Abstract
Breast cancer is the most common malignancy in United States women, accounting for >40,000 deaths each year. These breast tumors are comprised of phenotypically diverse populations of breast cancer cells. Using a model in which human breast cancer cells were grown in immunocompromised mice, we found that only a minority of breast cancer cells had the ability to form new tumors. We were able to distinguish the tumorigenic (tumor initiating) from the nontumorigenic cancer cells based on cell surface marker expression. We prospectively identified and isolated the tumorigenic cells as CD44+CD24−/lowLineage− in eight of nine patients. As few as 100 cells with this phenotype were able to form tumors in mice, whereas tens of thousands of cells with alternate phenotypes failed to form tumors. The tumorigenic subpopulation could be serially passaged: each time cells within this population generated new tumors containing additional CD44+CD24−/lowLineage− tumorigenic cells as well as the phenotypically diverse mixed populations of nontumorigenic cells present in the initial tumor. The ability to prospectively identify tumorigenic cancer cells will facilitate the elucidation of pathways that regulate their growth and survival. Furthermore, because these cells drive tumor development, strategies designed to target this population may lead to more effective therapies.
Despite advances in detection and treatment of metastatic breast cancer, mortality from this disease remains high because current therapies are limited by the emergence of therapy-resistant cancer cells (1, 2). As a result, metastatic breast cancer remains an incurable disease by current treatment strategies. Cancers are believed to arise from a series of sequential mutations that occur as a result of genetic instability and/or environmental factors (3, 4). A better understanding of the consequences of these mutations on the underlying biology of the neoplastic cells may lead to new therapeutic strategies.
In solid tumors, it has been demonstrated that only a small proportion of the tumor cells are able to form colonies in an in vitro clonogenic assay (5–11). Furthermore, large numbers of cells must typically be transplanted to form tumors in xenograft models. One possible explanation for these observations is that every cell within a tumor has the ability to proliferate and form new tumors but that the probability of an individual cell completing the necessary steps in these assays is small. An alternative explanation is that only a rare, phenotypically distinct subset of cells has the capacity to significantly proliferate and form new tumors, but that cells within this subset do so very efficiently (12). To distinguish between these possibilities, it is necessary to identify the clonogenic cells in these tumors with markers that distinguish these cells from other nontumorigenic cells. This identification has been accomplished in acute myelogenous leukemia, where it was demonstrated that a specific subpopulation of leukemia cells (that expressed markers similar to normal hematopoietic stem cells) was consistently enriched for clonogenic activity in nonobese diabetic/severe combined immunodeficient (NOD/SCID) immunocompromised mice, whereas other cancer cells were depleted of clonogenic activity (13–15). Such experiments have not been reported in solid cancers. If this model were also true for solid tumors, and only a small subset of cells within a tumor possess the capacity to proliferate and form new tumors, this finding would have significant implications for understanding the biology of and developing therapeutic strategies for these neoplasms.
To investigate the mechanisms of solid tumor heterogeneity, we developed a modification of the NOD/SCID mouse model in which human breast cancers were efficiently propagated in the mouse mammary fat pad (16). In the present study, we show that solid tumors contain a distinct population of cells with the exclusive ability to form tumors in mice. We refer to these cells as tumorigenic cells, or cancer-initiating cells, because they consistently formed tumors, whereas other cancer cell populations were depleted of cells capable of tumor formation. We identified cell surface markers that can distinguish between these cell populations. Our findings provide a previously uncharacterized model of breast tumor biology in which a defined subset of cells drives tumorigenesis, as well as generating tumor cell heterogeneity. The prospective identification of this tumorigenic population of cancer cells should allow for the identification of molecules expressed in these cells that could serve as targets to eliminate this critical population of cancer cells.
Materials and Methods
Mouse Preparation.
Eight-week-old female NOD/SCID mice were anesthetized by an i.p. injection of 0.2 ml of ketamine/xylazine (300 mg ketamine combined with 20 mg of xylazine in a 4-ml volume; 0.02 ml of the solution was used per 20-g mouse). Dilution to 200 μl was done by using Hanks' balanced salt solution (HBSS). Mice were then treated with VP-16 (etoposide) via an i.p. injection (30-mg etoposide dose per 1-kg mouse, diluted in serum-free HBSS for a final injection volume of 200 μl). At the same time, estrogen pellets were placed s.c. on the back of the mouse's neck by using a trocar. All tumor injections/implants were done 5 days after this procedure. In the following procedures, mice were always anesthetized as described above.
Primary Tumor Specimen Implantations.
