Abstract
MD-2 is a secreted glycoprotein that binds to the extracellular domain of Toll-like receptor 4 (TLR4) and is required for the activation of TLR4 by lipopolysaccharide (LPS). The protein contains seven Cys residues and consists of a heterogeneous collection of disulfide-linked oligomers. To investigate the role of sulfhydryls in MD-2 structure and function, we created 17 single and multiple Cys substitution mutants. All of the MD-2 mutant proteins, including one totally lacking Cys residues, were secreted and stable. SDS/PAGE analyses indicated that most Cys residues could participate in oligomer formation and that no single Cys residue was required for oligomerization. Of the single Cys substitutions, only C95S and C105S failed to confer LPS responsiveness on TLR4 when mutant and TLR4 were cotransfected into cells expressing an NF-κB reporter plasmid. Surprisingly, substitution of both C95 and C105 partially restored activity. Structural analyses revealed that C95 and C105 formed an intrachain disulfide bond, whereas C95 by itself produced an inactive dimer. In contrast to the cotransfection experiments, only WT MD-2 conferred responsiveness to LPS when secreted proteins were added directly to TLR4 reporter cells. Our data are consistent with a model in which most, possibly all sulfhydryls lie on the surface of a stable MD-2 core structure where they form both intra- and interchain disulfide bridges. These disulfide bonds produce a heterogeneous array of oligomers, including some species that can form an active complex with TLR4.
Mammalian cells respond to lipopolysaccharide (LPS), a major component of the outer membrane of Gram-negative bacterial cell walls via a collection of cell surface proteins, including Toll-like receptor 4 (TLR4), CD14, and MD-2 (1–4). Of these, only TLR4 is capable of transducing inflammatory signals and TLR4-deficient mice fail to respond to LPS (5, 6). The laboratories of both Golenbock and Miyake (7- 9) have demonstrated that TLR4 requires the presence of an accessory protein, MD-2, for responsiveness to LPS in vitro and in vivo. MD-2 binds to the extracellular domain of TLR4 (7) and to LPS (10, 11), and a potential LPS binding motif has been identified within the MD-2 molecule (12). Human and mouse MD-2/TLR4 complexes exhibit distinct specificities for taxol, lipid IVa, and Salmonella lipid A, and it has recently been shown that MD-2 contributes to these differences in specificity (13–15). It is currently thought that a serum protein, LPS binding protein, catalyzes the transfer of individual LPS molecules from micelles to CD14 on the cell surface (16), and that LPS is then transferred to the TLR4/MD-2 signaling complex (10). Whether MD-2 provides the LPS recognition site of the MD-2/TLR4 complex is not known, but it is clear that it plays an important role in LPS recognition.
The reduced form of MD-2 is a 20- to 25-kDa monomeric protein with seven Cys residues and two N-linked glycosylation sites. Differential glycosylation gives rise to three glycoforms distinguishable by SDS/PAGE, and glycosylation is required for MD-2 function (17, 18). We have previously shown that MD-2 can interact with TLR4 in the endoplasmic reticulum (ER) or that it can be secreted into the medium (19). Secreted MD-2 exists as a heterogeneous collection of disulfide linked oligomers, which can confer LPS responsiveness directly to TLR4 on the cell surface. In this paper we have created a series of single and multiple MD-2 Cys to Ser mutants to investigate the role of sulfhydryls in oligomer formation and LPS responsiveness.
Materials and Methods
Cell Lines.
Human embryonic kidney (HEK) 293, and HEK293T cell lines were obtained from the American Type Culture Collection (Manassas, VA). TLR4 reporter cells (HEK293 cells stably expressing hTLR4 and the NF-κB reporter gene, ELAM-1-luciferase) were a kind gift from Jesse Chow (Eisai Research Institute, Andover, MA) (20). Cell lines were maintained in DMEM containing 10% FBS, glutamine, and antibiotics.
