Abstract
Proteins with LIM domains have been implicated in transcriptional regulation. The four and half LIM domain (FHL) group of LIM-only proteins is composed of five members, some of which have been shown to have intrinsic activation function. Here we show that FHL2 is the only member of the family whose expression is inducible upon serum stimulation in cultured cells. Induction of FHL2 is coordinated in time with the increased levels of two early-response products, the oncoproteins Fos and Jun. FHL2 associates with both Jun and Fos, in vitro and in vivo. The FHL2-Jun interaction requires the Ser-63-Ser-73 JNK phosphoacceptor sites in c-Jun, but not their phosphorylation. FHL2 powerfully stimulates Fos- and Jun-dependent transcription, thereby acting as an inducible coactivator of AP-1 function. Moreover, we show that intracellular localization of FHL2 is controlled by signaling events and a Crm1-dependent active nuclear export mechanism. Thus, FHL2, as an inducible coactivator of AP-1, coordinately participates with Fos and Jun in the early transcriptional response to serum factors.
A number of proteins containing LIM domains have been shown to be involved in transcriptional regulation (1–3). LIM domains are constituted by a conserved cysteine- and histidine-rich structure shaped in two repeated zinc fingers (4) first identified in the proteins encoded by the Lin-11, Isl-1, and Mec-3 genes (5–7). The LIM domain has been shown to function as a protein–protein interaction domain (8), and has often been described in association with other functional protein motifs, such as homeobox and kinase domains (1–3). However, proteins containing only LIM domains exist, named LIM-only proteins (1, 2), some of which have been directly implicated in transcriptional control. One interesting example is LIM-only protein-2, a LIM-only protein required for erythropoietic differentiation, which binds to and regulates the activity of the GATA-1 and TAL-1 transcription factors (9, 10). Another example is the LIM-only protein MLP, an important regulator of muscle differentiation, which is able to interact with and modulate the function of MyoD (11).
An interesting subclass of LIM-only proteins is constituted by molecules containing four complete and one amino-terminal half LIM motif. Five proteins share the same structural organization and a high degree of sequence homology in this group: four and half LIM domain protein (FHL) 1, FHL2, FHL3, FHL4, and activator of cAMP-responsive element (CRE) modulator (CREM) in testis (ACT) (12). One distinctive hallmark of these proteins is their tissue-specific distribution (12–16). Among these proteins, ACT and FHL2 were found to carry an intrinsic activation function (12, 17). ACT is a specific coactivator of CREM in male germ cells where it elicits transcriptional activation in a CREB-binding protein- and phosphorylation-independent manner (17). FHL2 seems to be a more promiscuous coactivator because it was found to modulate the activity of androgen receptor (18), CRE-binding protein (CREB) (12), and WT1 (19), although some degree of specificity is present because it is unable to stimulate CREM- and Sp1-dependent transcription (12). FHL2 expression was originally described to be restricted to the heart (12, 13, 16) suggesting the existence of other physiologically relevant partners in cardiac cells. One transcription factor that has been shown to play an essential role in proliferation and differentiation of cardiomyocytes is AP-1 (20, 21). Because the constituents of AP-1, the oncoproteins Fos and Jun belong to the bZip class of transcription factors (22–26), as CREB and CREM (27, 28), the possibility that FHL2 could interact with Fos and Jun is appealing.
Here we show that the expression of the gene encoding FHL2 is inducible by serum. This characteristic is unique to FHL2 because all of the other members of the FHL family are not inducible. This feature prompted us to explore the possibility that FHL2 could indeed modulate the activity of the serum-inducible Jun and Fos proteins. We demonstrate that FHL2 associates with high efficacy and independently with Jun and Fos. Interestingly, the FHL2-Jun interaction requires the Ser 63-Ser 73 JNK phosphoacceptor sites in c-Jun, but not their phosphorylation. The association results in a powerful activation of AP-1-mediated transcription. Finally, we present evidence of signaling-regulated intracellular transport of FHL2 that appears to be based on a mechanism of active, Crm1/exportin-dependent nuclear export.
Materials and Methods
RNA Analysis.
Total RNA was prepared and analyzed by RNase protection as described (29). FHL1, FHL2, FHL3, FHL4, and ACT probes (12) were prepared by using an in vitro transcription kit (Promega). tRNA was used as a control. Mouse β-actin expression was the internal control for equal RNA loading; 4 μg of total RNA was used per assay.
GST Pull-Down Assay.
