Abstract
The tight junction (TJ) strand is a linear proteinaceous polymer spanning plasma membranes, and each TJ strand associates laterally with another TJ strand in the apposing membranes of adjacent cells to form “paired” TJ strands. Claudins have been identified as the major constituents of TJ strands, and when exogenously expressed in L fibroblasts, they polymerize into paired strands, which are morphologically similar to paired TJ strands in epithelia. Here, we show that a fusion protein of GFP with claudin-1 can also form similar paired strands in L fibroblasts, allowing us to directly observe individual paired claudin strands in live cells in real time. These paired strands showed more dynamic behavior than expected; they were occasionally broken and annealed, and dynamically associated with each other in both an end-to-side and side-to-side manner. Through this behavior of individual paired claudin strands, the network of strands was reorganized dynamically. Furthermore, fluorescence recovery after photobleaching analyses revealed that claudin molecules were not mobile within paired strands. Although these observations are not necessarily representative of TJ strands per se in epithelial cells, they provide important information on the structural and kinetic properties of TJ strands in situ with significant implications for barrier function of TJs.
The tight junction (TJ) is one mode of cell-to-cell adhesion in epithelial and endothelial cells. TJs seal the cells to create a primary barrier to the diffusion of solutes across the cellular sheet, and also function as a boundary between the apical and basolateral membrane domains to produce their polarization (1–5). On ultrathin section electron microscopy, TJs appear as a series of discrete sites of apparent fusion, involving the outer leaflets of the plasma membranes of adjacent cells (6). On freeze-fracture electron microscopy, TJs appear as a set of continuous, anastomosing intramembranous particle strands (TJ strands; ref. 7). These observations led to our current understanding of the three-dimensional structure of TJs; each TJ strand associates laterally with another TJ strand in apposing membranes of adjacent cells to form “paired” TJ strands, where the intercellular space is completely obliterated (reviewed in ref. 5).
To date, three distinct types of integral membrane proteins have been shown to be localized at TJs; occludin (8, 9), junctional adhesion molecule (10), and claudins (11). Occludin, an ≈65-kDa integral membrane protein with four transmembrane domains, was identified as the first component of TJ strands. However, several studies including gene knockout analyses revealed that TJ strands can be formed without occludin (12–15). JAM with a single transmembrane domain was recently shown to associate laterally with TJ strands, but not to constitute the strands per se (16). In contrast, claudin is now believed to be a major constituent of TJ strands (reviewed in refs. 5 and 17). Claudins with molecular masses of ≈23 kDa comprise a multigene family consisting of more than 20 members (11, 17–21). Claudins also bear four transmembrane domains, but do not show any sequence similarity to occludin.
Interestingly, when each claudin species was overexpressed in mouse L fibroblasts lacking endogenous claudins, exogenously expressed claudin molecules were polymerized within the plasma membrane to form “paired” strands at cell–cell contact regions (22). These paired claudin strands were morphologically indistinguishable from the in situ paired TJ strands at least at the electron microscopic level, although it remained elusive how claudin molecules are arranged within individual strands. In general, information on the dynamic behavior of proteinaceous polymers within plasma membranes as well as the molecular mechanism behind their lateral aggregation, i.e., polymerization, is still fragmentary. In this study, we successfully visualized individual paired claudin strands in live L cells using GFP technology, and pursued their dynamic behavior within plasma membranes. We believe that the results of this study provide important information on the physicochemical nature of proteinaceous polymers within plasma membranes, including TJ strands.
Materials and Methods
Antibodies and Cells.
Mouse anti-GFP mAb and mouse anti-FLAG mAb were purchased from Eastman Kodak and Chemicon, respectively. Mouse anti-ZO-1 mAb was raised and characterized previously (23). L transfectants expressing mouse claudin-1 (C1L), GFP-claudin-1 (C1GL) or FLAG-tagged claudin-1 (C1FL) were established previously (11). Briefly, claudin-1 was tagged with GFP or FLAG-peptide at its COOH terminus. The expression vectors for GFP-tagged and FLAG-tagged claudin-1 were previously reported as pBCL-1G and pCCL-1F, respectively (11). L transfectants were cultured in DMEM supplemented with 10% FCS.
Immunofluorescence Microscopy and Immuno-Replica Electron Microscopy.