Expression Vectors.
pEFBOS-MD-2-Flag-His (7) was kindly provided by Kensuke Miyake (Saga Medical School, Nabeshima, Japan). The hTLR4 expression vector was a kind gift from Ruslan Medzhitov (Yale University School of Medicine, New Haven, CT).
Site-Directed Mutagenesis.
Single Cys to Ser mutations were introduced into pEFBOS-MD-2-Flag-His by using the QuikChange site-directed mutagenesis kit (Stratagene). Multiple Cys to Ser mutations were introduced by using the QuikChange Multisite directed mutagenesis kit (Stratagene). All mutations were confirmed by automated DNA sequencing.
Immunoprecipitations and Western Blots.
Two million HEK293T cells in 10-cm tissue culture dishes were transfected with 5 μg of MD-2 (WT or mutant) plasmid by using calcium phosphate. Twenty-four hours after the medium was changed, supernatants were collected and adjusted to 25 mM iodoacetamide/100 mM Tris, pH 8.0. Six-milliliter samples were immunoprecipitated overnight with 12 μg of anti-FLAG M2 mAb (Sigma) and 60-μl packed Protein A Sepharose beads (Amersham Pharmacia) at 4°C. Samples were washed once with Hanks' balanced salt solution (HBSS), twice with PBS, and once with distilled water, and divided into three for further processing. Reduced (R) and nonreduced (NR) samples were boiled in SDS sample buffer with and without DTT, respectively. Samples were partially reduced (PR) under nondenaturing conditions by suspending them in 10 mM DTT/100 mM Tris/1 mM EDTA, pH 8.0 for 1 h at room temperature. The beads were pelletted and resuspended in 25 mM iodoacetamide/100 mM Tris, pH 8.0 and incubated for 30 min at room temperature, after which an equal volume of 2× SDS sample buffer (without DTT) was added followed by boiling. All samples were resolved by SDS/PAGE (9% for NR and PR, 13% for R), immunoblotted with anti-FLAG M2 mAb peroxidase conjugate (Sigma), and developed by using enhanced chemiluminescence (Amersham Pharmacia).
NF-κB Activity.
For cotransfection experiments HEK293 cells (1 × 104 cells per well) were transfected with 12 ng each of Ig-κB-luciferase and pRSV-β-galactosidase reporter plasmids (kindly provided by Ulrich Siebenlist, National Institute of Allergy and Infectious Diseases, Bethesda), 6 ng of TLR4 plasmid and 12 ng of vector encoding either WT or mutant MD-2. Total amounts of DNA were kept constant at 100 ng by the addition of empty vector. Cells were treated with or without 100 ng/ml LPS (phenol-extracted S. minnesota Re595, Sigma) for 8 h. After treatment, cells were lysed in Reporter Lysis buffer (Promega), and luciferase and β-galactosidase activities were determined by using Luciferase (Promega) and Tropix (PE Biosystems, Bedford, MA) Chemiluminescent assay kits. Relative luciferase units (RLU) were calculated by dividing the luciferase signal by the β-galactosidase signal. Triplicate RLU values from each point were normalized to the average RLU obtained from WT MD-2 after LPS stimulation. Normalized data from a minimum of three independent experiments (at least nine replicates per point) were averaged and SEM values were calculated. A two-sample t test (assuming unequal variance) was used to determine the significance of differences in NF-κB activation between two different data points.
In “add-back experiments,” TLR4 reporter cells (5 × 104 per well) were incubated for 8–16 h with 100 ng/ml LPS and 100 μl of serial dilutions of 20-fold concentrated culture supernatants containing WT or mutant MD-2, and NF-κB activities were measured in triplicate by using the Promega Luciferase Assay kit. MD-2 protein was quantified by immunoprecipitating 100 μl of 20-fold concentrated culture supernatants with anti-FLAG, resolving serial dilutions of the solubilized protein (in SDS reducing buffer) by 10% SDS/PAGE, and blotting with the anti-FLAG mAb. Protein was detected by enhanced chemiluminescence and quantified by using a 5500 imaging system (Alpha Innotech, San Leandro, CA) with imaging software from Scion (Frederick, MD). Intensities were compared with those from known concentrations of a C-terminal FLAG-tagged standard protein (FLAG-BAP fusion protein, Sigma), yielding an estimate of the molar concentration of MD-2 subunit.