For the implantation of fresh specimens, samples of human breast tumors were received within an hour after surgery. The tumors were cut up with scissors into small pieces, and the pieces were then minced with a blade to yield 2 × 2-mm pieces. Mincing was done in sterile RPMI medium 1640 under sterile conditions on ice. The tumor pieces were washed with serum-free HBSS before implantation. A 2-mm incision was then made in the mid abdomen area, and by using a trocar, one to two small tumor pieces were implanted in the region of the upper right and upper left mammary fat pads (right below the second nipple on both sides). A 6-0 suture was wrapped twice around the mammary fat pad nipple, allowing it to hold the implanted pieces in place. Sutures were removed after 5 days. Nexaban was used to seal the incision, and mice were monitored weekly for tumor growth.
Pleural Effusion Injections.
For the injection of the pleural effusions, cells were received shortly after thoracentesis and washed with serum-free HBSS. Cells were then suspended in serum free-RPMI/Matrigel mixture (1:1 volume) and then injected into the upper right and left mammary pads by using a 22-gauge needle. An amount equal to 0.2 ml, containing 1–2 million cells, was typically injected. The site of the needle injection was sealed with Nexaban to prevent any cell leakage.
Preparation of Single Cell Suspensions of Tumor Cells.
Before digestion with collagenase, xenograft tumors or primary human tumors were cut up into small pieces and then minced completely by using sterile blades. To obtain single cell suspensions, either pleural effusion cells or the resulting tumor pieces were then mixed with ultra-pure collagenase III in medium 199 (200–250 units of collagenase per ml) and allowed to incubate at 37°C for 3–4 h. Pipetting with a 10-ml pipette was done every 15–20 min. At the end of the incubation, cells were filtered through a 45-μl nylon mesh and washed with RPMI/20% FBS, then washed twice with HBSS. Cells to be injected were then suspended in HBSS/Matrigel mix (1:1 volume) and injected into the area of the mammary fat pad as described above. Nexaban was used to seal the injection site.
Cell Staining for Flow Cytometry.
Cells were counted and then transferred to a 5-ml tube, washed twice with HBSS with 2% heat-inactivated calf serum (HICS; 5 min at 1,000 rpm), then resuspended in 100 μl (per 106 cells) of HBSS with 2% HICS. Five microliters of Sandoglobin solution (1 mg/ml) was then added and incubated on ice for 10 min, after which the sample was washed twice with HBSS/2% HICS and resuspended in 100 μl (per 106 cells) of HBSS/2% HICS. Antibodies (appropriate dilution per antibody) were then added and incubated for 20 min on ice, and then washed twice with HBSS/2% HICS. When needed, a secondary antibody addition was conducted by resuspending in 100 μl (per 106 cells) of HBSS/2% HICS, and then adding 1–4 μl of secondary antibody (depending on the secondary antibody and its concentration), followed by a 20-min incubation. When a streptavidin step was used, cells were resuspended in 100 μl (per 106 cells) of HBSS/2% HICS, and then 1 μl of streptavidin, conjugated with the indicated fluorescent dye, was added, followed by a 20-min incubation. The cells were washed twice with HBSS/2% HICS and resuspended in 0.5 ml (per million cells) of HBSS/2% HICS that contained 7-aminoactinomycin D (7AAD, 1 μg/ml final concentration).
Flow Cytometry.
The antibodies used were anti-CD44 [allophycocyanin (APC), phycoerythrin (PE), or biotin], anti-CD24 (PE or FITC), anti-B38.1 (APC), anti-epithelial-specific antigen (ESA)–FITC (Biomeda, Foster City, CA), and anti-H2Kd, (PharMingen). Lineage marker antibodies were anti-CD2, -CD3 -CD10, -CD16, -CD18, -CD31, -CD64, and -CD140b. Unless noted, antibodies were purchased from PharMingen. Antibodies were directly conjugated to various fluorochromes, depending on the experiment. In all experiments, mouse cells and/or Lineage+ cells were eliminated by discarding H2Kd+ (mouse histocompatibility class I) cells or Lineage+ cells during flow cytometry. Dead cells were eliminated by using the viability dye 7AAD. Flow cytometry was performed on a FACSVantage (Becton Dickinson). Side scatter and forward scatter profiles were used to eliminate cell doublets. Cells were routinely sorted twice, and the cells were reanalyzed for purity, which typically was >95%.
Results and Discussion
Tumor Specimens and Engraftment Rate.
Human breast cancer specimens obtained from primary or metastatic sites in nine different patients [designated tumors 1–9 (T1–T9)] all engrafted in the NOD/SCID mice (Table 1). In one case, the cancer cells were obtained from a primary breast tumor (T2) whereas in other cases the cells were obtained from metastatic pleural effusions (T1 and T3–T9). Some experiments were conducted on cells after they had been passaged once or twice in mice (designated passage 1 and 2) whereas other experiments were conducted on unpassaged fresh or frozen tumor samples obtained directly from patients. During use of human cancer cells from tumors passaged in mice, contaminating mouse cells were removed by eliminating H2K+ cells (mouse histocompatibility class I).