Cells (1–2 × 105 cells per cm2) were cultured on coverslips for 24–48 h, washed with PBS, and fixed with 1% paraformaldehyde in PBS for 15 min. After washing with PBS, cells were permeabilized with 0.2% Triton X-100 in PBS for 15 min, and processed for immunofluorescence microscopy as described (11). Cy3-conjugated donkey anti-mouse IgG antibody (Jackson ImmunoResearch) was used as a secondary antibody.
For immuno-replica electron microscopy, C1GL cells were fixed with 1% paraformaldehyde in 0.1 M phosphate buffer (pH 7.3) for 5 min at room temperature, washed three times in 0.1 M phosphate buffer, immersed in 30% glycerol in 0.1 M phosphate buffer for 3 h, and then frozen in liquid nitrogen. Frozen samples were processed for immuno-replica electron microscopy as described (24). Goat anti-mouse IgG coupled with 10-nm gold (Amersham Pharmacia Biotech) was used as a secondary antibody.
Fluorescence Microscopy and Image Analysis.
To observe live cells, cells were cultured at low density (1 × 105 cells per cm2) on glass-bottomed dishes with No.1S cover glasses (Matsunami, Osaka) for 24–48 h. Images of cells were collected at 37°C with a DeltaVision optical sectioning microscope (Version 2.50; Applied Precision, Issaquah, WA) equipped with an Olympus IX70 (PlanApo ×100/1.40 N.A. oil immersion objective) through a cooled charge-coupled device camera (Series300 CH350, Photometrics, Tucson, AZ) with appropriate binning of pixels, exposure time, and time intervals. Fluorescence signals were visualized using a quad beamsplitter (Chroma Technology, Brattleboro, VT), a 490/20-nm excitation filter and a 528/38-nm emission filter (Chroma Technology). For time-lapse images, to reduce out-of-focus signals, two-dimensional deconvolution was calculated for all images at each time point. Time-lapse images of C1GL cells collected every 5 s for 10–95 frames were accumulated. All images, including movies, presented in this study were taken as single optical sections without any reconstruction along the z axis.
Fluorescence recovery after photobleaching analysis was performed with a Zeiss LSM510 confocal laser-scanning microscope, equipped with Zeiss Axioplan2 (Plan Apochromat ×100/1.40 N.A. differential interference contrast oil immersion objective) using a BP 505-50 nm filter. An argon laser with a maximum output of 25 mW was used at 75% capacity. During normal image capture, it was used at 0.1% transmission for GFP-claudin-1. After capture of the initial image, photobleaching was carried out at 7% laser transmission at a scan speed of 8.96 μs/pixel; then 15 iterations of laser irradiation were performed for the rectangular area of paired claudin strands. Live images after photobleaching were collected at intervals of 1 frame per 10 s, and 40–80 frames were accumulated.
Movies 1–7 are QuickTime files, which are published as supporting information on the PNAS web site, www.pnas.org.
Results and Discussion
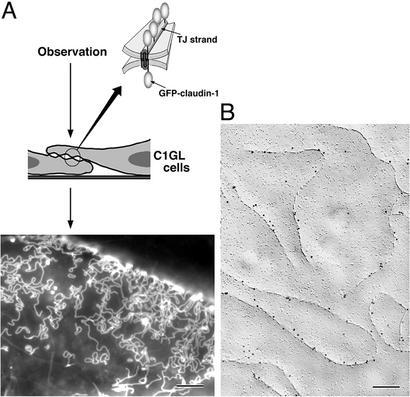
To examine the dynamic behavior of claudins using GFP technology in live cells, we used a transfection system with L fibroblasts. GFP was fused to the COOH terminus of mouse claudin-1, and cDNA encoding this construct was introduced into L fibroblasts (Fig. 1A). In stable transfectants (C1GL cells), GFP-claudin-1 was concentrated at the sites of cell–cell contact. When the contact plane was oriented obliquely or perpendicularly to the observation axis, as shown schematically in Fig. 1A, GFP-claudin-1 was clearly visualized to be concentrated in the contact plane not diffusely but as a strand-like structure. Freeze-fracture replica electron microscopy of C1GL cells identified well-developed networks of intramembranous particle strands ≈10 nm in thickness, as previously shown in L transfectants expressing claudin-1 (C1L cells; ref. 22). We then performed immuno-replica electron microscopy (24). Freeze-fracture replicas were obtained from C1GL cells, and then directly immunolabeled with an anti-GFP monoclonal antibody. As shown in Fig. 1B, GFP was detected exclusively from intramembranous particle strands, indicating that by fluorescence microscopy we detected individual intramembranous particle strands, i.e., reconstituted claudin strands, in live cells.