Results
Functional Consequences of Cys Substitutions.
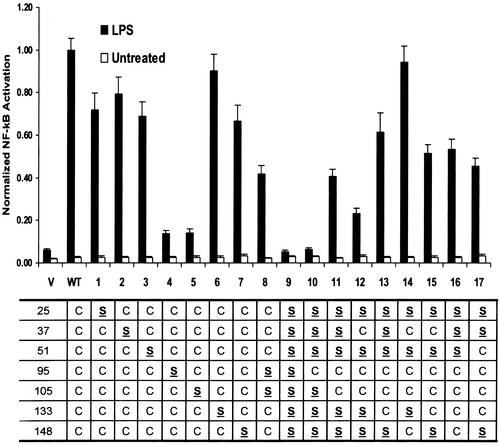
MD-2, an oligomeric protein with seven Cys residues, binds to the extracellular domain of TLR4 and is required for LPS signaling. To determine whether Cys residues are required for MD-2 function, we sequentially mutated each Cys to Ser and tested the mutants for the capacity to confer LPS responsiveness. In cotransfection experiments (Fig. 1), each MD-2 mutant was introduced into HEK293 cells along with TLR4 and NF-κB reporter plasmids, and NF-κB activation was measured in response to LPS. With the exceptions of C95S and C105S (mutants 4 and 5), single Cys substitutions (mutants 1–7) produced only minor losses (10–33%) in activity compared with WT MD-2. C95S and C105S showed >85% loss in activity when compared with WT MD-2 (P < 0.001), which however, was significantly greater than the empty vector control (P < 0.005). Surprisingly, substitution of both C95 and C105 produced a molecule (mutant 8) that was significantly more active than either single mutant alone (P < 0.001), suggesting that the presence of either C95 or C105 without the other is inhibitory. To further define the roles of C95 and C105 in MD-2 function, we generated a mutant lacking all Cys residues (mutant 9), one containing C95 only (mutant 10), and one containing only C95 and C105 (mutant 11). Although mutants 9 and 10 were totally inactive (Fig. 1), mutant 11 showed partial (41%) activity (P < 0.001 compared with either vector or WT). Thus, although C95 and C105 are not absolutely required for MD-2 function (mutant 8), they can, by themselves, yield partially active protein. In attempts to totally restore activity, additional sulfhydryls were introduced into mutant 11. Either C37 (mutant 12) or C133 (mutant 13) caused minor changes in activity, whereas a fourth sulfhydryl either totally restored activity (mutant 14), or had little effect on function (mutants 15, 16, and 17).
Figure 1.
Effects of Cys substitutions on MD-2 activity. HEK293 cells were cotransfected with reporter vectors, hTLR4, and empty vector (V), WT, or mutant MD-2. Transfectants were treated with (filled bars) or without (open bars) LPS, and NF-κB activities were determined and normalized to the activity of WT MD-2 in the presence of LPS. Each bar represents the mean and SEM of at least nine replicated points from three or more independent experiments. Residues that were mutated from Cys (C) to Ser (S) are indicated in the table below the graph, and mutant designations are specified on the x axis.