The human FHL2 ORF was subcloned in pGex-1T (Amersham Pharmacia). GST-fusion proteins were expressed in Escherichia coli and extracted in BCO (20 mM Tris⋅HCl, pH 8.0, 0.5 mM EDTA, 20% glycerol, 1 mM DTT, and 0.5 mM PMSF) containing 0.5 M KCl and 1% Triton. In vitro translation was performed with a TNT T7 quick coupled transcription-translation kit (Promega). GST-fusion proteins were purified on glutathione Sepharose beads (Amersham Pharmacia) and incubated with 5 μl of [35S]methionine-labeled in vitro translated product. Beads were washed three times. Bound proteins were eluted with 20 μl of SDS-loading buffer and separated by SDS-polyacrylamide gel electrophoresis. Ten percent of in vitro translated proteins were loaded as input. GST-c-Jun proteins bound to the glutathione Sepharose beads were phosphorylated in vitro with JNK enzyme (Calbiochem) for 30 min at 30°C. GST-Jun protein (1–10 μg) was incubated with 1 μl of JNK in a buffer containing 25 mM Hepes, pH 7.5, 2 mM MgCl2, 50 μM ATP, 0.01 mM vanadate, and 0.2 mM DTT. Beads were washed three times and then incubated with labeled in vitro translated FHL2 proteins.
Coimmunoprecipitation Assays.
The pCS2 Myc-FHL2 plasmid was described (12). COS cells were cotransfected with pCS2 Myc-FHL2 and pSVc-jun (30) or BK28 c-fos (30, 31) by using the calcium phosphate coprecipitation method. Cells were harvested 48 h after transfection in 1 ml of EBC (50 mM Tris⋅HCl, pH 8.0/170 mM NaCl/0.5% Nonidet P-40/50 mM NaF) containing 1 mM PMSF and 10 μg/ml aprotinin and leupeptin. Lysates were centrifugated at 14,000 × g for 10 min to pellet debris. After preclearing for 1 h with 50 μl of protein A-Sepharose (Amersham Pharmacia), the supernatants were incubated at 4°C for 3 h with 1 μl of anti-Myc 9-E-10 monoclonal antibody crosslinked to protein A-Sepharose beads. Beads were washed three times with EBC, resuspended in 20 μl of SDS loading buffer, and analyzed by Western blot with anti-c-Jun (Santa Cruz Biotechnology), anti-c-Fos (Upstate Biotechnology) and anti-Myc 9-E-10 antibodies. Coimmunoprecitation of FHL2 and c-Jun endogenous proteins was performed preparing a whole-cell extract from NIH 3T3 cells serum-starved during 24 h and induced with 20% serum for 2 h. Proteins were immunoprecipitated with 1 μl of c-Jun antibody (New England Biolabs) and analyzed by Western blot by using anti-FHL2 and anti-c-Jun (New England Biolabs) antibodies.
Cell Lines and Transfections.
COS cells were cultured in DMEM supplemented with 10% FCS. Cells were plated at a density of 5 × 105 per 6-cm plate. In transfections the total amount of expression vector DNA was kept constant by adding pSG5, and the total DNA amount was brought to 10 μg with pBluescript SK(−). The expression vectors for FHL2 (human FHL2 cDNA), GAL4-c-Jun AD (human c-Jun, amino acid 1–100), GAL4-c-JunAD A63/73 and GAL4-c-FosAD (human c-Fos, amino acid 212–380) are based on pSG5. A CMV-β-Gal plasmid (0.1 μg) was included to monitor transfection efficiency. For analysis of FHL expression under different stimuli, cells were treated with 20% serum, UV light (254 nm; 10 sec), 1 μM calcium ionophore A23187 (Sigma), and 10 μg/ml forskolin (Sigma).
Immunofluorescence Analysis.
NIH 3T3 cells were maintained in DMEM 10% calf serum. Cells were plated in 36-mm dishes and transfected with 4 μg of DNA (2 μg of pCS2 Myc FHL2 and 2 μg of pBluescript). Cells were fixed with 4% paraformaldehyde in PBS for 20 min and permeabilized with 0.2% Triton in PBS for 10 min. Nonspecific sites were blocked with 5% BSA in PBS for 1 h. Cells were incubated with anti-Myc monoclonal antibody 9-E-10 (diluted 1:1,000) overnight at 4°C, washed, and incubated 2 h at room temperature with secondary Cy3-conjugated anti-mouse serum antibody (diluted 1:1,000; Jackson ImmunoResearch). Cells were counterstained with 4,6-diamidino-2-phenylindole dihydrochloride (Roche Molecular Biochemicals) before microscopy.
Results and Discussion
FHL2 Is an Early-Response Protein.
Several physiological functions are ascribed to FHL proteins (12, 17–19). One question concerned whether their expression could be modulated by signaling inducing agents. We monitored RNA expression by RNase protection by using specific riboprobes designed for each FHL-encoding gene. After addition of serum to growth-arrested NIH 3T3 cells we found that expression of FHL2 is induced. Induction is significant with a peak 2 h after stimulation (Fig. 1A) and is also observed in other cell types (i.e., HeLa). Thus, FHL2 appears to function as an early-response gene.