Figure 1.
Live observation of individual paired claudin strands formed in cultured L fibroblasts. (A) Schematic drawing of the observation system. L cell transfectants expressing GFP-claudin-1 (C1GL cells) were used, and in these cells GFP-claudin-1 was polymerized into paired strands. When the cell–cell contact plane was oriented obliquely or perpendicularly to the observation axis, GFP-claudin-1 was clearly visualized to be concentrated as strand-like structures (Lower). This image was taken by conventional fluorescence microscopy. (B) Immuno-replica electron microscopy. Freeze-fracture replicas were obtained from C1GL cells and immunolabeled with anti-GFP monoclonal antibody (10-nm gold particles). GFP was detected exclusively on intramembranous particle strands. Bars = 4 μm (A) and 300 nm (B).
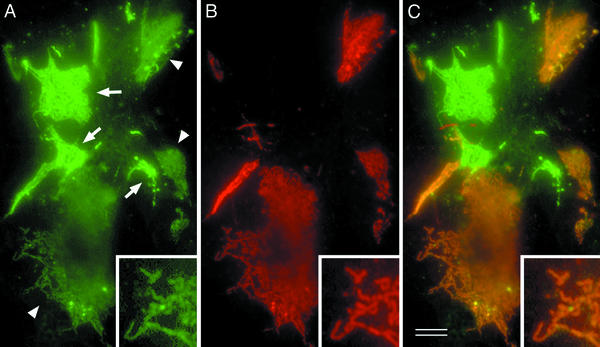
The question has naturally arisen whether these GFP-positive strands are representative of “single” strands on the free surface of cells or “paired” strands from two facing cells. To answer this question, we established L transfectants expressing the nonfluorescent FLAG-tagged claudin-1 (C1FL cells). When the GFP-tagged C1GL cells were cultured singly, all of the strand within each network showed the same bright intensity of GFP fluorescence. However, when C1GL cells were cocultured with C1FL cells, two types of GFP-positive networks were clearly distinguished in terms of the fluorescence intensity of all their strands: bright and dim (Fig. 2A). When these cocultured cells were immunofluorescently stained with anti-FLAG mAb, without exception the bright and dim networks were FLAG-negative and -positive, respectively (Fig. 2 B and C); that is, in the dim networks, the pattern of GFP-positive strands coincided precisely with that of FLAG-positive strands (Fig. 2 Inset). These findings indicated that the GFP-tagged strands in C1GL cells are always associated either with other GFP-tagged strands in adjacent C1GL cells (giving bright strands) or with FLAG-tagged strands in adjacent C1FL cells (giving dim, FLAG-tagged strands). We assume that the FLAG-tagged networks that do not coincide with GFP networks in the cocultures are formed by paired strands of FLAG-tagged protein (data not shown). The intensity of GFP fluorescence shows very small variations, indicating that pairing is quite complete.
Figure 2.
Coculture of C1GL cells with L transfectants expressing FLAG-claudin-1 (C1FL cells). Cocultured cellular sheets were fixed, permeabilized, and stained with anti-FLAG mAb in red. (A) GFP signals. Two types of GFP-positive networks, bright (arrows) and dim (arrowheads), were clearly distinguished in terms of their green fluorescence intensity. (B) FLAG staining. The GFP-bright networks lacked the FLAG signal, whereas the GFP-dim one was always positive for the FLAG signal, indicating that these two types of networks were formed at C1GL/C1GL and C1GL/C1FL cell contacts, respectively. (C) Merged image. Note that, in the GFP-dim networks, the pattern of GFP-positive strands coincided precisely with that of FLAG-positive strands (Inset). Bar = 5 μm.