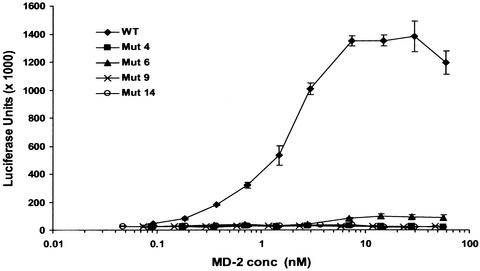
In striking contrast to the results from cotransfection experiments (Fig. 1), only WT MD-2 conferred responsiveness to LPS when supernatants were added directly to TLR4 reporter cells (Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). To see whether the mutants would be active at higher concentrations, supernatants containing WT and selected MD-2 mutants were concentrated 20-fold and serial dilutions added to TLR4 reporter cells. MD-2 concentrations were determined as described in Materials and Methods. As seen in Fig. 2, WT MD-2 gave a dose-response curve with an ED50 of 2 nM, and a significant response was still detected at a concentration of 200 pM. The mutant proteins, including the two that were most active in cotransfection experiments (6 and 14) were, however, inactive at the highest concentration achieved, (≈60 nM), indicating that the mutants were at least 300-fold less active than WT MD-2. Thus, all Cys residues are required for activity when MD-2 is added in the medium to TLR4 on the cell surface.
Figure 2.
Cys mutants are inactive in add-back experiments. TLR4 reporter cells were incubated with LPS and serial dilutions of 20-fold concentrated supernatants containing WT MD-2 or mutants 4, 6, 9, or 14. Concentrated supernatants from untransfected HEK293T cells did not confer LPS responsiveness (not shown).
MD-2 Cys Mutants Are Secreted, Stable Proteins.
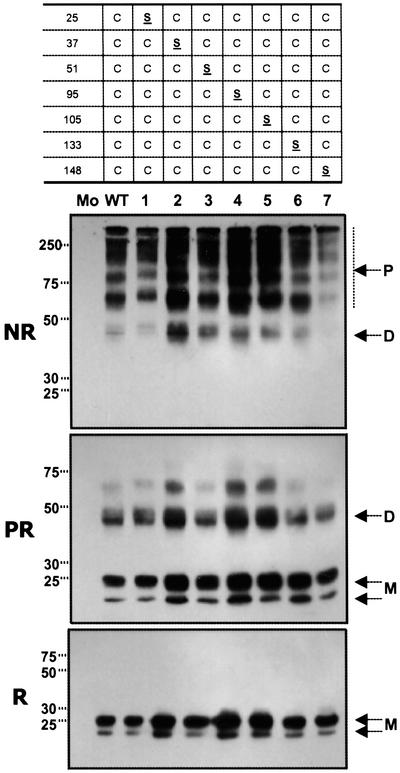
To correlate MD-2 function with structure, we wanted to examine secreted MD-2 WT and mutant proteins by SDS/PAGE. However, because disulfide bridges are often required for protein stability, we expected that some of the mutants would be structurally defective and would either fail to be secreted or would precipitate. This was especially true for mutant 9, which lacked Cys residues altogether. Surprisingly all mutants, including mutant 9, were secreted in amounts comparable to the WT (Figs. 3 and 4, reducing conditions) and were stable for at least 3 days in the medium. Mutant 9 was resistant to digestion with endoglycosidase H (data not shown), indicating that it had progressed through the normal secretion pathway. However, it eluted as intermediate molecular weight polymers by gel filtration under nondenaturing conditions (data not shown), possibly because of an intrinsic tendency for MD-2 to self-associate noncovalently (19) or to local instabilities in the mutant 9 protein, leading to nonspecific aggregation.
Figure 3.
Differential effects of single Cys substitutions on MD-2 oligomerization. WT or mutant MD-2 proteins were immunoprecipitated from transiently transfected HEK293T culture supernatants and analyzed by SDS/PAGE under nonreducing (NR and PR) or reducing (R) conditions. Samples designated PR were reduced and alkylated under nondenaturing conditions before analysis by SDS/PAGE. M, monomer; D, dimer; P, polymer. Mo refers to supernatant from mock-transfected HEK293T cells. In NR and PR panels, exposures were chosen to best show oligomer distributions. No MD-2 monomer was seen in WT or any single Cys mutant even at much higher exposure (not shown). In R, all samples were run at the same exposure and are the same samples used in add-back experiments (Fig. 6).