Table 1.
Engraftment of human breast cancers into NOD/SCID mice
| Tumor | Origin | Formation in mice | Passage in mice | Diagnosis |
|---|---|---|---|---|
| T1 | Metastasis | Yes | Yes | Infiltrating ductal carcinoma |
| T2 | Breast primary | Yes | Yes | Adenocarcinoma |
| T3 | Metastasis | Yes | Yes | Invasive lobular carcinoma |
| T4 | Metastasis | Yes | No | Invasive lobular carcinoma |
| T5 | Metastasis | Yes | Yes | Invasive lobular carcinoma |
| T6 | Metastasis | Yes | Yes | Inflammatory breast carcinoma |
| T7 | Metastasis | Yes | Yes | Invasive lobular carcinoma |
| T8 | Metastasis | Yes | Yes | Inflammatory breast carcinoma |
| T9 | Metastasis | Yes | Yes | Adenocarcinoma |
Mice were injected with unsorted T1 and T3 cells and a 2-mm piece of T2. Cells from T4–T9 were isolated by flow cytometry as described in Fig. 1. All nine tumors tested engrafted in our NOD/SCID mouse model. Except for T2, which was a primary breast tumor, all other tumors were metastases. All of the tumors were passaged serially in mice except for T4.
Identification of Tumorigenicity Markers.
Breast cancer cells were heterogeneous with respect to expression of a variety of cell surface markers (including CD44, CD24, and B38.1). CD24 and CD44 are adhesion molecules whereas B38.1 has been described as a breast/ovarian cancer-specific marker (17–19). To determine whether these markers could distinguish tumorigenic from nontumorigenic cells, flow cytometry was used to isolate cells that were positive or negative for each marker from passage 1 T1 or T2 cells. When 2 × 105 to 8 × 105 cells of each population were injected, all injections of CD44+ cells (8/8), B38.1+ cells (8/8), or CD24−/low cells (12/12) gave rise to visible tumors within 12 wk of injection, but none of the CD44− cell (0/8), or B38.1− cell (0/8) injections formed detectable tumors (Table 2). Although no tumors could be detected by palpation in the locations injected with CD24+ cells, 2 of 12 mice injected with CD24+ cells did contain small growths at the injection site that were detected on necropsy. These growths most likely arose from the 1–3% of CD24− cells that invariably contaminate the sorted CD24+ cells or alternatively from CD24+ cells with reduced proliferative capacity (Table 2). Because the CD44+ cells were exclusively B38.1+, we focused on the CD44 and CD24 markers in subsequent experiments.
Table 2.
Tumor formation ability of sorted cells
| Tumors/injections
|
|||
|---|---|---|---|
| 8 × 105 | 5 × 105 | 2 × 105 | |
| Passaged T1 | |||
| CD44− | 0/2 | 0/2 | — |
| CD44+ | 2/2 | 2/2 | — |
| B38.1− | 0/2 | 0/2 | — |
| B38.1+ | 2/2 | 2/2 | — |
| CD24+ | — | — | 1/6 |
| CD24− | — | — | 6/6 |
| Passaged T2 | |||
| CD44− | 0/2 | 0/2 | — |
| CD44+ | 2/2 | 2/2 | — |
| B38.1− | 0/2 | 0/2 | — |
| B38.1+ | 2/2 | 2/2 | — |
| CD24+ | — | — | 1/6 |
| CD24− | — | — | 6/6 |
Cells were isolated by flow cytometry as described in Fig. 1 based on expression of the indicated marker and assayed for the ability to form tumors after injection into the mammary fat pads of NOD/SCID mice at 8 × 105, 5 × 105, and 2 × 105 cells per injection. For 12 wk, mice were examined weekly for tumors by observation and palpation; then all mice were necropsied to look for growths at injection sites that were too small to palpate. The number of tumors that formed and the number of injections that were performed are indicated for each population. All tumors were readily apparent by visual inspection and palpation except for tumors from the CD24+ population, which were detected only upon necropsy.
Several antigens associated with normal cell types (Lineage markers: CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b) were found not to be expressed by the cancer cells based on analyses of tumors that had been passaged multiple times in mice. By eliminating Lineage+ cells from unpassaged or early passage tumor cells, normal human leukocytes, endothelial cells, mesothelial cells, and fibroblasts were eliminated. By microscopic examination, the Lineage− tumor cells consistently had the appearance of neoplastic cells (data not shown).