We then attempted to pursue the dynamic behavior of individual paired claudin strands using C1GL cells. One of the disadvantages of this observation system is the difficulty in keeping objects in focus for a long period, as live L transfectants are motile. When the cell–cell contacts were dynamically formed or destroyed, it was especially difficult to pursue the behavior of paired claudin strands. Therefore, we focused on the fairly stable cell–cell contact regions, and even in such regions we were usually only able to examine the dynamic behavior of individual strands for up to 15 min.
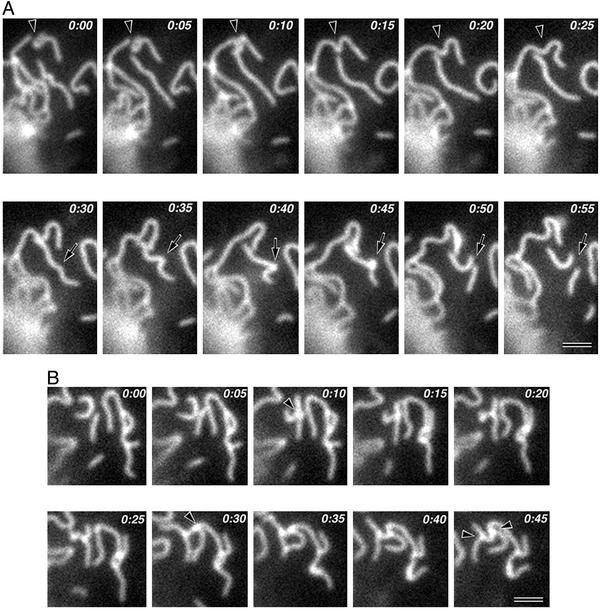
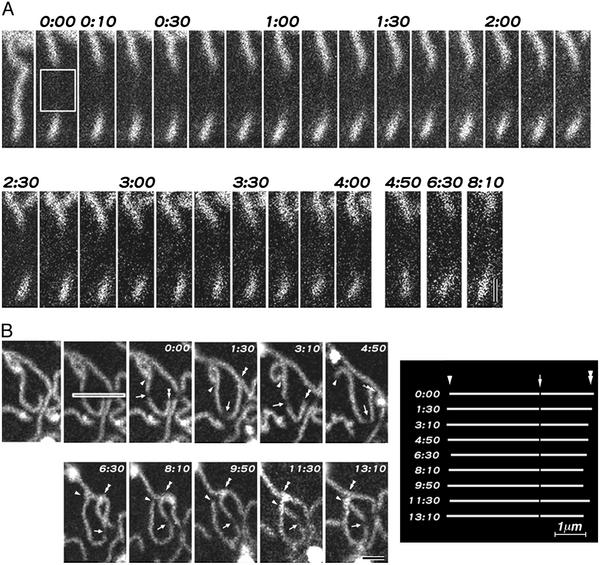
Within these limitations, we successfully observed the individual fluctuating strands within plasma membranes as time-lapse images. The strands did not elongate or shorten rapidly, in contrast to intracellular polymers such as microtubules (25). Thus, in this sense, the paired claudin strands were stable. However the strands showed very dynamic characteristics in three other respects. First, a paired claudin strand was occasionally broken into two shorter strands, and conversely the ends of two strands annealed to generate a new paired claudin strand (Fig. 3A and Movie 1). Secondly, the ends of strands interact with and bind to the sides of adjacent strands to form T-shaped junctions (Fig. 3B and Movie 2). The T-shaped junctions once formed appeared to be fairly stable, but they were sometimes dissociated. These junctions may correspond to the bifurcation of TJ strands observed by freeze-fracture replica electron microscopy (7). Third, adjacent paired claudin strands frequently interacted and associated with each other in a side-to-side manner (Movie 3). This association appeared to be more unstable than the end-to-side association. Movie 4 shows one typical time-lapse series indicating the above three types of dynamic characteristics of paired claudin strands.
Figure 3.
Time-lapse images of the dynamic behavior of paired claudin strands. Elapsed time is indicated at the top (in min:s). (A) Breaking and annealing. Two strands were annealed to generate a single strand (arrowheads), and another single strand was broken into two shorter strands (arrows). (B) End-to-side association. Three strands formed two T-shaped junctions by end-to-side association (arrowheads) (see Movies 1–4). Bars = 1 μm.