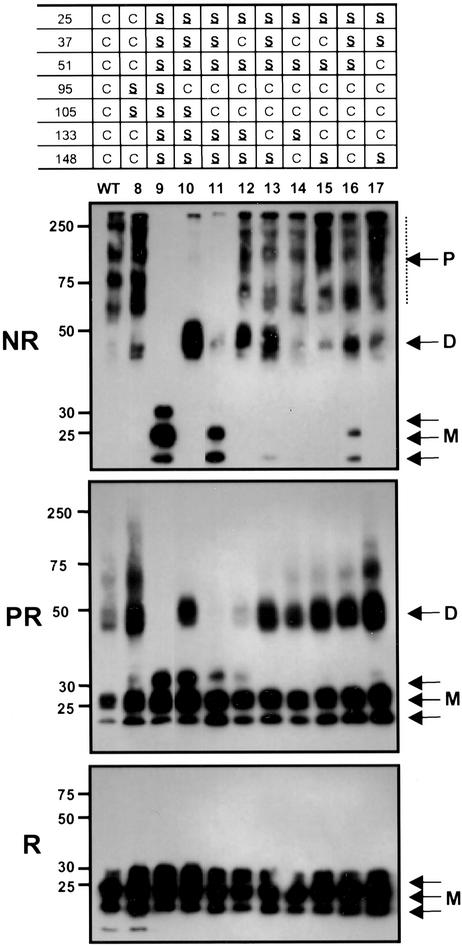
Figure 4.
Polymer distributions of MD-2 mutants containing multiple Cys substitutions. MD-2 protein containing multiple Cys to Ser substitutions were treated as in Fig. 3.
C95 and C105 Form an Intrachain Disulfide Bond.
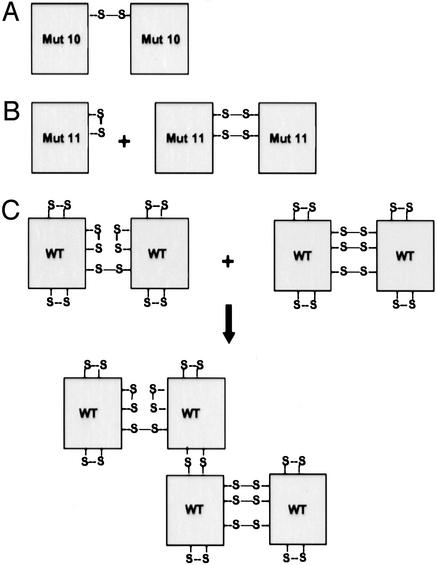
WT MD-2 is secreted as a heterogeneous array of disulfide linked polymers (Fig. 3). Because C95 and C105 are important for MD-2 function, we asked whether they might play a special role in polymerization of MD-2. SDS/PAGE analysis of secreted MD-2 mutant proteins (Figs. 3 and 4) showed this not to be the case. Thus all MD-2 mutants containing single Cys substitutions, including C95S and C105S (mutants 4 and 5, Fig. 3), as well as the C95/105S double mutant (mutant 8, Fig. 4) were similar to the WT protein in polymer distribution (NR) and stability to reduction under nondenaturing conditions (PR). To better understand how C95 and C105 influence MD-2 structure we examined variants that contained no Cys residues (mutant 9), C95 only (mutant 10), or C95 plus C105 (mutants 11). As expected, mutant 9 was monomeric. Introduction of C95 (mutant 10) gave a protein that was completely dimeric indicating that C95 efficiently formed an intermolecular disulfide bridge. Mutant 11 (C95 plus C105) was mainly monomeric, but also contained a small amount of dimer. The predominance of monomer in mutant 11 indicates that C95 and C105 preferentially form an intrachain disulfide bond. However, the presence of dimer in mutant 11 suggests that C95 and C105 are located on the external surface of MD-2 permitting the formation of intermolecular disulfide bridges (Fig. 4).