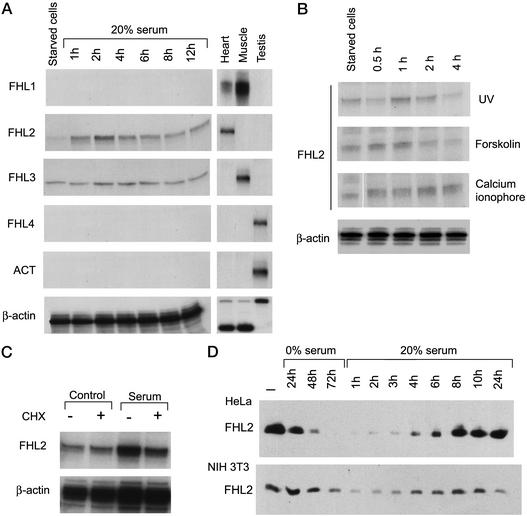
Figure 1.
FHL2 expression is induced by serum. (A) RNase protection analysis of FHL1, FHL2, FHL3, FHL4, and ACT transcripts in serum-stimulated NIH 3T3 fibroblasts. Cells were serum-starved (0.05% serum) 24 h before stimulation with 20% serum. (B) RNase protection analysis of FHL2 mRNA in NIH 3T3 cells serum-starved and then stimulated with 20% serum, UV light, forskolin, and calcium ionophore A23187. (C) RNase protection analysis of FHL2 transcript in NIH 3T3 cells starved with 0.05% serum during 24 h, treated or not with 15 μg/ml cycloheximide during 2 h, and then stimulated with 20% serum during 2 h. (D) Western blot analysis of the FHL2 protein in HeLa and NIH 3T3 fibroblasts serum-starved (0.05% serum) during 72 h and then stimulated with 20% serum during 24 h.
To determine whether FHL2 inducibility is specific to serum, we examined its response to UV light, calcium, and forskolin. FHL2 is induced after UV treatment, but not by calcium and forskolin (Fig. 1B). In comparison with serum, inducibility in response to UV light is of more modest magnitude and peaks earlier in time (1 h).
To investigate whether the induction of FHL2 by serum involves de novo protein synthesis, we pretreated cells with cycloheximide 2 h before the serum stimulation (Fig. 1C). Treatment with cycloheximide significantly reduces serum induction, indicating that synthesis of new proteins may participate in a positive regulatory loop required for FHL2 induction.
Finally, serum-induced expression of the FHL2 gene results in a remarkable increase in FHL2 protein synthesis in HeLa and NIH 3T3 cells (Fig. 1D). It is interesting that FHL2 protein levels in normally growing cells (Fig. 1D) are quite elevated, which indicates that the FHL2 gene is active in growing cells, and thereby, it may be specifically sensitive to serum deprivation.
Physical Interaction of FHL2 with AP-1.
The inducibility of the FHL2 gene (Fig. 1) and its pattern of expression (12, 13, 16) indicated that possible partners could include the products of the c-fos and c-jun oncogenes. Indeed, FHL2 is known to interact with bZip proteins, such as CREB (12, 17). We have used immunological and biochemical methods to examine if FHL2 and AP-1 physically interact. First, GST pull-down experiments were performed with GST-FHL2 fusion protein and in vitro translated [35S]methionine-labeled c-Jun and c-Fos proteins (Fig. 2A). Both c-Jun and c-Fos proteins bind specifically to GST-FHL2.
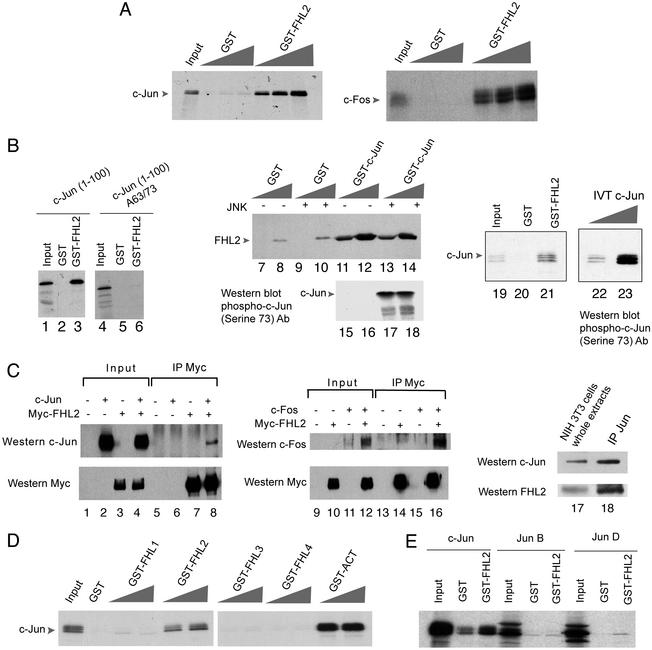
Figure 2.