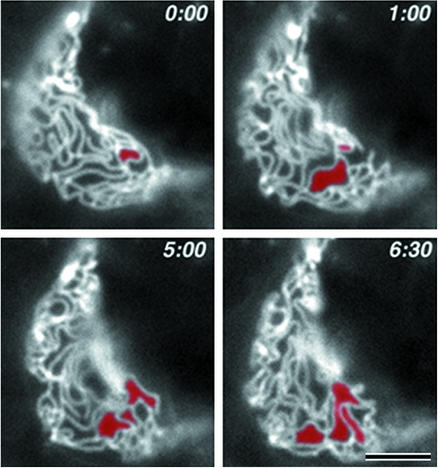
As mentioned above, for time-lapse observations it was necessary to select the fairly stable cell–cell contact planes. Therefore, the networks of paired claudin strands observed appeared to be stable as a whole. However, close inspection revealed that, through the dynamic behaviors of individual strands as shown in Movies 1–4, the network was reorganized continuously and dynamically, while maintaining the structural integrity of the network as a whole. Fig. 4 shows selected images of the network of strands in time-lapse series of Movie 5. To emphasize the structural changes, an arbitrary continuous membrane domain delineated by strands was colored red in the first frame of a time-lapse series, and when this domain became continuous to (merged with) an adjacent domain by breaking or dissociation of end-to-side/side-to-side junctions of paired claudin strands during the time-lapse observation, this adjacent domain was also colored red. Through this type of simple image processing, we were able to easily pursue the dynamic and continuous reorganization of the network of strands.
Figure 4.
Time-lapse images of the dynamic reorganization of networks of paired claudin strands. Elapsed time is indicated at the top (in min:s). In the first frame of time-lapse series, an arbitrary continuous membrane domain delineated by strands was colored red. When this domain became continuous to an adjacent domain during time-lapse observation, this adjacent domain was also colored red. The network was reorganized continuously and dynamically, while retaining the structural integrity of the network as a whole (see Movie 5). Bar = 2.5 μm.
Finally, we examined the fluorescence recovery after photobleaching in paired claudin strands. As shown in Fig. 5A and Movie 6, when a 0.9- and 0.7-μm-wide band of strands were photobleached in live cells, no recovery was detected during ≈8- and ≈7-min observations, respectively. Judging from the movements of non-bleached regions of strands, the structural integrity and continuity of strands did not appear to be affected at the bleached regions. Furthermore, even when a narrow band of a strand was bleached, the bleached mark remained detectable, and its relative position within the strand did not change over 10 min after photobleaching (Fig. 5B and Movie 7). These findings suggested that the paired claudin strands were fairly stable in terms of the arrangement of claudin molecules; claudin molecules would not be inserted into or pull out from the sides of strands, and would not move within strands.
Figure 5.
Time-lapse images of fluorescence recovery after photobleaching of reconstituted strands. Elapsed time is indicated at the top (in min:s). The bleached zone is outlined by a white box. (A) A band of strand 0.9 μm in width was photobleached, and no recovery was detected. The length of the nonbleached strand segment did not appear to change. (B) A narrow (0.15-μm-wide) band of a strand was bleached. The bleached mark (arrows) remained detectable, and its relative position within the strand did not change for 13 min after photobleaching. The positions of the bleached point (arrows) relative to both ends (arrowheads and double arrowheads) were measured and summarized schematically in Right (see Movies 6 and 7). Bars = 0.5 μm (A) and 1 μm (B).
Almost 30 yr have passed since the TJ strand was first identified in epithelial/endothelial cells by freeze-fracture replica electron microscopy (7, 26, 27). Because these structures have been observed by freeze-fracture replica electron microscopy, it was not known whether they represent a rigid or a dynamic structure. In this study, the behavior of the paired claudin strands reconstituted in L fibroblasts and forming a structure identical to the TJ strands of epithelial cells was directly observed in live cells. Interestingly, the paired claudin strands showed very dynamic behavior, moving in the plane of the membrane, breaking and annealing and associating with each other both in an end-to-side and a side-to-side manner, while at the same time showing a strong stability of the molecular array within the strand. Considering that two apposing membranes of adjacent cells were involved in the formation of individual paired claudin strands, this dynamic strand behavior was surprising. As previously shown (28), the paired claudin strands correspond to the so-called kissing points of apposing membranes in ultra-thin sectional images, where the extracellular space was completely obliterated. These observations therefore indicated that these kissing points moved, fused, disappeared, and reappeared dynamically in L transfectants.