Three Sulfhydryl Residues Promote Oligomerization.
To determine the number of additional Cys residues required to generate the WT oligomer profile, we introduced a third Cys, either C37 (mutant 12) or C133 (mutant 13) into mutant 11 (C95, C105). As seen in Fig. 4, mutants 12 and 13 are largely dimeric under nonreducing conditions, suggesting C37 and C133 each form an interchain disulfide bridge, whereas C95 and C105 retain their intrachain pairing. However, the presence of large polymers in these mutants indicates that C95 and C105 can alternatively form intermolecular disulfide bridges, as they do in mutant 11. Mutant 12 was more susceptible to reduction under native conditions (Fig. 4, PR) than mutant 13, suggesting that the C37 interchain disulfide bridge was more exposed to the medium than the C133 bridge. Introduction of a fourth Cys to mutants 12 and 13 produces wild-type distributions of oligomers (mutants 14–17, Fig. 4) with the exception of mutant 16 (C95, 105, 133, 148), which contains monomer. The presence of monomer in mutant 16 suggests that C133 and C148 form a second intrachain disulfide bond. By contrast, the absence of monomer in mutants 15 (C37, 95, 105, 133) and 17 (C51, 95, 105, 133) suggests that C37 and C51 are unable to form an intrachain disulfide bond with C133.
Discussion
MD-2 consists of a relatively short polypeptide chain of about 145 aa with seven conserved Cys residues. In this paper, we show that these Cys residues play an important role in the generation of an active TLR4-MD-2 LPS signaling complex, and that most are located on the surface of the MD-2 molecule where they can form either inter- or intrachain disulfide bridges. The add-back experiments (Figs. 2 and 6) demonstrate that all Cys residues are required for secreted MD-2 to confer LPS responsiveness to TLR4. By contrast, the MD-2/TLR4 co-transfection experiments (Fig. 1) show that as few as two Cys residues (C95 and C105, mutant 11) can confer at least partial LPS responsiveness. One possible explanation for this discrepancy is that under cotransfection, but not add-back conditions, chaperones compensate for minor alterations in the MD-2 structure resulting from the Cys substitutions. High local concentrations of MD-2 and TLR4 in the ER probably do not account for the discrepancy, because we were unable to restore function of the MD-2 mutants by increasing their concentrations 20-fold in add-back experiments. Our observation that all Cys residues are required for activity when MD-2 interacts with TLR4 at the cell surface, but are not required when they interact in the ER, together with data showing that all seven Cys residues are completely conserved in MD-2 from the five currently available species, suggests that the interaction of MD-2 with TLR4 at the cell surface may play an important role in maintaining a functional TLR4 complex in vivo.
C95 and C105 were of special interest because mutation of either of these residues resulted in a near total loss in function, a finding that had also been previously reported by others (9, 21). Schromm et al. (9) showed that the C95Y mutant retained its capacity to bind to TLR4 in a coprecipitation experiment, and similarly we observed that of the mutants tested (mutants 1–12 and 14) all coprecipitated with TLR4 (M.N.K. and A.V., unpublished result). The interpretation that C95 or C105 is required for MD-2 function is negated by the observation that deletion of both these residues (mutant 8, Fig. 1) results in an increase in activity relative to either C95S or C105S alone. Removal of C95, C105, or both had no effect on MD-2 oligomerization (Figs. 3 and 4), indicating that losses in activity were not caused by a loss in polymer formation. To correlate function with structure, we created mutants that contained only C95, or C95 and C105 (mutants 10 and 11, respectively). Because mutant 11 was predominantly monomeric (Fig. 4), we reasoned that C95 and C105 formed an intrachain disulfide bond, as illustrated in Fig. 5B. The alternative possibility, that mutant 11 was monomeric because it failed to form any disulfide bonds, was highly unlikely because C95 quantitatively formed an interchain disulfide bridge in the absence of C105, as demonstrated by the dimeric structure of mutant 10 (Fig. 5A). In addition to monomer, mutant 11 also contained a small amount of dimer, indicating that C95 and C105 can form both inter- and intrachain disulfide bonds (Fig. 5B). The capacity of C95 and presumably C105 to form interchain disulfide bonds may provide an explanation for why the C95S/C105S double mutant is more active than either C95S or C105S, i.e., in the absence of C105, C95 may form an inappropriate interchain disulfide bond that inhibits MD-2 function and vice versa.
