Abstract
Retinitis pigmentosa is a photoreceptor degenerative disease leading to blindness in adulthood. Leber congenital amaurosis (LCA) describes a more severe condition with visual deficit in early childhood. Defects in the retinitis pigmentosa GTPase regulator (RPGR) and an RPGR-interacting protein (RPGRIP) are known causes of retinitis pigmentosa and LCA, respectively. Both proteins localize in the photoreceptor connecting cilium (CC), a thin bridge linking the cell body and the light-sensing outer segment. We show that RPGR is absent in the CC of photoreceptors lacking RPGRIP, but not vice versa. Mice lacking RPGRIP elaborate grossly oversized outer segment disks resembling a cytochalasin D-induced defect and have a more severe disease than mice lacking RPGR. Mice lacking both proteins are phenotypically indistinguishable from mice lacking RPGRIP alone. In vitro, RPGRIP forms homodimer and elongated filaments via interactions involving its coiled-coil and C-terminal domains. We conclude that RPGRIP is a stable polymer in the CC where it tethers RPGR and that RPGR depends on RPGRIP for subcellular localization and normal function. Our data suggest that RPGRIP is also required for disk morphogenesis, putatively by regulating actin cytoskeleton dynamics. The latter hypothesis may be consistent with a distant homology between the C-terminal domain of RPGRIP and an actin-fragmin kinase, predicted by fold recognition algorithms. A defect in RPGRIP encompasses loss of both functions, hence the more severe clinical manifestation as LCA.
Genetic defects in the retinitis pigmentosa GTPase regulator (RPGR) and the RPGR-interacting protein (RPGRIP) are known causes of RP and Leber congenital amaurosis (LCA), respectively (1, 2). The function of RPGR is not fully understood. Its N-terminal half has sequence homology to RCC1, a guanine nucleotide exchange factor for Ran GTPase. RPGRIP was isolated as a protein that interacted with RPGR in yeast two-hybrid screens (3–5). RPGRIP binds to the RCC1 homology domain of RPGR via its C-terminal portion. Toward its N-terminal portion, there is a region of 330 residues predicted to form a coiled-coil structure. In contrast to RPGR, which is expressed widely, RPGRIP expression is confined to retinal photoreceptors.
Both proteins were found to localize in the connecting cilium (CC) of photoreceptors. On the basis that RPGRIP exhibited a stronger association with the ciliary axoneme and that the CC localization of RPGRIP remained unchanged in mice lacking RPGR, it was proposed that RPGRIP was likely the primary resident protein in the CC, whereas RPGR localized in this cellular compartment via binding to RPGRIP (5, 6). The CC localization of these two proteins provides clues about their in vivo functions. The photoreceptor CC is the only link between the cell body and the outer segment. It is structurally analogous to the transitional zone of motile cilia (7). The CC, together with the outer segment, can be considered a modified cilium in which the distal portion elaborates stacks of photosensitive disk membranes. The outer segment is renewed daily, a process in which new membranes are added at the base to form new disks and older ones are shed at the tip (8, 9). Proteins destined for the outer segments must pass through the CC against steep concentration gradients. Thus the CC must regulate active protein transport and restrict their redistribution. A second role of the CC relates to disk morphogenesis. Nascent disks are formed by evagination of the plasma membranes at the distal CC (10). This process depends on an F-actin network located at the distal end of the CC (11), which appears during photoreceptor maturation just before the discs form (12). This actin network is seemingly unique to photoreceptors, because it is absent from motile cilia or flagella, suggesting that photoreceptors have a unique mechanism of using F-actin in elaborating disk membranes (13). Indeed, interference with actin filament polymerization by cytochalasin D inhibits initiation of membrane evagination and new disk formation (14, 15). As resident proteins of the CC, RPGRIP and RPGR may therefore participate in aspects of protein trafficking through the CC and/or disk morphogenesis.
To investigate the in vivo function of RPGRIP and the in vivo relevance of the physical interaction between RPGRIP and RPGR, we analyzed mice carrying a targeted disruption in the RPGRIP gene. Our data show that RPGRIP is essential for RPGR function and separately is also required for normal disk morphogenesis.
Materials and Methods
Generation of RPGRIP-Targeted Mice.
Genomic fragments spanning exons 5–6 and exons 14–15 were amplified by PCR from 129/Sv mouse genomic DNA. These fragments, 1.6 and 4.5 kb in length, respectively, were cloned into the vector pGT-N29 flanking the neor gene to generate the targeting vector. The targeting vector was linearized and electroporated into J1 embryonic stem (ES) cells, and neomycin-resistant colonies were selected. Two ES clones were identified in which the targeting vector was inserted between exons 14 and 15 of the RPGRIP gene. Both targeted clones were microinjected into C57BL/6 blastocysts to generate chimeras, which were crossed with C57BL/6 mice, and two independent lines of mutant mice were derived. Subsequent analyses showed that RPGRIP expression was ablated in both lines of mutants, and that their early retinal phenotype was identical. Therefore, only one of the lines was expanded and used for detailed phenotype analyses. The genotype of mice was determined by PCR. PCR primers for the targeted allele were P1 (5′-CTGGAGCGGCTGAATCACCTC) and P2 (5′-GGTCTCAGAGATTTACCTACCGTCTC). PCR primers for the WT allele were P1 and P3 (5′-GAGATCTGTGTGCCCCTGCCTC). Mice lacking both RPGRIP and RPGR (6) were generated by crossing them for two generations to obtain doubly homozygous mutants.