FHL2 associates with AP-1. (A) FHL2 binds to c-Jun and c-Fos in vitro. GST pull-down assays were performed with in vitro translated radiolabeled c-Jun and c-Fos together with GST-FHL2 fusion protein. GST protein was used as a control. (B) For the interaction between c-Jun and FHL2, the c-Jun activation domain is sufficient, whereas the presence of Ser-63 and Ser-73 residues is necessary but their phosphorylation is not. GST pull-down experiments were performed by using GST-FHL2 together with in vitro translated c-Jun (1–100) and c-Jun (1–100) A63/A73. The binding between a GST-c-Jun protein, phosphorylated in vitro by JNK or not, and an in vitro translated FHL2 protein was assayed by GST pull-down. GST pull-down assay representing the interaction between GST-FHL2 and in vitro translated c-Jun was reproduced in parallel with a Western blot analysis by using a anti-phospho-c-Jun (Ser 73) antibody, to distinguish the binding of each phosphorylated form of c-Jun to GST-FHL2 protein. (C) FHL2 interacts with c-Jun and c-Fos in vivo. FHL2 coimmunoprecipitates with c-Jun and c-Fos, after cotransfection in mammalian cells of c-Jun, c-Fos, and Myc-FHL2 expression vectors (lanes 1–16). Immunoprecipitation was performed with the anti-Myc antibody. Whole extracts (Left) and immunocomplexes (Right) were analyzed by Western blot by using anti-c-Jun, anti-c-Fos, and anti-Myc antibodies. Endogenous FHL2 and c-Jun proteins from serum-stimulated NIH 3T3 cells coimmunoprecipitate (lanes 17–18). Immunoprecipitation was performed with an anti-c-Jun antibody and immunocomplexes were analyzed by Western blot with anti-c-Jun and anti-FHL2 antibodies. (D) GST pull-down assays show that in vitro translated c-Jun does not bind to GST-FHL1, GST-FHL3, GST-FHL4 proteins, but binds to GST-ACT protein. (E) GST pull-down experiments show that FHL2 is not interacting with other members of the Jun family, JunB and JunD. Equivalent amounts of the three Jun proteins were used.
To characterize further the interaction between FHL2 and c-Jun, we tested a construct containing the minimal transactivation domain of c-Jun (amino acids 1–100), and found that it is sufficient for FHL2 binding (Fig. 2B, lane 3). Next, we wanted to determine the contribution made by Ser-63 and Ser-73, the two phosphoacceptor sites for JNK in c-Jun (32–36). Mutation of the two serines into alanine completely abolished interaction with FHL2 (Fig. 2B, lane 6). Moreover, a JNK-phosphorylated form and a nonphosphorylated form of GST-c-Jun bind with equal efficacy to FHL2 (Fig. 2B, lanes 11–14). This result is confirmed by GST pull-down experiments with GST-FHL2 and in vitro translated c-Jun. In vitro translated c-Jun migrates into three bands that represent different levels of phosphorylation, as revealed by the anti-phospho Ser-73 antibody (Fig. 2B, lanes 22 and 23). Our GST pull-down assay demonstrates that all forms of c-Jun, independently from the levels of phosphorylation, associate equally to FHL2 (Fig. 2B, lane 21). Thus, binding to FHL2 requires the Ser-63 and Ser-73, but not their phosphorylation. This finding is reminiscent of the c-Jun/CREB-binding protein interaction, which is also dependent on Ser-63-Ser-73, but not on their phosphorylation (37). c-Jun and CREB seem to have structurally distinct modes of recognition of the KIX domain of CREB-binding protein (38). Similarly, FHL coactivators seem to operate on a specificity code for different types of activators.
To prove that the interaction between FHL2 and AP-1 occurs in vivo, we performed coimmunoprecipitation experiments. First, we used mammalian cells that express ectopically the two proteins. Cells were cotransfected with expression vectors for Myc-tagged FHL2, c-Jun, and c-Fos. Protein extracts were subjected to immunoprecipitation by using an anti-Myc antibody. c-Jun and c-Fos efficiently associate with FHL2 in immunoprecipitated complexes from cotransfected cells (Fig. 2C, lanes 8 and 16). Moreover, endogenous c-Jun and FHL2 proteins coimmunoprecipitate in serum-stimulated NIH 3T3 cells (Fig. 2C, lane 18), showing that this interaction occurs in a physiological context.
Thus, we conclude that FHL2 efficiently associates with both c-Jun and c-Fos in vitro and in vivo. By using a number of assays we have found no evidence for an effect of FHL2 on AP-1-binding activity to DNA (not shown).