Figure 5.
Diagram of proposed MD-2 structures. (A) Mutant 10 is observed as a dimer, indicating that C95 is able to form an interchain disulfide. (B) The formation of an intrachain disulfide bond between C95 and C105 explains why mutant 11 is mainly monomeric. This intrachain disulfide prevents the formation of the C95-C95 interchain disulfide bridge observed in mutant 10. However, some dimer is observed and can be explained by the formation of either two C95-C105 or C95-C95 and C105-C105 interchain disulfide bonds. (C) Possible structures for dimeric and tetrameric WT MD-2. MD-2 dimers contain mixtures of inter- and intrachain disulfide bonds. The intrachain bonds from two dimers can alternatively form interchain bridges leading to tetramer formation. A similar process could generate the distribution of polymers seen in the WT lanes of Figs. 3 and 4.
Recently Re and Strominger (21) showed that a monomeric fraction of WT MD-2 was active in add-back experiments. We observed that WT MD-2 samples from transfected HEK293T and HeLa cells rarely yielded monomeric MD-2 (WT Figs. 3 and 4, and Fig. 7A, which is published as supporting information on the PNAS web site). However, under serum-free conditions (used by Re and Strominger), both of these cell types do secrete a minimal amount of monomeric MD-2. In add-back experiments, no difference in LPS responsiveness was observed, regardless of whether monomeric MD-2 was present (Fig. 7). Thus, although monomeric MD-2 may be active, it is not required for LPS responsiveness and is therefore not likely to be physiologically relevant.
MD-2 exhibits several unusual structural features. Secreted proteins are typically stabilized by intrachain disulfide bonds and when these are disrupted by mutation or chemical manipulation, quality control mechanisms prevent the proteins from leaving the ER (22, 23). Any defective molecules that are secreted into the medium are rapidly degraded and/or precipitate. By contrast, all of our MD-2 mutants, including mutant 9 that lacks Cys residues altogether, are secreted as mature, soluble, and stable proteins, in quantities similar to WT MD-2. Thus, MD-2 appears to adopt a stable, but inactive, tertiary structure in the absence of disulfide bonds. In addition, most secreted proteins that contain intermolecular disulfide bridges are homogenous in size and are cross linked by distinct Cys residues. MD-2, on the other hand, consists of a heterogeneous collection of oligomers where no single Cys residue is necessary for oligomer formation, and where individual Cys residues can pair with multiple partners. Electrophoretic analyses of the mutants created in this study suggest that several Cys residues can form intermolecular disulfide bridges, indicating that they lie on the MD-2 surface. These include C37, C51, C95, C133, and C148, and it is possible that all are on the surface. An interesting feature of the MD-2 structure is the capacity of C95 and C105, and probably C133 and C148 to form both intrachain and interchain disulfide bonds.
Taken together, our results are consistent with the model for MD-2 shown schematically in Fig. 5C. According to this model, the MD-2 polypeptide forms a stable core structure that does not require intrachain disulfide residues for stability. Most if not all of the seven Cys residues point to the exterior of the molecule where they can form either inter- or intrachain disulfide bonds leading to the formation of oligomers of varying size, each cross-linked by a heterogeneous collection of disulfide bridges. How this unusual protein interacts with TLR4 to form an active signaling complex remains to be determined.
Supplementary Material
Acknowledgments
We thank Dr. Seth Steinberg for aid with statistical analyses and Dr. Paul Roche for helpful suggestions during the preparation of the manuscript.