Antibodies, Immunoblotting, Immunofluorescence, and Retinal Phenotype Analyses.
A His-tagged fusion protein encompassing residues 2–222 of mouse RPGRIP was produced in Escherichia coli and used to immunize a rabbit. A polyclonal RPGRIP antibody targeting the C terminus of RPGRIP was described previously (6). The RPGR antibody (RPGR-S1) targets residues 494–563 of mouse RPGR (GenBank accession no. NP_035415), common to all known splice variants of RPGR. Mouse blue and green cone opsin antibodies were raised in chicken against the peptide sequences CRKPMADESDVSGSQKT and FGKKVDDSSELSSTSKT, respectively. The monoclonal anti-Rhodopsin antibody rho 1D4 and the chicken anti-RP1 antibody were described (16, 17). Immunoblotting and immunofluorescence staining were performed as described (5). Retinal phenotypes were examined by histology and electroretinogram (ERG), performed as described (18).
Yeast Two-Hybrid Assays.
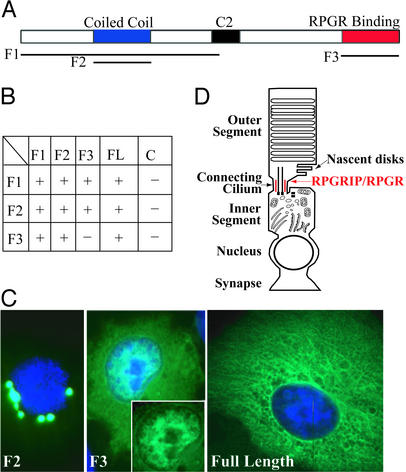
Yeast two-hybrid screening was performed by using the GAL4 system 3 (CLONTECH) as described (5). Four baits were constructed. F1 consisted of residues 1–820 of RPGRIP. F2, predicted to form a coiled-coil structure, consisted of residues 214–550. F3 spanned residues 1002–1345, which included the RPGR-binding region. The full-length RPGRIP was also constructed into a bait plasmid (FL).
Transient Expression in COS-7 Cells.
COS-7 cells were maintained in DMEM supplemented with 10% FBS at 37°C in 5% CO2. Transfection was carried out by using the Geneshuttle 40 reagent (Quantum, Durham, NC) according to the manufacturer's instructions. RPGRIP fragments matching the F1, F2, and F3 baits and the full-length RPGRIP sequences were inserted into the pEGFP-C2 vector (CLONTECH) to generate the expression constructs. After transient transfection, recombinant proteins were visualized with the aid of the enhanced GFP tag.
Bioinformatics.
Amino acid sequence of the C-terminal domain of RPGRIP was submitted to the ffas (19) fold prediction server and compared with a database of sequence profiles representing proteins with known 3D structures. The best-matching family was that of actin-fragmin kinase (AFK) from slime mold Physarum polycephalum (20). The statistical significance of the prediction (e value, 0.48 in our internal tests of the ffas server) indicates an ≈95% fold assignment accuracy. The plausibility of the alignment was further corroborated by the agreement in the predicted secondary structure of RPGRIP as well as the reasonable quality of the 3D model (data not shown).
Results
Generation of RPGRIP Mutant Mice.
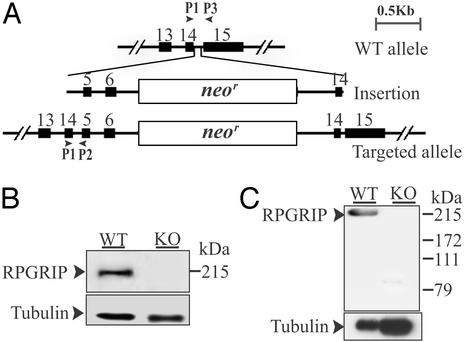
The murine RPGRIP gene was disrupted by gene targeting (Fig. 1A). From among 800 drug-resistant clones screened, two targeted clones were identified. Closer examination found that in these clones the 3′ arms underwent homologous recombination, whereas the 5′ arm was inserted (Fig. 1A). Because this RPGRIP allele was disrupted by a large insert containing three duplicated exons and the neor gene, it was deemed most likely to be an insertionally inactivated allele. Both embryonic stem clones were injected into C57BL/6 blastocysts, and both were transmitted through the germ lines. Mice homozygous for the targeted allele were produced at the expected Mendelian ratio from heterozygous crosses. RT-PCR and sequencing analyses of the homozygous mutant retinas found a premature stop in the mRNA by using the primer pair P1 and P3 (Fig. 1A). Ablation of RPGRIP was also demonstrated by immunoblotting (Fig. 1 B and C) and by immunofluorescence (refer to Fig. 2C) analyses. It was possible that an N-terminal truncated RPGRIP polypeptide could be synthesized (≈500 residues out of the 1,331-residue full length protein); immunoblotting analysis by using the N-terminal antibody (Fig. 1C) suggests that a truncated RPGRIP polypeptide does not substantially accumulate in photoreceptors. Given that mice heterozygous for the mutant allele do not exhibit a retinal phenotype (see below), this allele is likely to lead to a loss of function. It is noted that human RPGIRP mutant alleles in patients with recessive LCA (2) have nonsense mutations located further downstream from the site of disruption in the mouse mutant allele described here.