We next wished to establish the specificity of the interactions observed. For that purpose, we tested by GST pull-down assay the interaction of c-Jun with all of the members of the FHL family (Fig. 2D). We found that c-Jun does not bind to FHL1, FHL3, and FHL4, whereas it is able to bind to ACT. This interaction is easily explained by the very high similarity between ACT and FHL2 proteins (80% at the amino acid levels). However, the physiological relevance of this observation is limited because of the unique, highly specific expression of ACT in male germ cells (17). Specificity of FHL2 interactive potential is strikingly demonstrated by the lack of efficient association with highly similar proteins, the other members of the c-Jun family. GST pull-down results indeed show that neither Jun B nor Jun D interact efficiently with FHL2 (Fig. 2E).
FHL2 Stimulates AP-1 Transcriptional Activity.
To evaluate whether the ability of FHL2 to interact with c-Jun and c-Fos results in a modulation of their transcriptional activity, we performed transfection experiments in mammalian cells. Cotransfection of increasing amounts of FHL2 with a c-Jun expression vector with or without c-Fos expression vector into COS cells results in a dose-dependent potentiation of transactivation of a TRE-CAT reporter (Fig. 3A). FHL2 consistently stimulates 4-fold the transcriptional activity of a c-Jun/c-Jun homodimer and stimulates 5-fold the transcriptional activity of a c-Jun/c-Fos heterodimer (Fig. 3A).
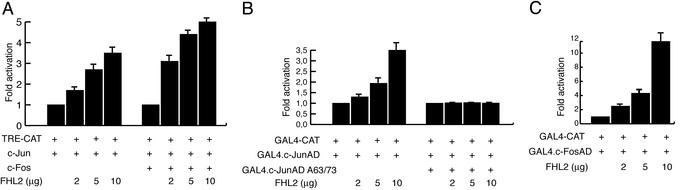
Figure 3.
(A) FHL2 stimulates c-Jun activity from an AP-1 site. COS cells were transiently transfected with 0.25 μg of TRE-TK-CAT, 0.1 μg of pSV c-Jun, 0.1 μg of BK28 c-Fos, and increasing pSV FHL2 amounts (2, 5, and 10 μg). The activity of the TRE-CAT reporter cotransfected with c-Jun expression vector is normalized to a value of 1. (B) c-Jun residues Ser-63 and Ser-73 are essential for efficient stimulation by FHL2. COS cells were cotransfected with 0.25 μg of a GAL4-CAT reporter (pG5Ad MLCAT), 0.1 μg of pSV GAL4-c-JunAD or pSV GAL4-c-JunAD A63/73, and increasing amounts of pSV FHL2 (2, 5, and 10 μg). The activity of the GAL4-CAT reporter transfected with GAL4-c-JunAD or GAL4-c-JunAD A63/73 expression vectors is normalized to a value of 1. (C) FHL2 potentiates transactivation by GAL4-c-FosAD. GAL4-CAT (0.25 μg), pSV GAL4-c-FosAD (0.5 μg), and increasing amounts of pSV FHL2 (2, 5, and 10 μg) were cotransfected into COS cells. After a chloramphenicol acetyl transferase (CAT) assay, results were quantified by using a PhosphorImager. The activity of the GAL4-CAT reporter cotransfected with GAL4-c-FosAD expression vector is normalized to a value of 1. The results represent the average of at least four experiments.
To avoid interference with cellular factors, we performed transient cotransfection assays with GAL4-c-Jun constructs and a FHL2 expression vector together with a GAL4-CAT reporter. The activation domain of c-Jun (amino acids 1–100) is sufficient for effective interaction with FHL2 (Fig. 2B) and also to elicit FHL2-dependent powerful stimulation of GAL4-c-Jun transcriptional activity (Fig. 3B). In keeping with the results of physical interaction (Fig. 2B), mutation of serines 63–73 into alanine completely abolishes FHL2-mediated potentiation (Fig. 3B).
Finally, we examined the ability of FHL2 to modulate the transcriptional activity of a GAL4-c-FosAD fusion protein on a GAL4-CAT reporter. Coexpression of FHL2 leads to a powerful, dose-dependent increase in the transactivation elicited by the GAL4-c-FosAD fusion protein (Fig. 3C).
Thus, FHL2 operates as a powerful coactivator of both Fos and Jun. Because of its early-response kinetics of inducibility, it seems that FHL2 functions in a time-coordinated manner to elicit activation of the AP-1 complex.
FHL2 Translocates into the Nucleus on Serum and UV-Light Stimulation.
Recent results have shown that FHL2 binds to integrins and is recruited to focal adhesion complexes (39). Probably through this route FHL2 is able to transmit Rho signals from the cell membrane to the nucleus. Yet, the transcriptional coactivator function of FHL2 and its association with Fos and Jun indicated a specific role in the nucleus. Thus, we wanted to assess the intracellular localization of FHL2 and identify the mechanism implicated in its translocation into the nucleus.