It is likely that the GFP-tagged strands in our cells are not associated with the subcortical cytoskeleton. Most claudin species end in valine at their COOH termini (reviewed in ref. 5), which bind directly to PDZ domains (reviewed in refs. 29 and 30). Therefore, the cytoplasmic surface of TJ strands strongly attracts and recruits many PDZ domain-containing proteins, including ZO-1, -2, and -3 in epithelial/endothelial cells (5, 31–33). However, the paired claudin strands visualized in this study were expected to lack the ability to bind to PDZ-containing proteins, because the COOH terminus of claudin-1 was blocked with GFP. Indeed, endogenous ZO-1 was recruited to the claudin-1-positive cell–cell contact sites in L transfectants expressing non-tagged claudin-1 (C1L cells), but not in C1GL cells (data not shown). Considering that the cytoplasmic tail of claudin-1 is fairly short, the paired claudin strands visualized in this study could be regarded as “naked” strands in terms of their association with cytoplasmic proteins. In describing the dynamic behavior of the “naked” paired claudin strands in C1GL cells, we realize that these observations are not necessarily fully representative of TJ strands in epithelial cells. We also fused GFP to the NH2 terminus of claudin-1, which reconstituted paired strands in L cells, but, again these strands did not recruit ZO-1, probably due to the steric hindrance (data not shown).
If the dynamic characteristics of the reconstituted strands observed in L transfectants are shared with the TJ strands in situ, the dynamic reorganization of the network of strands as shown in Fig. 4 (Movie 5) is also expected to occur in epithelial/endothelial cells. This reorganization would allow materials to pass across the belt-like TJs, while retaining the structural integrity of the TJ strand network as a whole. Indeed, it has been shown that various materials, including growth factors (34), are transported across the epithelial/endothelial cellular sheets through the paracellular pathway, i.e., across the belt-like TJs, under various physiological as well as pathological conditions (1–4, 35). Of course, we attempted to visualize individual TJ strands in polarized epithelial cells using similar GFP technology, but it was technically very difficult (or impossible), partly because in these cells, the plane of the TJ strand network is parallel to the observation axis, and partly because the density of strands is very high. In future studies, it should be determined to what extent the behavior of the in situ TJ strands is dynamic as compared with the reconstituted paired claudin strands, and how the dynamic characteristics of the naked strands are modified and/or regulated in epithelial/endothelial cells. We believe that this study marks an important step in answering these interesting questions.
Supplementary Material
Acknowledgments
We thank Dr. H. Yamauchi (KAN Research Institute) for continuous encouragement. This study was supported in part by a Grant-in-Aid for Cancer Research and a Grant-in-Aid for Scientific Research (A) from the Ministry of Education, Science and Culture of Japan (to S.T.), and by the Japan Society for the Promotion of Science Research for the Future Program (to M.F.).
Abbreviations
- C1GL
L transfectants expressing GFP-claudin-1
- TJ
tight junction
References
- 1.Gumbiner B. Am J Physiol. 1987;253:C749–C758. doi: 10.1152/ajpcell.1987.253.6.C749. [DOI] [PubMed] [Google Scholar]
- 2.Schneeberger E E, Lynch R D. Am J Physiol. 1992;262:L647–L661. doi: 10.1152/ajplung.1992.262.6.L647. [DOI] [PubMed] [Google Scholar]
- 3.Anderson J M, van Itallie C M. Am J Physiol. 1995;269:G467–G475. doi: 10.1152/ajpgi.1995.269.4.G467. [DOI] [PubMed] [Google Scholar]
- 4.Balda M S, Matter K. J Cell Sci. 1998;111:541–547. doi: 10.1242/jcs.111.5.541. [DOI] [PubMed] [Google Scholar]
- 5.Tsukita S, Furuse M, Itoh M. Nat Rev Mol Cell Biol. 2001;2:285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 6.Farquhar M G, Palade G E. J Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staehelin L A. Int Rev Cytol. 1974;39:191–283. doi: 10.1016/s0074-7696(08)60940-7. [DOI] [PubMed] [Google Scholar]
