading) and stacks with the n-1 base pair at the postinsertion site (gray shading). Formation of the closed conformation involves rearrangement of the O and O1 helices, which simultaneously blocks the template preinsertion site and unblocks the insertion site. These rearrangements move the acceptor template base (n) to the insertion site, where it pairs with an incoming dNTP (green). Nucleotide incorporation occurs on formation of a cognate base pair and proper assembly of the catalytic site (orange shading). The cycle is completed with translocation of the DNA by one base pair position. The polymerase resets to the open conformation in preparation for the next round of DNA synthesis.
Insertion and preinsertion sites.
A characteristic feature of A-family polymerase⋅DNA primer template binary complexes (open conformation) is that the complementary base for the incoming nucleotide, the acceptor template base (n), is not located in the insertion site adjacent to the terminal base pair at the 3′ end of the primer (3, 9). A conserved tyrosine (Tyr-714 in BF) stacks against the n-1 template base, blocking access of the acceptor template base to the insertion site (Fig. 3; and see Fig. 5, which is published as supporting information on the PNAS web site). Additionally, in the open BF⋅DNA structures, the DNA template turns sharply away from the helical axis of the DNA duplex, placing the acceptor base in an additional pocket located in the interface between the O and O1 helices of the fingers (the template preinsertion site; Fig. 2). The floor of the preinsertion site is formed by a flexible loop, connecting these two helices, that can adjust to accommodate differences in purine and pyrimidine stereochemistry (Fig. 6, which is published as supporting information on the PNAS web site); all four nucleotides are observed placed at this preinsertion site. No equivalent preinsertion site has been recognized in previously determined structures of polymerases complexed with substrates. In other open complexes, the single-stranded template overhang either is not present in the DNA (8) or differs in detail from the conformation of the BF structure (9). In the closed BF, Taq (9), and T7 (11) ternary complexes, the preinsertion site is blocked (see below).
Although the acceptor template base is sequestered at the preinsertion site in the open BF structures, template-directed DNA polymerization still takes place in the BF crystals. This implies that conformational changes must occur in the crystal to position the acceptor base into the insertion site for catalysis. A superposition of the open and closed conformations of the BF polymerase shows that the transition from the open to the closed state is primarily confined to rigid-body motions of the finger subdomain (Fig. 2) and conformational changes in three hinge regions in the fingers. The conformational change results in a 40° rotation of the O helix (Fig. 2), but there are no obvious contacts in the wild-type BF crystal lattice that would prevent such a large motion and inhibit catalysis in the crystals.
Although this conformational change between open and closed states following dNTP binding has been described (4, 9, 25), identification of the template preinsertion site provided here allows for a more complete description of this transition. The nature of the conformational changes dictates that the insertion and preinsertion sites be mutually exclusive. In the open state, Tyr-714 blocks the insertion site [as seen previously (6, 9)], and the preinsertion site is open to bind the acceptor base of the template strand (this study). Conversely, in the closed state, the insertion site is not blocked by Tyr-714 and the loop connecting the O and O1 helices blocks the preinsertion site. Tyr-714 is located at the base of the O helix; the preinsertion site is formed by one face of the O helix and the loop connecting the O and O1 helices. The insertion site is formed by the opposite face of the O helix. Global motions of the O helix that correspond to the exchange between the open and closed structures of the polymerase, therefore, not only control Tyr-714 (as described in ref. 9) but also serve to couple the local conformational changes at the insertion and the preinsertion sites (this study).
Catalytic site.
In the open conformation, the 3′ hydroxyl of the primer strand forms a hydrogen bond with Asp-830 (Fig. 5), a residue that is universally conserved in all polymerases (12) and essential for activity (26). This interaction was not observed in the closed BF complex or in other A-family polymerases because those structures were obtained by using a primer terminus that lacks the 3′ hydroxyl group. Formation of the phosphodiester bond is catalyzed by a two-metal mechanism (4). Two metal sites are observed in the closed conformation. However, in the open BF crystal structures there is only one Mg2+, coordinated by Asp-830 and Asp-653. The other metal, located closely to the 3′ terminus, is absent. This finding supports recent observations made on the terminal transferase human pol β that the catalytically competent form of the active site assembles with the addition of the catalytic metal, either concurrent with or subsequent to dNTP binding at the insertion site (27).
One consequence of the presence of a labile metal coordination sphere is that the Asp-830 performs a dual role: in the absence of metal, it forms a hydrogen bond with the 3′ hydroxyl that is subsequently replaced by interactions with the newly introduced Mg2+ ion. An accompanying change in sugar pucker at the 3′ terminus of the primer strand is sufficient to position the 3′ hydroxyl for an in-line attack on the α-phosphate of the nucleotide (see below).