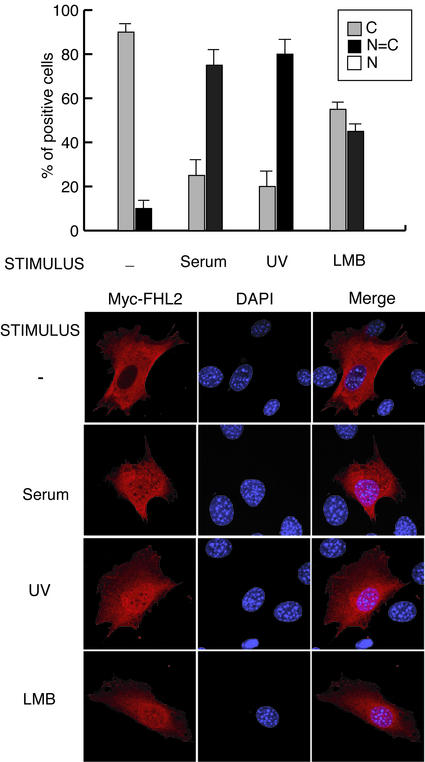
We performed immunofluorescence analyses of NIH 3T3 cells transfected with a Myc-FHL2 expression vector. Under conditions of serum deprivation and by using an anti-Myc antibody we observed a predominantly cytoplasmic FHL2 localization in 80% of the cells. In these cells, confocal laser microcopy shows FHL2 staining at focal adhesion complexes as well as diffusely in the cytoplasm. Stimulation of cells with 20% serum or UV light induces a significant translocation of FHL2 into the nucleus. Indeed, 3 h after serum-stimulation, FHL2 staining is equally localized between nucleus and cytoplasm in 75% of the cells.
We next wanted to assess if the cytoplasmic localization of FHL2 in absence of signal is due to a sequestration of FHL2 into the cytoplasm or to an exclusion from the nucleus. To answer this question, we treated the cells with leptomycin B, an inhibitor of active nuclear export. After leptomycin B treatment, 45% of the cells presented an FHL2-positive signal both in the nucleus and in the cytoplasm instead of the 20% in basal conditions (Fig. 4). We conclude that active nuclear export Crm1/exportin-dependent is, at least in part, responsible for the cytoplasmic localization of FHL2. These observations favor a scenario where FHL2 interacts with Fos and Jun, and thus modulates AP-1 function, by translocating into the nucleus when cells are stimulated.
Figure 4.
Stimulation by serum and UV light induces nuclear translocation of FHL2. NIH 3T3 cells were transiently transfected with a Myc-FHL2 expression vector. Cells were serum-starved during 24 h, treated with cycloheximide (15 μg/ml) for 2 h, and then stimulated with 20% serum or UV light, or treated with leptomycin B (LMB; 20 ng/ml) and as a control left untreated. Cells were fixed 3 h after serum and UV stimulation and 6 h after addition of leptomycin B and then stained with the anti-Myc antibody. 4,6-Diamidino-2-phenylindole dihydrochloride (DAPI) was used to stain nuclei. More than 100 positive cells were analyzed and experiments were independently reproduced at least three times. Images were acquired by using a confocal laser scanning microscope.
Conclusion
Many cofactors have been described to modulate AP-1 activity. These include proteins that appear to act at the level of DNA-binding and/or transcriptional activation. Although DNA binding was shown to be positively regulated by the nuclear redox factor Ref-1 (40, 41), it has been suggested that transcriptional activity is under the control of at least two types of molecules, Jem-1, that is expressed in acute promyelocytic leukemia cells in response to retinoids (42) and p52/54, a group of factors found to induce AP-1 function in vitro (43). In addition, CREB-binding protein/p300 participate in c-Jun-mediated transcriptional activation by functioning as signaling integrators and chromatin-remodeling agents through their histone acetyltransferase activity (37, 44). Although it is likely that all these molecules operate in different manners, and possibly at different times and locations, it important to stress the various unique features of FHL2. First, differently from the other cofactors, FHL2 is inducible and thereby acts in a physiological manner in concert with the induced products of the c-fos and c-jun oncogenes. Second, FHL2 expression was originally described to be tissue-specific (12, 13, 16, 17), suggesting that its function could be linked to distinct pathways of proliferation and/or differentiation of distinct cell types. However, our findings indicate that FHL2 expression is not restricted because it can be induced in noncardiac cells. These findings support a more general role for this cofactor and strengthen previous observations in which FHL2 was found to coactivate noncardiac transcription factors, such as the androgen receptor and WT-1 (18, 19).