Abbreviations
- LPS
lipopolysaccharide
- TLR4
Toll-like receptor 4
- ER
endoplasmic reticulum
- HEK
human embryonic kidney
References
- 1.Akira S, Takeda K, Kaisho T. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 2.Imler J L, Hoffmann J A. Trends Cell Biol. 2001;11:304–311. doi: 10.1016/s0962-8924(01)02004-9. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 4.Dziarski R, Gupta D. J Endotoxin Res. 2000;6:401–405. doi: 10.1179/096805100101532243. [DOI] [PubMed] [Google Scholar]
- 5.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 6.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. J Immunol. 1999;162:3749–3752. [PubMed] [Google Scholar]
- 7.Shimazu R, Akashi S, Ogata H, Nagai Y, Fukudome K, Miyake K, Kimoto M. J Exp Med. 1999;189:1777–1782. doi: 10.1084/jem.189.11.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, Akira S, Kitamura T, Kosugi A, Kimoto M, Miyake K. Nat Immunol. 2002;3:667–672. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 9.Schromm A B, Lien E, Henneke P, Chow J C, Yoshimura A, Heine H, Latz E, Monks B G, Schwartz D A, Miyake K, et al. J Exp Med. 2001;194:79–88. doi: 10.1084/jem.194.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.da Silva Correia J, Soldau K, Christen U, Tobias P S, Ulevitch R J. J Biol Chem. 2001;276:21129–21135. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 11.Viriyakosol S, Tobias P S, Kitchens R L, Kirkland T N. J Biol Chem. 2001;276:38044–38051. doi: 10.1074/jbc.M105228200. [DOI] [PubMed] [Google Scholar]
- 12.Mancek M, Pristovsek P, Jerala R. Biochem Biophys Res Commun. 2002;292:880–885. doi: 10.1006/bbrc.2002.6748. [DOI] [PubMed] [Google Scholar]
- 13.Akashi S, Nagai Y, Ogata H, Oikawa M, Fukase K, Kusumoto S, Kawasaki K, Nishijima M, Hayashi S, Kimoto M, et al. Int Immunol. 2001;13:1595–1599. doi: 10.1093/intimm/13.12.1595. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki K, Akashi S, Shimazu R, Yoshida T, Miyake K, Nishijima M. J Endotoxin Res. 2001;7:232–236. [PubMed] [Google Scholar]
- 15.Muroi M, Ohnishi T, Tanamoto K. Infect Immun. 2002;70:3546–3550. doi: 10.1128/IAI.70.7.3546-3550.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenton M J, Golenbock D T. J Leukocyte Biol. 1998;64:25–32. doi: 10.1002/jlb.64.1.25. [DOI] [PubMed] [Google Scholar]
- 17.da Silva C J, Ulevitch R J. J Biol Chem. 2002;277:1845–1854. doi: 10.1074/jbc.M109910200. [DOI] [PubMed] [Google Scholar]
- 18.Ohnishi T, Muroi M, Tanamoto K. J Immunol. 2001;167:3354–3359. doi: 10.4049/jimmunol.167.6.3354. [DOI] [PubMed] [Google Scholar]
- 19.Visintin A, Mazzoni A, Spitzer J A, Segal D M. Proc Natl Acad Sci USA. 2001;98:12156–12161. doi: 10.1073/pnas.211445098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chow J C, Young D W, Golenbock D T, Christ W J, Gusovsky F. J Biol Chem. 1999;274:10689–10692. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 21.Re F, Strominger J L. J Biol Chem. 2002;277:23427–23432. doi: 10.1074/jbc.M202554200. [DOI] [PubMed] [Google Scholar]
- 22.Ellgaard L, Molinari M, Helenius A. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- 23.Wickner S, Maurizi M R, Gottesman S. Science. 1999;286:1888–1893. doi: 10.1126/science.286.5446.1888. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.