Postinsertion site and DNA duplex binding region.
Each of the four Watson–Crick base pairs has been captured at the postinsertion site of BF. The complementarity of the site to Watson–Crick base pairs and the conserved geometry of minor groove hydrogen bond acceptors agree with observations made for analogous complexes in Taq (10). Additionally, successive translocations of the DNA duplex observed in the BF structures presented here allow all four Watson–Crick base pairs to be seen at each position along the DNA duplex binding region. These structures highlight the sequence-independent nature of minor groove recognition (Fig. 7, which is published as supporting information on the PNAS web site) and the A to B form DNA conformational transition that extends out to the n-5 base pair.
Replication Fidelity Is Achieved by a Series of Conformational Interlocks.
It has been established that the integration of polymerase functions (catalysis, DNA translocation, and high-fidelity base pair recognition) is achieved by conformational coupling mechanisms that link global conformational changes to the behavior of individual sites and kinetic steps (2, 28). Here, we extend this model and propose that frameshifts are also prevented by a conformational coupling mechanism.
BF polymerase has the advantage that processive, accurate replication takes place directly in the crystal. Consequently, any conformations that are observed are likely to be “on-path.” The high-resolution observations of successive replication events allows us to confirm, refine, and extend aspects of the previously described conformational coupling mechanisms. The conformation of the open BF complexes reported here must occur at least one step before nucleotide incorporation because the incoming nucleotide is absent, the 3′ primer terminus is occluded, and the acceptor template base is sequestered. These structures, therefore, define a state additional to those described elsewhere for DNA polymerases. In this state, the acceptor base on the template strand occupies a binding site, the preinsertion site, before being positioned in the insertion site, which is occluded by Tyr-714 in the open conformation. We also provide structural evidence that the incoming nucleotide, in addition to pairing with the acceptor base, also completes the assembly of the metal center necessary for the catalytic two-metal mechanism. Although this concept has been proposed previously, the only structural evidence has been obtained on pol β human terminal transferase, rather than a member of the A-family (27). This new information, together with previous studies, refines the model in which fidelity of DNA replication arises from an intricate series of interlocking conformational changes (Fig. 3). The interlocks function as checkpoints that halt progression of the catalytic cycle when base mispairing or strand register errors (frameshifts) impose geometries that hinder procession to the next step, thereby setting up a series of checks and balances that allow the reaction to reverse without incorporation or pause to allow nucleotide excision. Here, we summarize the current model, pointing out the new information.
Nucleotide-mediated assembly of the two-metal mechanism (4) provides the first steric checkpoint at which the reaction can stall. The incoming nucleotide completes the coordination sphere for the catalytic Mg2+ ion and displaces the hydrogen bond between the 3′ primer hydroxyl and Asp-830. Chemically productive assembly of the catalytic site is likely to occur only on formation of cognate base pairs because mispairs will form less favorable geometries (11, 14). Such coupling of the assembly of catalytic functionality with base pair readout amplifies the discrimination conferred by base pair readout alone.
The movement of the acceptor template base between the preinsertion site and insertion sites provides a mechanism by which the reaction can readily reverse itself when stalled at the first checkpoint. In this model, on stalling, rapid formation of the preinsertion site provides a binding site for the acceptor template base to move back into, out of the insertion site, thereby facilitating the release of the unincorporated, mismatched nucleotide. The rigid-body motion of the O helix couples the movement of Tyr-714 in and out of the insertion site with the concerted opening and closing of the preinsertion site, and corresponds to the exchange of the open and closed conformations of the polymerase. In this model there is, therefore, no obligate linkage between nucleotide binding and formation of the closed state. Indeed, we and others have frequently observed nucleotide triphosphates bound along the O helix in the open conformation with (S.J.J. and L.S.B., unpublished results) and without primer⋅template DNA (23, 24). The expectation is that the conformational exchange mechanism occurs on a faster time scale than nucleotide incorporation; this conformational change should, therefore, not be a rate-limiting step. A recently identified mutant in pol β that increases the affinity for correct dNTP binding in the ground state also suggests that the rate-limiting step occurs after transition to the closed conformation (27). The prediction is that the rate-limiting step observed to occur after dNTP binding, but before catalysis (29, 30), corresponds either to binding of the second Mg2+ or breaking of the hydrogen bond between Asp-830 and the 3′ hydroxyl.
The second main checkpoint is provided by the postinsertion site (3, 10, 11, 15). The hydrogen bonds between the protein and the minor groove are expected to form only on correctly formed Watson–Crick base pairs (31). DNA replication in the presence of a mismatch at this position is, therefore, likely to stall because of decreased binding to the postinsertion site or because of a change in the geometry of the primer 3′ hydroxyl. Stalling at the postinsertion site promotes excision rather than continued synthesis (2, 28).