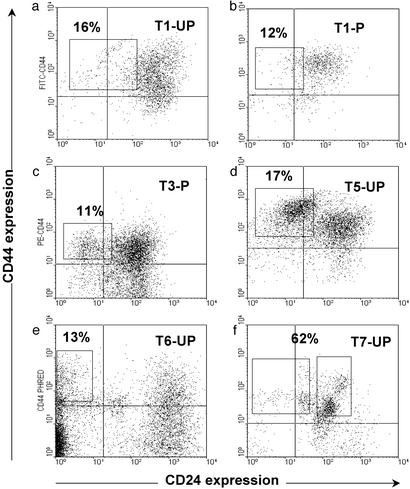
Depending on the tumor, 11–35% of the Lineage− cancer cells in tumors or pleural effusions were CD44+CD24−/low (Fig. 1). CD44+CD24−/lowLineage− cells or other populations of Lineage− cancer cells that had been isolated from nine patients were injected into the mammary fat pads of mice (Table 3). When injecting unsorted, passaged T1 or T2 cells, 5 × 104 cells consistently gave rise to tumors, but 104 cells gave rise to tumors in only a minority of cases. In contrast, as few as 103 T1 or T2 CD44+CD24−/lowLineage− cells gave rise to tumors in all cases (Table 3). In T1 and T2, up to 2 × 104 cells that were CD44+Lineage− but CD24+ failed to form tumors. These data suggest that the CD44+CD24−/lowLineage− population is 10- to 50-fold enriched for the ability to form tumors in NOD/SCID mice relative to unfractionated tumor cells. Whether the CD44+CD24−/lowLineage− cells were isolated from passaged tumors (T1, T2, and T3) or from unpassaged cancer cells obtained directly from patients (T1, T4–T6, T8, and T9), they were enriched for tumorigenic activity. Note that T7 was the only one of nine cancers studied that did not fit this pattern (Fig. 1f). Other than T7, CD24+Lineage− cancer cells in both unpassaged and passaged tumors were unable to form new tumors (Table 3). Therefore, the xenograft and unpassaged patient tumors were composed of similar populations of phenotypically diverse cancer cell types, and in both cases only the CD44+CD24−/lowLineage− cells had the capacity to proliferate to form new tumors (P < 0.001).
Figure 1.
Isolation of tumorigenic cells. Flow cytometry was used to isolate subpopulations of T1 (a and b), T3 (c), T5 (d), T6 (e), and T7 (f) cells that were tested for tumorigenicity in NOD/SCID mice. T1 (b) and T3 (c) had been passaged (P) once in NOD/SCID mice, whereas the rest of the cells were frozen or unfrozen samples obtained directly after removal from a patient (UP). Cells were stained with antibodies against CD44, CD24, Lineage markers, and mouse-H2K (for passaged tumors obtained from mice), and 7AAD. Dead cells (7AAD+), mouse cells (H2K+), and Lineage+ normal cells were eliminated from all analyses. Each plot depicts the CD24 and CD44 staining patterns of live human Lineage− cancer cells, and the frequency of the boxed tumorigenic cancer population as a percentage of cancer cells in each specimen is shown.
Table 3.
Tumorigenic breast cancer cells were highly enriched in the ESA+CD44+CD24−/low population
| Tumors/injections
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 × 105 | 105 | 5 × 104 | 2 × 104 | 104 | 5 × 103 | 103 | 500 | 200 | 100 | |
| Mouse passage 1 | ||||||||||
| Unsorted | 8/8 | 8/8 | 10/10 | 3/12 | 0/12 | — | — | — | ||
| CD44+CD24+ | — | — | — | 0/10 | 0/10 | 0/14 | 0/10 | — | — | — |
| CD44+CD24−/low | — | — | — | 10/10 | 10/10 | 14/14 | 10/10 | — | — | — |
| CD44+CD24−/lowESA+ | — | — | — | — | — | — | 10/10* | 4/4 | 4/4 | 1/6 |
| CD44+CD24−/lowESA− | — | — | — | — | — | — | 0/10* | 0/4 | 0/4 | 0/6 |
| Mouse passage 2 | ||||||||||
| CD44+CD24+ | — | — | — | — | 0/9 | — | — | — | — | — |
| CD44+CD24−/low | — | — | — | — | 9/9 | — | — | — | — | — |
| Patients' tumor cells | ||||||||||
| CD44+CD24+ | — | 0/3 | 0/4 | 0/8 | 1/13 | 0/2 | — | — | — | — |
| CD44+CD24−/low | — | 3/3 | 4/4 | — | 11/13 | 1/1 | — | — | — | — |
| CD44+CD24−/lowESA+ | — | — | — | — | — | 2/2 | 2/2 | — | — | — |
| CD44+CD24−/lowESA− | — | — | — | — | — | 2/2† | 0/2 | — | — | — |
Cells were isolated from passage 1 (Mouse passage 1) T1, T2, and T3, passage 2 (Mouse passage 2) T3, and unpassaged (Patients' tumor cells) T1, T4, T5, T6, T8, and T9. CD44+CD24+Lineage− populations and CD44+CD24−/lowLineage− cells were isolated by flow cytometry as described in Fig. 1. The indicated number of cells of each phenotype was injected into the breast of NOD/SCID mice. The frequency of tumorigenic cells calculated by the modified maximum likelihood analysis method is ≈5/105 if single tumorigenic cells were capable of forming tumors, and every transplanted tumorigenic cell gave rise to a tumor (33). Therefore, this calculation may underestimate the frequency of the tumorigenic cells because it does not take into account cell–cell interactions and local environmental factors that may influence engraftment. In addition to the markers that are shown, all sorted cells in all experiments were Lineage−, and the tumorigenic cells from T1, T2, and T3 were further selected as B38.1+. The mice were observed weekly for 4–6.5 mo, or until the mice became sick from the tumors.