- 8.Furuse M, Hirase T, Itoh M, Nagafuchi A, Yonemura S, Tsukita Sa, Tsukita Sh. J Cell Biol. 1993;123:1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ando-Akatsuka Y, Saitou M, Hirase T, Kishi M, Sakakibara A, Itoh M, Yonemura S, Furuse M, Tsukita Sh. J Cell Biol. 1996;133:43–47. doi: 10.1083/jcb.133.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, et al. J Cell Biol. 1998;142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita Sh. J Cell Biol. 1998;141:1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balda M S, Whitney J A, Flores C, González S, Cereijido M, Matter K. J Cell Biol. 1996;134:1031–1049. doi: 10.1083/jcb.134.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saitou M, Fujimoto K, Doi Y, Itoh M, Fujimoto T, Furuse M, Takano H, Noda T, Tsukita S. J Cell Biol. 1998;141:397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou M, Furuse M, Sasaki H, Takano H, Noda T, Tsukita S. Mol Biol Cell. 2000;11:4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moroi S, Saitou M, Fujimoto K, Sakakibara A, Furuse M, Yoshida O, Tsukita S. Am J Physiol. 1998;274:C1708–C1717. doi: 10.1152/ajpcell.1998.274.6.C1708. [DOI] [PubMed] [Google Scholar]
- 16.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. J Cell Biol. 2001;154:491–497. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukita S, Furuse M. Trends Cell Biol. 1999;9:268–273. doi: 10.1016/s0962-8924(99)01578-0. [DOI] [PubMed] [Google Scholar]
- 18.Morita K, Furuse M, Fujimoto K, Tsukita S. Proc Natl Acad Sci USA. 1999;96:511–516. doi: 10.1073/pnas.96.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morita K, Sasaki H, Fujimoto K, Furuse M, Tsukita S. J Cell Biol. 1999;145:579–588. doi: 10.1083/jcb.145.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morita K, Sasaki H, Furuse M, Tsukita S. J Cell Biol. 1999;147:185–194. doi: 10.1083/jcb.147.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simon D B, Lu Y, Choate K A, Velazquez H, Al-Sabban E, Praga M, Casari G, Bettinelli A, Colussi G, Rodriguez-Soriano J, et al. Science. 1999;285:103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 22.Furuse M, Sasaki H, Fujimoto K, Tsukita S. J Cell Biol. 1998;143:391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh M, Nagafuchi A, Yonemura S, Kitani-Yasuda T, Tsukita S, Tsukita S. J Cell Biol. 1993;121:491–502. doi: 10.1083/jcb.121.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujimoto K. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- 25.Desai A, Mitchison T J. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 26.Staehelin L A, Mukherjee T M, Williams A A. Protoplasma. 1969;67:165–184. doi: 10.1007/BF01248737. [DOI] [PubMed] [Google Scholar]
- 27.Goodenough D A, Revel J P. J Cell Biol. 1970;45:272–290. doi: 10.1083/jcb.45.2.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota K, Furuse M, Sasaki H, Sonoda N, Fujita K, Nagafuchi A, Tsukita S. Curr Biol. 1999;9:1035–1038. doi: 10.1016/s0960-9822(99)80452-7. [DOI] [PubMed] [Google Scholar]
- 29.Ponting C P, Phillips C, Davies K E, Blake D J. BioEssays. 1997;19:469–479. doi: 10.1002/bies.950190606. [DOI] [PubMed] [Google Scholar]
- 30.Ranganathan R, Ross E M. Curr Biol. 1997;7:R770–R773. doi: 10.1016/s0960-9822(06)00401-5. [DOI] [PubMed] [Google Scholar]
- 31.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. J Cell Biol. 1999;147:1351–1367. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fanning A S, Anderson J M. Curr Opin Cell Biol. 1999;11:432–439. doi: 10.1016/S0955-0674(99)80062-3. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Mariscal L, Betanzos A, Avila-Flores A. Semin Cell Dev Biol. 2000;11:315–324. doi: 10.1006/scdb.2000.0178. [DOI] [PubMed] [Google Scholar]
- 34.Mullin J M, Ginanni N, Laughlin K V. Cancer Res. 1998;58:1641–1645. [PubMed] [Google Scholar]
- 35.Tsukita S, Furuse M. J Cell Biol. 2000;149:13–16. doi: 10.1083/jcb.149.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.