Figure 1.
Targeted disruption of the RPGRIP gene. (A) The WT RPGRIP allele was disrupted by the insertion of the targeting construct. PCR primers P1, P2, and P3 are indicated by arrowheads. (B) Ablation of RPGRIP was confirmed by probing WT and mutant (KO) retinal homogenates with the C-terminal (B) and the N-terminal (C) RPGRIP antibodies on immunoblots. The blots were re-probed with anti-acetylated α tubulin as a loading control.
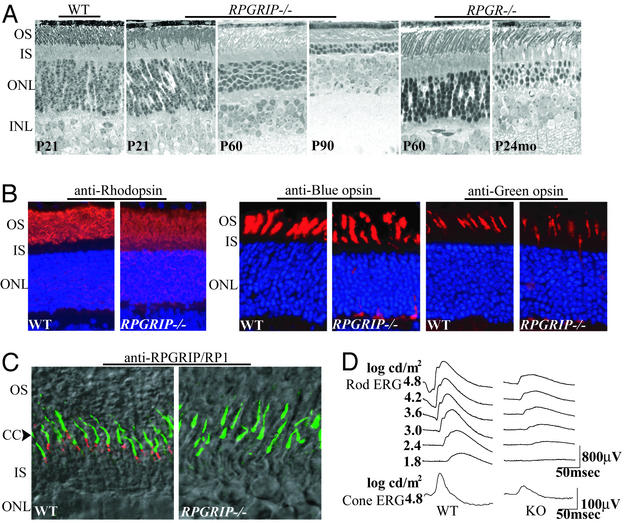
Figure 2.
Retinal phenotype of the RPGRIP−/− mice. (A) Photomicrographs of retinal sections from WT, RPGRIP−/−, and RPGR−/− mice. (B) Partial mislocalization of rhodopsin, blue and green cone opsins in the RPGRIP−/− photoreceptors. Staining for the opsins (red) is normally found in the outer segments but appears in other layers of the retinas as well in the mutant photoreceptors. Cell nuclei were stained blue with the nuclear dye Hoechst 33342. (C) Normal localization of the RP1 protein (green) in the RPGRIP−/− photoreceptors. Interestingly, the bulk of RP1 signal (green) is seen just distal to RPGRIP (red), suggesting a distribution along microtubules in the proximal outer segments. Overlain confocal immunofluorescence and Nomarski images are shown. (D) Rod and cone ERGs from WT and RPGRIP−/− mice at P22. OS, outer segment; IS, inner segment; ONL, outer (photoreceptor) nuclear layer; INL, inner nuclear layer.
Retinal Phenotype of RPGRIP−/− Mice.
The RPGRIP−/− mice appeared healthy, were fertile, and were similar in general growth characteristics to their WT and heterozygous littermates. Mice heterozygous for the targeted allele, followed up to 6 mo of age both by ERG and by histology (data not shown), did not show any photoreceptor abnormality. The RPGRIP−/− mice initially developed a full complement of photoreceptors, as judged by the thickness of the photoreceptor nuclear layer (Fig. 2A). The photoreceptors, however, were never fully normal even at the earliest age tested [postnatal day (P)15]. The outer segments appeared disorganized, and there were pyknotic nuclei, indicative of ongoing cell death. Photoreceptor cell loss was near completion by 3 mo of age (Fig. 2A). This course of disease was more severe than that seen in mice lacking RPGR, which lost nearly all of their cones (not shown) and two-thirds of their rods by 2 yr of age (Fig. 2A). The disease courses of the mouse models are consistent with the differential disease severity in human patients carrying RPGRIP or RPGR mutations (2, 21), indicating conserved physiological functions and pathogenic mechanisms between the murine and human genes.