Recent observations indicate that ACT, a protein closely related to FHL2, interacts with a cell-specific kinesin that actively controls its nuclear-cytoplasmic export (45). Although it is unclear whether nuclear-cytoplasmic shuttling of FHL2 implicates a kinesin-based mechanism, it is intriguing to note that a mitotic kinesin-like protein has recently been found to be intimately connected to Rho-dependent signaling and to be required for microtubule bundling (46). Because FHL2 has been shown to be implicated in the transmission of Rho-induced signals (47), it will be important to establish the specific physiological function of the FHL2 regulation of AP-1 activity and whether this activity may be coupled to the stimulation of the Rho-based signaling system. Finally, the recent finding that FHL2 may operate as an adaptor protein to couple metabolic enzymes to sites of high-energy consumption in the cardiac sarcomere (48) indicates that this LIM-only protein may have pleiotropic functions depending on its subcellular localization. Our experiments provide an important clue on the physiological significance of the nuclear-cytoplasmic shuttling of FHL2.
Acknowledgments
We thank P. Angel, R. Schule, G. M. Fimia, D. De Cesare, E. Heitz, S. Roux, and all of the members of the Sassone–Corsi laboratory for help, reagents, and discussion. A.M. was supported by a long-term Ph.D. fellowship from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. This work was supported by grants from Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation de la Recherche Médicale, Université Louis Pasteur, Human Frontier Science Program (RG-240), Organon (Akzo/Nobel), and Association pour la Recherche sur le Cancer.
Abbreviations
- FHL
four and half LIM domain protein
- CRE
cAMP-responsive element
- CREB
CRE-binding protein
- CREM
CRE modulator
- ACT
activator of CREM in testis
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Curtiss J, Heilig J S. BioEssays. 1998;20:58–69. doi: 10.1002/(SICI)1521-1878(199801)20:1<58::AID-BIES9>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 2.Dawid I B, Breen J J, Toyama R. Trends Genet. 1998;14:156–162. doi: 10.1016/s0168-9525(98)01424-3. [DOI] [PubMed] [Google Scholar]
- 3.Bach I. Mech Dev. 2000;91:5–17. doi: 10.1016/s0925-4773(99)00314-7. [DOI] [PubMed] [Google Scholar]
- 4.Jurata L W, Gill G N. Curr Top Microbiol Immunol. 1998;228:75–113. doi: 10.1007/978-3-642-80481-6_4. [DOI] [PubMed] [Google Scholar]
- 5.Freyd G, Kim S K, Horvitz H R. Nature. 1990;344:876–879. doi: 10.1038/344876a0. [DOI] [PubMed] [Google Scholar]
- 6.Pfaff S L, Mendelsohn M, Stewart C L, Edlund T, Jessell T M. Cell. 1996;84:309–320. doi: 10.1016/s0092-8674(00)80985-x. [DOI] [PubMed] [Google Scholar]
- 7.Way J C, Chalfie M. Cell. 1988;54:5–16. doi: 10.1016/0092-8674(88)90174-2. [DOI] [PubMed] [Google Scholar]
- 8.Schmeichel K L, Beckerle M C. Cell. 1994;79:211–219. doi: 10.1016/0092-8674(94)90191-0. [DOI] [PubMed] [Google Scholar]
- 9.Osada H, Grutz G G, Axelson H, Forster A, Rabbitts T H. Leukemia. 1997;11, Suppl. 3:307–312. [PubMed] [Google Scholar]
- 10.Wadman I A, Osada H, Grutz G G, Agulnick A D, Westphal H, Forster A, Rabbitts T H. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong Y, Flick M J, Kudla A J, Konieczny S F. Mol Cell Biol. 1997;17:4750–4760. doi: 10.1128/mcb.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fimia G M, De Cesare D, Sassone-Corsi P. Mol Cell Biol. 2000;20:8613–8622. doi: 10.1128/mcb.20.22.8613-8622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan K K, Tsui S K, Lee S M, Luk S C, Liew C C, Fung K P, Waye M M, Lee C Y. Gene. 1998;210:345–350. doi: 10.1016/s0378-1119(97)00644-6. [DOI] [PubMed] [Google Scholar]
- 14.Morgan M J, Madgwick A J. Biochem Biophys Res Commun. 1999;255:251–255. doi: 10.1006/bbrc.1999.0180. [DOI] [PubMed] [Google Scholar]
- 15.Morgan M J, Madgwick A J. Biochem Biophys Res Commun. 1999;255:245–250. doi: 10.1006/bbrc.1999.0179. [DOI] [PubMed] [Google Scholar]
- 16.Chu P H, Ruiz-Lozano P, Zhou Q, Cai C, Chen J. Mech Dev. 2000;95:259–265. doi: 10.1016/s0925-4773(00)00341-5. [DOI] [PubMed] [Google Scholar]