There is no clear linkage between stalling at the postinsertion site and the exchange mechanism of the preinsertion and insertion sites, because the reaction reversal involves exonuclease-dependent excision rather than a microscopic reversal of the chemical step. However, the post- and preinsertion sites are adjacent to each other (Fig. 3). Structural defects at the postinsertion site resulting from a mismatch could, therefore, interfere with proper formation of the preinsertion site. This, in turn, would interfere with the template feeding into the insertion site, slowing down the polymerization reaction and thereby providing a clear structural mechanism for stalling. Structural studies of mismatched bases at the postinsertion site are needed to test this hypothesis.
The regulated transfer of the template from the preinsertion site to the insertion site proposed here could confer significant advantages to the polymerase with respect to frameshift fidelity. The A-family polymerases discriminate strongly against frameshift errors, with an error rate of ≤10−5 for KF (32). This strategy eliminates competition for the active site by other template overhang bases located further upstream because a base cannot enter the insertion site without first passing through the preinsertion site. This site is positioned such that only the base immediately adjacent to the primer terminal base pair is able to bind properly. Continuous regulation of the template position may also reduce frameshift errors by discouraging template release and rebinding, which has been proposed to initiate strand slippage (2).
The preinsertion site is likely to be conserved among the A-family of DNA polymerases. The critical sequences at the end of the O helix, including Tyr-714 and flanking glycine residues, form part of a sequence motif (motif B) that is highly conserved within the A-family (12). Furthermore, mutagenesis studies on one member of the A-family, KF, strongly support the critical role that the model assigns to Tyr-714. Mutation of Tyr-714 to a phenylalanine in KF has a negligible effect (33); mutation to serine or alanine results in a higher frequency of both point and frameshift mutations (34, 35). This is entirely consistent with the notion that regulated movement of the template depends on concerted blocking and unblocking of the insertion and preinsertion sites. Mutation to smaller residues, but not to phenylalanine, destroys the ability to block access to the insertion site.
A preinsertion site may be conserved among polymerases from other families. T7 RNA polymerase exhibits an essentially identical preinsertion structure in complexes at both initiation and elongation stages of RNA synthesis (36, 37). Current structural data from the B-family eukaryotic pol α and archeal DNA polymerases suggest that this family may also use a preinsertion site. In the closed ternary complex of RB69 (15), a 90° rotation along the template backbone places the template in the center of a helix–loop–helix motif bounded by a conserved tyrosine and two conserved glycines. This motif is also observed in each of the available B-family apo structures (16–19). Verification of such a binding site must await structural determination of a catalytically competent open binary complex.
It is interesting to note the lack of a template preinsertion site in eukaryotic pol β (X-family polymerase) and HIV reverse transcriptase, both of which have lower DNA replication fidelity than the A- and B-family polymerases (38, 39), although the abrupt turn in the template strand on entering the insertion site is retained. Recently published structures of the error-prone Y-family lesion-bypass polymerases provide an even more striking contrast to the high-fidelity A-family polymerase structures. The replication site of the closed ternary Dpo4 complex (40) is large enough to accommodate a second template base. The O helix is replaced by a β-strand and an extended loop. The fingers appear to lack the necessary architecture to form a preinsertion site. Instead of a sharp turn in the template backbone, the DNA template assumes a more linear conformation as it enters the active site. All of these features suggest the lack of a preinsertion site and may contribute to the low processivity and fidelity of this class of enzymes.
Supplementary Material
Acknowledgments
Research was carried out (in whole or in part) at the National Synchrotron Light Source, Brookhaven National Laboratory, which is supported by the U.S. Department of Energy, Division of Materials Sciences and Division of Chemical Sciences, under Contract No. DE-AC02-98CH10886. We thank Joel Tuttle for purification of the Bacillus protein, Lawrence Forsberg for assistance with data collection and crystallization, and H. W. Hellinga for critical discussions and extensive assistance with this manuscript. This work was supported by grants to L.S.B. from the American Cancer Society, the Searle Scholars Program, and the Human Frontier Science Program (Grant RG0351/1998-M).
Abbreviations
- BF
Bacillus fragment
- KF
Klenow fragment
- Taq
Thermus aquaticus
- T7
bacteriophage T7
Footnotes
Data deposition: The coordinates and structure factors for the Bacillus polymerase⋅DNA complexes have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1L3S, 1L3T, 1L3U, 1L5U, 1L3V, and 1LV5).