Being a highly polarized neuron, the photoreceptor outer segment and the cell body have vastly different protein distributions. To determine whether protein localization was affected in photoreceptors lacking RPGRIP, we carried out immunofluorescence staining for a battery of photoreceptor-specific proteins in young (≈P20) RPGRIP−/− mouse retinas. These included rod and cone opsins, arrestin, transducin, cGMP phosphodiesterase, cGMP-gated cationic channel, peripherin/RDS, and Rom-1. With the exception of rod and cone opsins, these proteins were found correctly localized. Rod and cone opsins, normally segregated to the outer segments, were partially mislocalized in the cell bodies (Fig. 2B). The degree of cone opsin mislocalization was similar to that of RPGR−/− mice at the same age (6) and therefore might be explained by the loss of RPGR function (see below). Given the grossly abnormal disks at an even younger age (P15; see Fig. 3) and the propensity of rhodopsin to lose its polarized distribution in degenerating photoreceptors regardless of the primary defect (22), the rhodopsin mislocalization in the RPGRIP−/− photoreceptors could be attributed at least in part to the disruption of the outer segment disks.
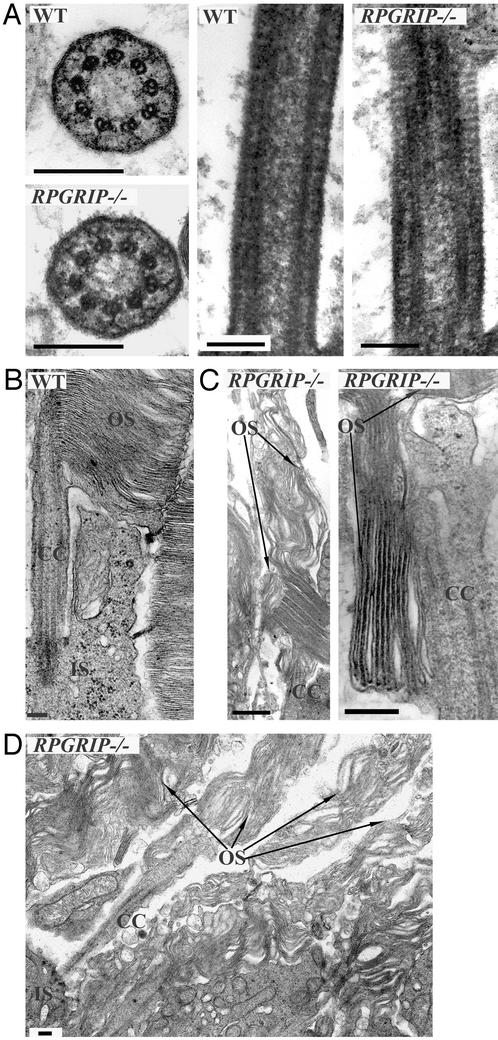
Figure 3.
Ultrastructural examination of RPGRIP−/− photoreceptors. (A) Transverse and longitudinal profiles of the connecting cilia. Mutant and the WT CC appear similar in the microtubule arrays, transmembrane assemblages, and ciliary necklaces. (B) Longitudinal section through a WT photoreceptor, illustrating the normal morphology of the apical inner segment, CC, and outer segment. (C) In the mutant, disk diameters are greatly expanded and orientation is altered. Enlarged disks appear bending upward (Left) or downward (Right). (D) A lower-magnification image of the mutant retina showing several photoreceptors. OS, outer segment; IS, inner segment. (Bars = 0.2 μm.)
The RP1 protein, required for photoreceptor viability, was reported to localize in the CC (17). We therefore examined whether loss of RPGRIP disrupted the subcellular localization of RP1. We found that the staining pattern of RP1 was comparable in WT and the RPGRIP−/− retinas (Fig. 2C; green), thus excluding RP1 mislocalization as part of the pathogenic process. Interestingly, our finding also suggests that RP1 is primarily localized distal to the CC and most likely along microtubules in the proximal outer segments.
A number of photoreceptor proteins also exhibit light-dependent movement between the inner and outer segments (23, 24). We asked whether this process might be impaired due to loss of RPGRIP. By immunofluorescence, we found that in RPGRIP−/− mice, both arrestin and α-transducin moved light-dependently with a comparable kinetics as in WT mice (data not shown), indicating that RPGRIP does not participate in this process. Together our data suggest that RPGRIP is not essential for transporting proteins across the CC, whether constitutive or light-triggered, nor is it required for restricting protein redistribution across the CC.
Although morphologically abnormal, both rod and cone photoreceptors were able to respond to light as assayed by ERG. At 20–30 days of age, dark-adapted (rod) ERG analysis showed both a greatly reduced maximum amplitude and a reduced sensitivity in the RPGRIP−/− mice (Fig. 2D). Cone ERGs also showed reduced amplitude. Because RPGRIP does not reside in the outer segments, where phototransduction takes place, it is unlikely that RPGRIP directly participates in this process. Disruption of the outer segment structure and loss of photoreceptors, on the other hand, could account for the impaired ERG responses.
Grossly Expanded Disks in RPGRIP−/− Mice.