Two thousand cells were injected in these experiments.
Tumor formation by 5,000 T5 ESA+CD44+CD24−/lowLineage− cells was detected 8–9 wk after injections, whereas tumor formation by 5,000 T5 ESA−CD44+CD24−/lowLineage− cells was detected 10–12 wk after injections.
In three of the tumors, further enrichment of tumorigenic activity was possible by isolating the ESA+ subset of the CD44+CD24−/low population. ESA has been used in the past to distinguish epithelial cancer cells from benign reactive mesothelial cells (20). When ESA+CD44+CD24−/low Lineage− cells were isolated from passaged T1, as few as 200 cells consistently formed tumors of ≈1 cm between 5 and 6 mo after injection, whereas 2,000 ESA−CD44+CD24−/lowLineage− cells or 20,000 CD44+CD24+ cells always failed to form tumors (Table 3). Ten thousand unsorted cells formed tumors in only 3 of 12 mice. This result suggests that the ESA+CD44+CD24−/low Lineage− population was >50-fold enriched for the ability to form tumors relative to unfractionated tumor cells (Table 3). The ESA+CD44+CD24−/lowLineage− population accounted for 2–4% of passage 1 T1 cells (2.5–5% of cancer cells). The ESA+CD44+CD24−/lowLineage− population (0.6% of cancer cells) from unpassaged T5 cells was also enriched for tumorigenic activity, compared with ESA−CD44+CD24−/low Lineage− cells, but both the ESA+ and ESA− fractions had some tumorigenic activity (Table 3). Among unpassaged T5 cells, as few as 1,000 ESA+CD44+CD24−/lowLineage− cells consistently formed tumors.
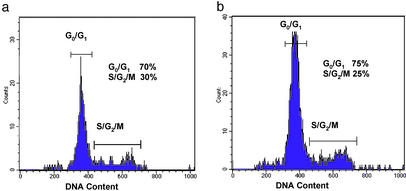
To determine whether the difference in tumorigenicity of the cell populations was due to differences in cell cycle, we analyzed these populations by flow cytometry. Comparison of the cell cycle status of tumorigenic and nontumorigenic cancer cells from T1 revealed that both exhibited a similar cell cycle distribution (Fig. 2). Therefore, neither population was enriched for cells at a particular stage of the cell cycle, and the nontumorigenic cells were able to undergo at least a limited number of divisions in the xenograft model.
Figure 2.
DNA content of tumorigenic and nontumorigenic breast cancer cells. The cell cycle status of the ESA+CD44+CD24−/lowLineage− tumorigenic cells (a) and the remaining Lineage− nontumorigenic cancer cells (b) isolated from T1 were determined by Hoechst 33342 staining of DNA content (20). The tumorigenic and nontumorigenic cell populations exhibited similar cell cycle distributions.
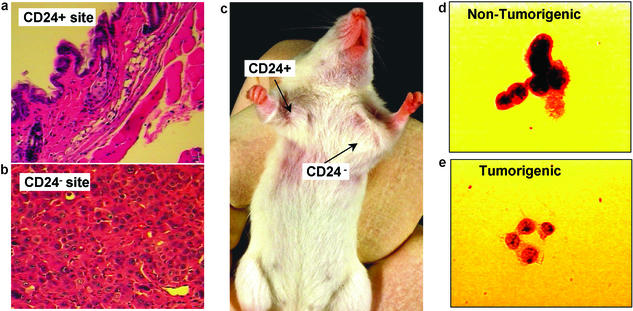
Six months after injection, the injection sites of 20,000 tumorigenic CD44+CD24−/lowLineage− cells and 20,000 nontumorigenic CD44+CD24+Lineage− cells were examined by histology. The CD44+CD24−/lowLineage− injection sites contained tumors ≈1 cm in diameter, whereas the CD44+CD24+Lineage− injection sites contained no detectable tumors (Fig. 3c). Only normal mouse mammary tissue was seen by histology at the sites of the CD44+CD24+Lineage− injections (Fig. 3a), whereas the tumors formed by CD44+CD24−/lowLineage− cells contained malignant cells as judged by hematoxylin and eosin-stained sections (Fig. 3b). Even when CD44+CD24+Lineage− injection sites from 58 mice (each administered 1,000–50,000 cells) were examined after 16–29 wk, no tumors were detected. Furthermore, the tumorigenic and nontumorigenic populations were indistinguishable morphologically. Both the tumorigenic and nontumorigenic subsets of Lineage− cells from passaged and unpassaged tumors contained >95% cancer cells as judged by Wright staining or Papanicolaou staining and microscopic analysis. By histology, the CD44+CD24−/low Lineage− cells and the rest of the Lineage− cells had the appearances of epithelial cancer cells (Fig. 3 d and e).