- 17.Fimia G M, De Cesare D, Sassone-Corsi P. Nature. 1999;398:165–169. doi: 10.1038/18237. [DOI] [PubMed] [Google Scholar]
- 18.Muller J M, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. EMBO J. 2000;19:359–369. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du X, Hublitz P, Gunther T, Wilhelm D, Englert C, Schule R. Biochim Biophys Acta. 2002;1577:93–101. doi: 10.1016/s0167-4781(02)00414-1. [DOI] [PubMed] [Google Scholar]
- 20.Eriksson M, Leppa S. J Biol Chem. 2002;277:15992–16001. doi: 10.1074/jbc.M107340200. [DOI] [PubMed] [Google Scholar]
- 21.Omura T, Yoshiyama M, Yoshida K, Nakamura Y, Kim S, Iwao H, Takeuchi K, Yoshikawa J. Hypertension. 2002;39:81–86. doi: 10.1161/hy0102.100783. [DOI] [PubMed] [Google Scholar]
- 22.Ransone L J, Verma I M. Annu Rev Cell Biol. 1990;6:539–557. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 23.Vogt P K, Bos T J. Adv Cancer Res. 1990;55:1–35. doi: 10.1016/s0065-230x(08)60466-2. [DOI] [PubMed] [Google Scholar]
- 24.Angel P, Karin M. Biochim Biophys Acta. 1991;1072:129–157. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 25.Karin M, Liu Z, Zandi E. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 26.Vogt P K. Oncogene. 2001;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- 27.De Cesare D, Fimia G M, Sassone-Corsi P. Trends Biochem Sci. 1999;24:281–285. doi: 10.1016/s0968-0004(99)01414-0. [DOI] [PubMed] [Google Scholar]
- 28.Mayr B, Montminy M. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Lamph W W, Wamsley P, Sassone-Corsi P, Verma I M. Nature. 1988;334:629–631. doi: 10.1038/334629a0. [DOI] [PubMed] [Google Scholar]
- 31.Sassone-Corsi P, Sisson J C, Verma I M. Nature. 1988;334:314–319. doi: 10.1038/334314a0. [DOI] [PubMed] [Google Scholar]
- 32.Pulverer B J, Kyriakis J M, Avruch J, Nikolakaki E, Woodgett J R. Nature. 1991;353:670–674. doi: 10.1038/353670a0. [DOI] [PubMed] [Google Scholar]
- 33.Smeal T, Binetruy B, Mercola D A, Birrer M, Karin M. Nature. 1991;354:494–496. doi: 10.1038/354494a0. [DOI] [PubMed] [Google Scholar]
- 34.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 35.Pulverer B J, Hughes K, Franklin C C, Kraft A S, Leevers S J, Woodgett J R. Oncogene. 1993;8:407–415. [PubMed] [Google Scholar]
- 36.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature. 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 37.Bannister A J, Oehler T, Wilhelm D, Angel P, Kouzarides T. Oncogene. 1995;11:2509–2514. [PubMed] [Google Scholar]
- 38.Campbell K M, Lumb K J. Biochemistry. 2002;41:13956–13964. doi: 10.1021/bi026222m. [DOI] [PubMed] [Google Scholar]
- 39.Wixler V, Geerts D, Laplantine E, Westhoff D, Smyth N, Aumailley M, Sonnenberg A, Paulsson M. J Biol Chem. 2000;275:33669–33678. doi: 10.1074/jbc.M002519200. [DOI] [PubMed] [Google Scholar]
- 40.Xanthoudakis S, Miao G, Wang F, Pan Y C, Curran T. EMBO J. 1992;11:3323–3335. doi: 10.1002/j.1460-2075.1992.tb05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xanthoudakis S, Curran T. EMBO J. 1992;11:653–665. doi: 10.1002/j.1460-2075.1992.tb05097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tong J H, Duprez E, Lanotte M. Leukemia. 1999;13:1982–1992. doi: 10.1038/sj.leu.2401560. [DOI] [PubMed] [Google Scholar]
- 43.Oehler T, Angel P. Mol Cell Biol. 1992;12:5508–5515. doi: 10.1128/mcb.12.12.5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez-Balbas M A, Bannister A J, Martin K, Haus-Seuffert P, Meisterernst M, Kouzarides T. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macho B, Brancorsini S, Fimia G M, Setou M, Hirokawa N, Sassone-Corsi P. Science. 2002;298:2388–2390. doi: 10.1126/science.1077265. [DOI] [PubMed] [Google Scholar]
- 46.Mishima M, Kaitna S, Glotzer M. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 47.Muller J M, Metzger E, Greschik H, Bosserhoff A K, Mercep L, Buettner R, Schule R. EMBO J. 2002;21:736–748. doi: 10.1093/emboj/21.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lange S, Auerbach D, McLoughlin P, Perriard E, Schafer B W, Perriard J C, Ehler E. J Cell Sci. 2002;115:4925–4936. doi: 10.1242/jcs.00181. [DOI] [PubMed] [Google Scholar]