Because RPGRIP was a resident protein in the CC, we examined by electron microscopy whether loss of RPGRIP led to a defective CC structure (Fig. 3A). The known ultrastructural details (7), such as the array of the 9+0 microtubule doublets, transmembrane assemblages, and ciliary necklaces, were all found to be comparable in the mutant and WT photoreceptors. These data suggest that RPGRIP is not essential for the development or maintenance of the core CC structure.
Ultrastructural examination of the outer segment disks proved informative (Fig. 3 B–D). In the RPGRIP mutant photoreceptors at P15, disk diameters were greatly expanded to several-fold the normal size. Stacks of disk membranes were frequently arranged parallel to the long axis of the outer segments instead of the normal perpendicular orientation. The altered disk orientation presumably resulted from the inability of the interphotoreceptor space to accommodate the oversized disks. Such disk defect was not seen in the RPGR mutant. The disk morphology of the RPGRIP mutant is reminiscent of the abnormal disk morphogenesis induced by cytochalasin D (14), which inhibits actin filament polymerization. There was no accumulation of extracellular vesicles in the RPGRIP−/− retinas such as those reported in several retinal disease models (25–27).
Dependence of RPGR on RPGRIP for Localization in the CC.
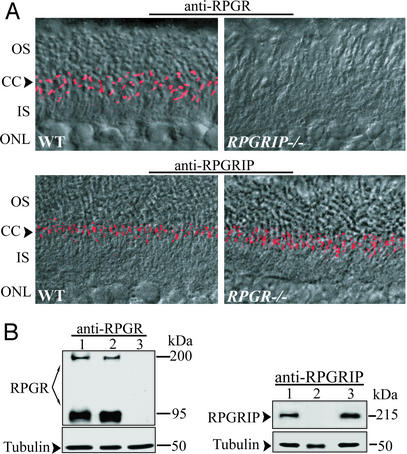
Availability of the RPGRIP−/− and RPGR−/− mutant models provided an excellent opportunity to test their functional relationship in vivo. To determine which protein was the primary CC resident, or whether these proteins localized here independently, we examined the subcellular localization of RPGRIP and RPGR in mice lacking either protein (Fig. 4A). We found that the CC localization of RPGR was abolished in mice lacking RPGRIP but not vice versa. These data demonstrate that RPGRIP binds to the core structure of the CC, whereas RPGR localizes to the CC through binding to RPGRIP.
Figure 4.
Dependence of RPGR on RPGRIP for localization in the CC. (A) Retina sections probed with RPGR (Upper) or RPGRIP (Lower) antibodies. Overlain immunofluorescence and Nomarski confocal images are shown. In the WT retina (Left), both RPGR and RPGRIP are localized in the CC. In the RPGRIP−/− retina, the CC localization of RPGR is abolished (Upper Right). RPGRIP localization is unaffected by loss of RPGR (Lower Right). OS, outer segment; IS, inner segment; ONL, outer (photoreceptor) nuclear layer. (B) Immunoblotting of retinal homogenates for RPGR and RPGRIP indicates that neither protein is reduced in abundance on loss of its interacting partner. Lane 1, WT; lane 2, RPGRIP−/−; lane 3, RPGR−/−. Blots were reprobed with antiacetylated α-tubulin antibody as loading controls.
Disruption of stable protein interactions frequently leaves components of the complex destabilized. Because the CC is a minute structure relative to the entire cell, the released RPGR was not expected to be of sufficient abundance to produce ectopic staining, nor did we see ectopic RPGR in the RPGRIP−/− photoreceptors. This, however, left open the possibility that absence of RPGR in the CC might be due to reduced RPGR synthesis or stability. To rule out this scenario, mutant and WT retinal homogenates were analyzed by immunoblotting (Fig. 4B). RPGR from WT and RPGRIP−/− retinas was found to be at similar abundance. Therefore, RPGR was present but mislocalized in RPGRIP−/− photoreceptors. Similarly, RPGRIP was also found in comparable amounts in WT and RPGR−/− retinas. Thus in the case of RPGR-RPGRIP interaction, loss of either component has little effect on the accumulation of the other. This could be interpreted as indicating that these two proteins do not cofold or exist in a permanent complex. Instead, the RPGRIP–RPGR interaction may be transient or dynamically regulated.
Loss of RPGRIP Is Inclusive of an RPGR Defect.
Alternative mechanisms could be envisioned to explain the greater disease severity associated with loss of RPGRIP. First, RPGRIP may be required both for RPGR function and for a second cellular process. Second, RPGRIP may be required only for RPGR function, but the free RPGR migrates to an ectopic location (e.g., the outer segment), where it disrupts disk formation. Third, RPGRIP and RPGR may perform fully independent functions despite their physical proximity. As an initial test of these hypotheses, we generated mice doubly homozygous for the RPGRIP and RPGR mutations and examined them by histology and ERG at ≈20 days of age and by histology at 45 days of age (data not shown). The double mutant had a disease phenotype indistinguishable from the RPGRIP single mutant. This outcome is consistent with the first hypothesis. The second hypothesis predicted a less severe disease because a deleterious effect of free RPGR would have been removed. Had the third hypothesis been correct, the disease might have been more severe, because two independent insults are likely to produce an additive effect.