Figure 3.
Histology from the CD24+ injection site (a; ×20 objective magnification) revealed only normal mouse tissue, whereas the CD24−/low injection site (b; ×40 objective magnification) contained malignant cells. (c) A representative tumor in a mouse at the CD44+CD24−/lowLineage− injection site, but not at the CD44+CD24+Lineage− injection site. T3 cells were stained with Papanicolaou stain and examined microscopically (×100 objective). Both the nontumorigenic (d) and tumorigenic (e) populations contained cells with a neoplastic appearance, with large nuclei and prominent nucleoli.
The Tumorigenic Population Is Capable of Generating the Phenotypic Heterogeneity Found in the Initial Tumor.
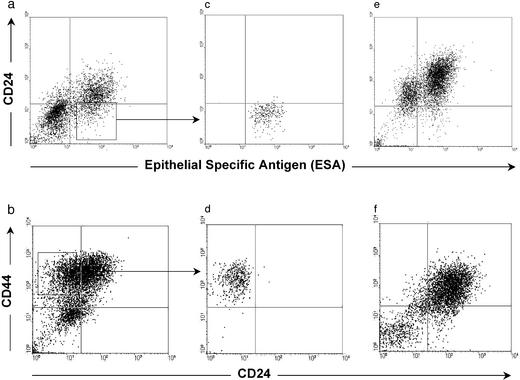
The ability of small numbers of CD44+CD24−/lowLineage− tumorigenic cells to give rise to new tumors was reminiscent of the organogenic capacity of normal stem cells. Normal stem cells self-renew and give rise to phenotypically diverse cells with reduced proliferative potential. To test whether tumorigenic breast cancer cells also exhibit these properties, tumors arising from 200 ESA+CD44+CD24−/low Lineage− T1 or 1,000 CD44+CD24−/lowLineage− T2 cells were dissociated and analyzed by flow cytometry. The heterogeneous expression patterns of ESA, CD44, or CD24 in the secondary tumors resembled the phenotypic complexity of the tumors from which they were derived (Fig. 4 a and b vs. e and f). Within these secondary tumors, the CD44+CD24−/lowLineage− cells remained tumorigenic, whereas other populations of Lineage− cancer cells remained nontumorigenic (Table 3). Thus, tumorigenic cells gave rise to both additional CD44+CD24−/lowLineage− tumorigenic cells as well as to phenotypically diverse nontumorigenic cells that recapitulated the complexity of the primary tumors from which the tumorigenic cells had been derived. These CD44+CD24−/low Lineage− tumorigenic cells from T1, T2, and T3 have now been serially passaged through four rounds of tumor formation in mice, yielding similar results in each passage with no evidence of decreased tumorigeneity (data not shown). These observations suggest that CD44+CD24−/lowLineage− tumorigenic cancer cells undergo processes analogous to the self-renewal and differentiation of normal stem cells.
Figure 4.
Phenotypic diversity in tumors arising from CD44+CD24−/lowLineage− cells. The plots depict the CD24 and CD44 or ESA staining patterns of live human Lineage− cancer cells from T1 (a, c, and e) or T2 (b, d, and f). T1 CD44+Lineage− cells (a) or T2 Lineage− cells (b) were obtained from tumors that had been passaged once in NOD/SCID mice. ESA+CD44+CD24−/lowLineage− tumorigenic cells from T1 (c) or CD44+CD24−/lowLineage− tumorigenic cells from T2 (d) were isolated and injected into the breasts of NOD/SCID mice; e and f depict analyses of the tumors that arose from these cells. In both cases, the tumorigenic cells formed tumors that contained phenotypically diverse cells similar to those observed in the original tumor.
Our results demonstrate that heterogeneous populations of cells in breast cancers consist of a phenotypically distinct tumorigenic population, as well as a much larger population that lacks this tumorigenic potential. It is known that breast cancer cells are genetically unstable, and thus individual breast cancer cells from the tumorigenic population may sometimes be unable to proliferate as a consequence of chromosomal aberrations acquired during mitosis (21–23). Nevertheless, the observation that in eight of nine tumor specimens the tumorigenic subpopulation displayed a common phenotype that allowed for their identification suggests that common pathways may drive this tumorigenic population.