References
- 1.Echols H, Goodman M F. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 2.Kunkel T A, Bebenek K. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 3.Kiefer J R, Mao C, Braman J C, Beese L S. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- 4.Brautigam C A, Steitz T A. Curr Opin Struct Biol. 1998;8:54–63. doi: 10.1016/s0959-440x(98)80010-9. [DOI] [PubMed] [Google Scholar]
- 5.Ollis D L, Brick P, Hamlin R, Xuong N G, Steitz T A. Nature. 1985;313:762–766. doi: 10.1038/313762a0. [DOI] [PubMed] [Google Scholar]
- 6.Kiefer J R, Mao C, Hansen C J, Basehore S L, Hogrefe H H, Braman J C, Beese L S. Structure (London) 1997;5:95–108. doi: 10.1016/s0969-2126(97)00169-x. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y, Eom S H, Wang J, Lee D S, Suh S W, Steitz T A. Nature. 1995;376:612–616. doi: 10.1038/376612a0. [DOI] [PubMed] [Google Scholar]
- 8.Eom S H, Wang J, Steitz T A. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Korolev S, Waksman G. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Waksman G. Protein Sci. 2001;10:1225–1233. doi: 10.1110/ps.250101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublié S, Tabor S, Long A M, Richardson C C, Ellenberger T. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 12.Delarue M, Poch O, Tordo N, Moras D, Argos P. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 13.Pelletier H, Sawaya M R, Kumar A, Wilson S H, Kraut J. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 14.Sawaya M R, Prasad R, Wilson S H, Kraut J, Pelletier H. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- 15.Franklin M C, Wang J, Steitz T A. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Sattar A K, Wang C C, Karam J D, Konigsberg W H, Steitz T A. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez A C, Park H W, Mao C, Beese L S. J Mol Biol. 2000;299:447–462. doi: 10.1006/jmbi.2000.3728. [DOI] [PubMed] [Google Scholar]
- 18.Hopfner K P, Eichinger A, Engh R A, Laue F, Ankenbauer W, Huber R, Angerer B. Proc Natl Acad Sci USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Y, Jeruzalmi D, Moarefi I, Leighton L, Lasken R, Kuriyan J. Struct Fold Des. 1999;7:1189–1199. doi: 10.1016/s0969-2126(00)80053-2. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Chopra R, Verdine G L, Harrison S C. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 21.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 22.Jacobo-Molina A, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beese L S, Friedman J M, Steitz T A. Biochemistry. 1993;32:14095–14101. doi: 10.1021/bi00214a004. [DOI] [PubMed] [Google Scholar]
- 24.Li Y, Kong Y, Korolev S, Waksman G. Protein Sci. 1998;7:1116–1123. doi: 10.1002/pro.5560070505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doublié S, Sawaya M R, Ellenberger T. Struct Fold Des. 1999;7:R31–R35. doi: 10.1016/S0969-2126(99)80017-3. [DOI] [PubMed] [Google Scholar]
- 26.Polesky A H, Steitz T A, Grindley N D F, Joyce C M. J Biol Chem. 1990;265:14579–14591. [PubMed] [Google Scholar]
- 27.Vande Berg B J, Beard W A, Wilson S H. J Biol Chem. 2001;276:3408–3416. doi: 10.1074/jbc.M002884200. [DOI] [PubMed] [Google Scholar]
- 28.Johnson K A. Annu Rev Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 29.Kuchta R D, Benkovic P, Benkovic S J. Biochemistry. 1988;27:6716–6725. doi: 10.1021/bi00418a012. [DOI] [PubMed] [Google Scholar]
- 30.Wong I, Patel S S, Johnson K A. Biochemistry. 1991;30:526–537. doi: 10.1021/bi00216a030. [DOI] [PubMed] [Google Scholar]
- 31.Seeman N C, Rosenberg J M, Rich A. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bebenek K, Joyce C M, Fitzgerald M P, Kunkel T A. J Biol Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 33.Carroll S S, Cowart M, Benkovic S J. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 34.Minnick D T, Bebenek K, Osheroff W P, Turner R M, Jr, Astatke M, Liu L, Kunkel T A, Joyce C M. J Biol Chem. 1999;274:3067–3075. doi: 10.1074/jbc.274.5.3067. [DOI] [PubMed] [Google Scholar]
- 35.Bell J B, Eckert K A, Joyce C M, Kunkel T A. J Biol Chem. 1997;272:7345–7351. doi: 10.1074/jbc.272.11.7345. [DOI] [PubMed] [Google Scholar]
- 36.Tahirov T H, Temiakov D, Anikin M, Patlan V, McAllister W T, Vassylyev D G, Yokoyama S. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 37.Yin Y W, Steitz T A. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 38.Roberts J D, Kunkel T A. In: DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. Pamphilis M D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 217–247. [Google Scholar]
- 39.Bebenek K, Kunkel T A. In: Reverse Transcriptase. Skalka A M, Goff S P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 85–102. [Google Scholar]
- 40.Ling H, Boudsocq F, Woodgate R, Yang W. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.