Formation of Stable RPGRIP Polymer.
To explore how RPGRIP itself was targeted to the core CC structure, we carried out yeast two-hybrid screens by using several regions of RPGRIP as baits (Fig. 5A). These screens did not identify known constituents of the CC or centrosome. However, RPGRIP itself was identified by all three baits as a binding partner. On further analysis by cotransformation in yeast, the coiled-coil region of RPGRIP exhibited homotypic binding, whereas the C-terminal tail (RPGR-binding domain) interacted with the coiled-coil region (Fig. 5B). These observations were followed up by transient expression in COS cells. The coiled-coil region produced large dots (Fig. 5C Left), indicating formation of homopolymer but no elongated fibers. The tail domain contained a tripartite nuclear localization signal, which may explain its partial nuclear localization (Fig. 5C Center). Only the full-length RPGRIP formed detergent-insoluble fibers (Fig. 5C Right). These data suggest that the coiled-coil region of RPGRIP mediates homodimer formation, whereas interaction between the coiled-coil and the tail domains may account for the formation of elongated higher-order polymers. By immunoelectron microscopy, RPGRIP was seen external to the microtubule arrays in the CC (5). We therefore propose that RPGRIP exists as elongated homopolymers situated between the axonemal microtubules and the plasma membrane. How RPGRIP is tethered to the core CC structure remains unresolved.
Figure 5.
Formation of RPGRIP polymers. (A) Domain diagram of RPGRIP. Lines underneath denote fragments that were used as baits in yeast two-hybrid screens and in transient expression experiments. (B) Summary of yeast two-hybrid assays. “+” indicates growth of cotransformants in selection media and protein binding. (C) Transient expression of RPGRIP or its fragments in COS cells. Green, recombinant proteins; blue, nuclear dye. Both F1 (not shown) and F2 (Left) fragments appeared as dots surrounding the nuclei. (Center) F3 was enriched in the nucleus. (Inset) The same nucleus without the blue channel to illustrate nuclear localization of the recombinant protein. Full-length RPGRIP (Right) appeared as insoluble fibers. (D) Schematic diagram of a photoreceptor. Nascent disks are added by evagination of the plasma membrane directly above the CC, where RGPRIP and RPGR are housed. The RPGRIP polymer (red) is situated between the axonemal cytoskeleton and plasma membrane based on observations by immunoelectron microscopy (5).
RPGRIP May Be Distantly Related to an AFK.
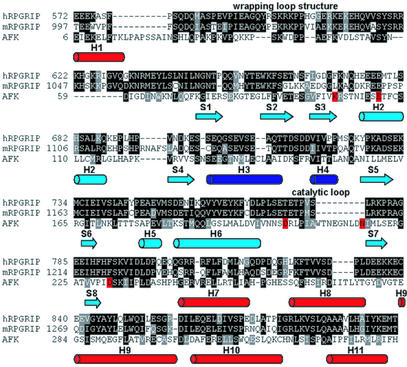
Using recently updated fold recognition algorithm ffas (19), we identified a possible distant homology between the C-terminal domain of RPGRIP and an AFK from the slime mold P. polycephalum. A homology alignment between the C-terminal domain of RPGRIP and AFK is shown in Fig. 6. The strongest conservation patterns were found in regions typical for AFK only but not for kinases in general. Even in the catalytic region (blue), shared by a broader group of kinases, the strongest homology resides in the additional α-helical region present only in AFK. Interestingly other regions typical for AFK are conserved as well, such as the exterior helices (red) and the wrapping loop structure situated on the other side of the structure away from the catalytic region.
Figure 6.
A distant homology between RPGRIP and an AFK. Homology alignment between the C-terminal domain of RPGRIP (human and mouse) and AFK is shown. The 3D structure of AFK was described previously (20). The strongest conservation patterns are found in regions typical for AFK only (red). Even in the catalytic region (light blue), shared by a broader group of kinases, the strongest homology resides in the additional alpha helical region present only in AFK (dark blue). Interestingly, other regions typical for AFK are conserved as well, such as the exterior helices (red) and the wrapping loop structure situated on the other side of the structure from the catalytic region. Cylinders represent α-helices, whereas arrows represent β-strands. GenBank accession nos.: human RPGRIP, 9652036; mouse RPGRIP, 11496167; AFK, 5542182. The human sequence is not full-length at the N terminus, hence the discrepancy in residue numbers between the human and mouse RPGRIPs in the alignment.
Discussion
This study demonstrates two in vivo roles of RPGRIP. First, RPGRIP retains RPGR in the CC. In the absence of RPGRIP, RPGR is diffusely mislocalized in photoreceptors and is unlikely to execute its normal function. Thus the consequence of a defective RPGRIP includes the loss of RPGR function as well. This interpretation is consistent with the RPGRIP/RPGR double homozygotes being phenotypically indistinguishable from the RPGRIP single mutant.