The tumorigenic CD44+CD24−/lowLineage− population shares with normal stem cells the ability to proliferate extensively, and to give rise to diverse cell types with reduced developmental or proliferative potential (24). The extensive proliferative potential of the tumorigenic population was demonstrated by the ability of as few as 200 passaged or 1,000 unpassaged ESA+CD44+CD24−/low Lineage− cells to give rise to tumors (>1 cm in diameter) that could be serially transplanted in NOD/SCID mice. The tumorigenic population from T1, T2, and T3 has now been purified and serially passaged four times through NOD/SCID mice. This extensive proliferative potential contrasts with the bulk of CD44− and/or CD24+ cancer cells that lacked the ability to form detectable tumors. Not only was the CD44+CD24−/lowLineage− population of cells able to give rise to additional tumorigenic CD44+CD24−/lowLineage− cells, but they were also able to give rise to phenotypically diverse nontumorigenic cells that composed the bulk of the tumors. This result was the case even after two rounds of serial passaging. Thus, CD44+CD24−/lowLineage− cells from most tumors seem to exhibit properties of cancer stem cells. Unequivocal demonstration of the stem cell capacity of these cells will require development of model systems capable of tumor generation from a single cell (25). Nonetheless, our results demonstrate that there is a hierarchy of breast cancer cells in which some cells have the ability to proliferate extensively, whereas the majority of tumor cells that can be derived from this population have only limited proliferative potential in vivo. It has previously been shown that the phenotype of acute myelogenous leukemia leukemogenic cells is similar to that of early hematopoietic progenitor stem cells (14). Our results suggest that this result may also be true for tumorigenic breast cancer cells, because early multipotent epithelial progenitor cells also have been reported to express ESA and CD44 (26–28).
The ability to separate tumorigenic and nontumorigenic populations of tumor cells should allow the molecular characterization of these cells and elucidation of the pathways that account for their tumorigenic potential. Furthermore, the existence of a subset of tumorigenic cells within a tumor would provide an explanation for a number of clinical observations in breast cancer patients. For instance, it has been demonstrated that up to 30% of breast cancer patients may show micrometastatic disease in their bone marrow at the time of presentation. However, after 5 yr, only ≈50% of these patients will demonstrate clinically evident metastases. This finding has been speculated to be due to tumor dormancy. However, an alternative explanation consistent with a cancer stem cell model is that cancer cells in the bone marrow of patients may arise from the spread of either tumorigenic or nontumorigenic cancer cells, and only when tumorigenic cells metastasize will frank tumors that are clinically significant develop. This second explanation suggests that the development of diagnostic reagents that allow for the prospective identification of tumorigenic cells may have prognostic significance for patients with breast cancer.
The identification of tumorigenic and nontumorigenic breast cancer cells also has important therapeutic implications. Traditionally, drug therapies have been developed based on the ability of these agents to cause tumor regression in animal models. Because we have shown that the majority of cancer cells within a tumor are nontumorigenic, therapies directed at these cells would cause tumor regression. However, if therapies fail to target the tumorigenic cells, then these cells would persist after therapy and be able to regenerate the tumor, resulting in tumor relapse. It is known that normal stem cells have mechanisms that make them relatively resistant to chemotherapy, such as increased expression of BCL-2 family proteins, increased expression of membrane transporters like breast cancer drug resistance protein, and multiple drug resistance (29–32). The expression of such proteins in tumorigenic breast cancer cells may make them inherently more resistant to current therapies. The prospective identification of the tumorigenic population of cancer cells should allow the identification of molecules expressed in these cells that could be targeted to eliminate this crucial population of cancer cells, leading to more effective cancer therapies.
Acknowledgments
We thank Mark Kukaruga and Ann Marie Deslaurier for flow cytometry, Steve Ethier for tumor specimens, and Brian Clarke for the mathematical calculation of the frequency of tumorigenic cancer cells. This work was supported by National Cancer Institute Grant CA-075136. Flow cytometry was supported by National Cancer Institute Grant CA-46592.
Abbreviations
- NOD/SCID
nonobese diabetic/severe combined immunodeficient
- HICS
heat-inactivated calf serum
- ESA
epithelial-specific antigen
- Tn
tumor n
Footnotes
See commentary on page 3547.