Second, loss of RPGRIP leads to a profound defect in disk formation, indicating a role for RPGRIP in disk morphogenesis. Disk morphogenesis can be disrupted by different mechanisms. Several structural proteins are required that include peripherin/RDS, ROM-1, and rhodopsin, the latter because of its sheer abundance. Inactivation or reduced dosage involving these genes leads to characteristic defects or a complete lack of disk formation (28–32). A disruption in protein trafficking can also lead to an insufficient amount of these proteins reaching the outer segments and thus to abnormal disk formation. Often, when protein trafficking is the primary target of a gene mutation in rodent models, accumulation of rhodopsin-bearing vesicles is seen transiently at the early stage of disease (25–27, 33). This has been suggested to result from accumulation of rhodopsin at the inner-segment plasma membranes and from aberrant budding. Accumulation of rhodopsin-bearing vesicles is not seen in the RPGRIP−/− retinas. In contrast, the cytochalasin D-induced defect (13–15), characterized by cessation of new disk evagination but continued addition of membranes to existing disks, reflects perturbation of actin cytoskeleton dynamics. In vivo, these mechanistic differences can be difficult to distinguish on the basis of phenotype analyses, in part due to the secondary effects of degeneration. In many instances, overlapping mechanisms may be at work. Despite these potential pitfalls in inferring disease mechanism from photoreceptor phenotypes, it is worth noting that RPGRIP−/− photoreceptors exhibit profoundly distorted disks with comparatively modest protein mislocalization, some of which could be attributed to a loss of RPGR function alone. The morphological defect in the RPGRIP−/− photoreceptors is most consistent with new disks being initiated at a reduced rate so that fewer but larger disks are formed. Although somewhat speculative, these observations implicate the cellular machinery at the CC controlling cytoskeleton dynamics as being affected by the loss of RPGRIP.
The above interpretation is supported by additional evidence suggesting a link between RPGRIP and the actin cytoskeletons, i.e., a distant homology between the C-terminal domain of RPGRIP and AFK (Fig. 6). Actin–fragmin complex is the in vivo substrate for AFK, and phosphorylation by AFK controls the F-actin capping activity of the complex. Fragmin is a member of the gelsolin family and has been implicated in cellular processes requiring rapid actin cytoskeleton reorganization. A gelsolin-like protein (the mammalian homolog of Drosophila flightless I) is highly enriched in the CC of mammalian photoreceptors (34), which in complex with actin could serve as a substrate for an AFK-like kinase activity. We propose that RPGRIP may be part of the multiprotein complex that regulates actin cytoskeleton reorganization during new disk formation. Molecular identification of this complex will await future studies.
Acknowledgments
We thank Drs. V. Arshavsky (Harvard Medical School), R. Molday (University of British Columbia, Vancouver), and E. Pierce (University of Pennsylvania, Philadelphia) for antibodies. This work was supported by National Institutes of Health Grants EY10309, EY10581 (to T.L.), and GM60049 (to M.G. and A.G.), and by the Foundation Fighting Blindness.
Abbreviations
- RPGR
retinitis pigmentosa GTPase regulator
- RPGRIP
RPGR-interacting protein
- LCA
Leber congenital amaurosis
- CC
connecting cilium
- ERG
electroretinogram
- Pn
postnatal day n
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Meindl A, Dry K, Herrmann K, Manson F, Ciccodicola A, Edgar A, Carvalho M R, Achatz H, Hellebrand H, Lennon A, et al. Nat Genet. 1996;13:35–42. doi: 10.1038/ng0596-35. [DOI] [PubMed] [Google Scholar]
- 2.Dryja T P, Adams S M, Grimsby J L, McGee T L, Hong D H, Li T, Andreasson S, Berson E L. Am J Hum Genet. 2001;68:1295–1298. doi: 10.1086/320113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boylan J P, Wright A F. Hum Mol Genet. 2000;9:2085–2093. doi: 10.1093/hmg/9.14.2085. [DOI] [PubMed] [Google Scholar]
- 4.Roepman R, Bernoud-Hubac N, Schick D E, Maugeri A, Berger W, Ropers H-H, Cremers F P M, Ferreira P A. Hum Mol Genet. 2000;9:2095–2105. doi: 10.1093/hmg/9.14.2095. [DOI] [PubMed] [Google Scholar]
- 5.Hong D H, Yue G, Adamian M, Li T. J Biol Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 6.Hong D H, Pawlyk B S, Shang J, Sandberg M A, Berson E L, Li T. Proc Natl Acad Sci USA. 2000;97:3649–3654. doi: 10.1073/pnas.060037497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Besharse J C, Horst C J. In: Ciliary and Flagellar Membranes. Bloodgood R A, editor. New York: Plenum; 1990. pp. 389–417. [Google Scholar]
- 8.Young R W. J Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young R W, Bok D. J Cell Biol. 1969;42:392–403. doi: 10.1083/jcb.42.2.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg R H, Fisher S K, Anderson D H. J Comp Neurol. 1980;190:501–508. doi: 10.1002/cne.901900307. [DOI] [PubMed] [Google Scholar]
- 11.Chaitin M H, Schneider B G, Hall M O, Papermaster D S. J Cell Biol. 1984;99:239–247. doi: 10.1083/jcb.99.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaitin M H, Carlsen R B, Samara G J. Exp Eye Res. 1988;47:437–446. doi: 10.1016/0014-4835(88)90054-1. [DOI] [PubMed] [Google Scholar]
- 13.Hale I L, Fisher S K, Matsumoto B. J Comp Neurol. 1996;376:128–142. doi: 10.1002/(SICI)1096-9861(19961202)376:1<128::AID-CNE8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 14.Williams D S, Linberg K A, Vaughan D K, Fariss R N, Fisher S K. J Comp Neurol. 1988;272:161–176. doi: 10.1002/cne.902720202. [DOI] [PubMed] [Google Scholar]
- 15.Vaughan D K, Fisher S K. Invest Ophthalmol Visual Sci. 1989;30:339–342. [PubMed] [Google Scholar]
- 16.Molday R. In: Progress in Retinal Research. Osborne N, Chader G, editors. Vol. 8. New York: Pergamon; 1988. pp. 174–209. [Google Scholar]
- 17.Liu Q, Zhou J, Daiger S P, Farber D B, Heckenlively J R, Smith J E, Sullivan L S, Zuo J, Milam A H, Pierce E A. Invest Ophthalmol Visual Sci. 2002;43:22–32. [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Sandberg M A, Pawlyk B S, Rosner B, Hayes K C, Dryja T P, Berson E L. Proc Natl Acad Sci USA. 1998;95:11933–11938. doi: 10.1073/pnas.95.20.11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaroszewski L, Rychlewski L, Godzik A. Protein Sci. 2000;9:1487–1496. doi: 10.1110/ps.9.8.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinbacher S, Hof P, Eichinger L, Schleicher M, Gettemans J, Vandekerckhove J, Huber R, Benz J. EMBO J. 1999;18:2923–2929. doi: 10.1093/emboj/18.11.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharon D, Bruns G A, McGee T L, Sandberg M A, Berson E L, Dryja T P. Invest Ophthalmol Visual Sci. 2000;41:2712–2721. [PubMed] [Google Scholar]
- 22.Nir I, Papermaster D. Prog Clin Biol Res. 1989;314:251–264. [PubMed] [Google Scholar]
- 23.Whelan J P, McGinnis J F. J Neurosci Res. 1988;20:263–270. doi: 10.1002/jnr.490200216. [DOI] [PubMed] [Google Scholar]
- 24.Sokolov M, Lyubarsky A L, Strissel K J, Savchenko A B, Govardovskii V I, Pugh E N, Jr, Arshavsky V Y. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- 25.Hagstrom S A, Adamian M, Scimeca M, Pawlyk B S, Yue G, Li T. Invest Ophthalmol Visual Sci. 2001;42:1955–1962. [PubMed] [Google Scholar]
- 26.Marszalek J R, Liu X, Roberts E A, Chui D, Marth J D, Williams D S, Goldstein L S. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 27.Pazour G J, Baker S A, Deane J A, Cole D G, Dickert B L, Rosenbaum J L, Witman G B, Besharse J C. J Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Travis G H, Sutcliffe J G, Bok D. Neuron. 1991;6:61–70. doi: 10.1016/0896-6273(91)90122-g. [DOI] [PubMed] [Google Scholar]
- 29.Molday R, Hicks D, Molday L. Invest Ophthalmol Visual Sci. 1987;28:50–60. [PubMed] [Google Scholar]
- 30.Hawkins R K, Jansen H G, Sanyal S. Exp Eye Res. 1985;41:701–720. doi: 10.1016/0014-4835(85)90179-4. [DOI] [PubMed] [Google Scholar]
- 31.Clarke G, Goldberg A F, Vidgen D, Collins L, Ploder L, Schwarz L, Molday L L, Rossant J, Szel A, Molday R S, et al. Nat Genet. 2000;25:67–73. doi: 10.1038/75621. [DOI] [PubMed] [Google Scholar]
- 32.Lem J, Krasnoperova N V, Calvert P D, Kosaras B, Cameron D A, Nicolo M, Makino C L, Sidman R L. Proc Natl Acad Sci USA. 1999;96:736–741. doi: 10.1073/pnas.96.2.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Snyder W K, Olsson J E, Dryja T P. Proc Natl Acad Sci USA. 1996;93:14176–14181. doi: 10.1073/pnas.93.24.14176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitt A, Wolfrum U. Exp Eye Res. 2001;73:837–849. doi: 10.1006/exer.2001.1086. [DOI] [PubMed] [Google Scholar]