References
- 1.Stockler M, Wilcken N R, Ghersi D, Simes R J. Cancer Treat Rev. 2000;26:151–168. doi: 10.1053/ctrv.1999.0161. [DOI] [PubMed] [Google Scholar]
- 2.Schultz L B, Weber B L. Curr Opin Oncol. 1999;11:429–434. doi: 10.1097/00001622-199911000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Aubele M, Werner M. Anal Cell Pathol. 1999;19:53–58. doi: 10.1155/1999/960923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golub T R. N Engl J Med. 2001;344:601–602. doi: 10.1056/NEJM200102223440809. [DOI] [PubMed] [Google Scholar]
- 5.Heppner G H. Cancer Res. 1984;44:2259–2265. [PubMed] [Google Scholar]
- 6.Wodinsky I, Swiniarski J, Kensler C J. Cancer Chemother Rep. 1967;51:415–421. [PubMed] [Google Scholar]
- 7.Southam C, Brunschwig A. Cancer. 1961;14:971–978. [Google Scholar]
- 8.Hamburger A W, Salmon S E. Science. 1977;197:461–463. doi: 10.1126/science.560061. [DOI] [PubMed] [Google Scholar]
- 9.Fialkow P J. Birth Defects Orig Artic Ser. 1976;12:123–132. [PubMed] [Google Scholar]
- 10.Bergsagel D E, Valeriote F A. Cancer Res. 1968;28:2187–2196. [PubMed] [Google Scholar]
- 11.Weisenthal L, Lippman M E. Cancer Treat Rep. 1985;69:615–632. [PubMed] [Google Scholar]
- 12.Reya T, Morrison S J, Clarke M F, Weissman I L. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 13.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri M, Dick J. Nature. 1994;17:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 14.Bonnet D, Dick J. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 15.Larochelle A, Vormoor J, Hanenberg H, Wang J C, Bhatia M, Lapidot T, Moritz T, Murdoch B, Xiao X L, Kato I, et al. Nat Med. 1996;2:1329–1337. doi: 10.1038/nm1296-1329. [DOI] [PubMed] [Google Scholar]
- 16.Sakakibara T, Xu Y, Bumpers H, Chen F, Bankert R, Arredondo M, Edge S, Repasky E. Cancer J Sci Am. 1996;2:291–300. [PubMed] [Google Scholar]
- 17.Ahrens T, Sleeman J, Schempp C, Howells N, Hofmann M, Ponta H, Herrlich P, Simon J. Oncogene. 2001;20:3399–3408. doi: 10.1038/sj.onc.1204435. [DOI] [PubMed] [Google Scholar]
- 18.Uchida N, Buck D, He D, Reitsma M, Masek M, Phan T, Tsukamoto A, Gage F, Weissman I L. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kufe D, Nadler L, Sargent L, Shapiro H, Hand P, Austion F, Colcher D, Schlom J. Cancer Res. 1983;43:851–857. [PubMed] [Google Scholar]
- 20.Packeisen J, Kaup-Franzen C, Knieriem H. Hybridoma. 1999;18:37–40. doi: 10.1089/hyb.1999.18.37. [DOI] [PubMed] [Google Scholar]
- 21.Murphy K, Dennis A, Rosen J. FASEB J. 2000;14:2291–2302. doi: 10.1096/fj.00-0128com. [DOI] [PubMed] [Google Scholar]
- 22.Xu X. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 23.Fukasawa K, Weiner F, Mai S. Oncogene. 1997;11:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 24.Morrison S J, Weissman I L. Immunity. 1994;1:661–673. doi: 10.1016/1074-7613(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 25.Lagasse E, Connors H, Al-Dhalimy M, Reitsma M, Dohse M, Osborne L, Wang X, Finegold M, Weissman I L, Grompe M. Nat Med. 2000;6:1229–1234. doi: 10.1038/81326. [DOI] [PubMed] [Google Scholar]
- 26.Stingl J, Eaves C, Kuusk U, Emerman J. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- 27.Liu A, True L, LaTray L, Nelson P, Ellis W, Vessella R, Lange P, Hood L, van den Engh G. Proc Natl Acad Sci USA. 1997;94:10705–10710. doi: 10.1073/pnas.94.20.10705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudjonsson T, Villadsen R, Nielsen H L, Ronnov-Jessen L, Bissell M J, Petersen O W. Genes Dev. 2002;16:693–706. doi: 10.1101/gad.952602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lagasse E, Weissman I L. Cell. 1997;89:1021–1031. doi: 10.1016/s0092-8674(00)80290-1. [DOI] [PubMed] [Google Scholar]
- 30.Lagasse E, Weissman I L. J Exp Med. 1994;179:1047–1052. doi: 10.1084/jem.179.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domen J, Gandy K L, Weissman I L. Blood. 1998;91:2272–2282. [PubMed] [Google Scholar]
- 32.Zhou S, Schuetz J D, Bunting K D, Colapietro A M, Sampath J, Morris J J, Lagutina I, Grosveld G C, Osawa M, Nakauchi H, Sorrentino B P. Nat Med. 2001;7:1028–1034. doi: 10.1038/nm0901-1028. [DOI] [PubMed] [Google Scholar]
- 33.Porter E H, Berry R J. Br J Cancer. 1964;17:583–601. doi: 10.1038/bjc.1963.78. [DOI] [PMC free article] [PubMed] [Google Scholar]