Abstract
The liver plays several critical roles in the metabolic adaptation to fasting. We have shown previously that the transcriptional coactivator peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) is induced in fasted or diabetic liver and activates the entire program of gluconeogenesis. PGC-1α interacts with several nuclear receptors known to bind gluconeogenic promoters including the glucocorticoid receptor, hepatocyte nuclear factor 4α (HNF4α), and the peroxisome proliferator-activated receptors. However, the genetic requirement for any of these interactions has not been determined. Using hepatocytes from mice lacking HNF4α in the liver, we show here that PGC-1α completely loses its ability to activate key genes of gluconeogenesis such as phosphoenolpyruvate carboxykinase and glucose-6-phosphatase when HNF4α is absent. It is also shown that PGC-1α can induce genes of β-oxidation and ketogenesis in hepatocytes, but these effects do not require HNF4α. Analysis of the glucose-6-phosphatase promoter indicates a key role for HNF4α-binding sites that function robustly only when HNF4α is coactivated by PGC-1α. These data illustrate the involvement of PGC-1α in several aspects of the hepatic fasting response and show that HNF4α is a critical component of PGC-1α-mediated gluconeogenesis.
A crucial function of the liver is the regulation of systemic fuel availability. During prolonged fasting, the liver activates gluconeogenesis to maintain blood glucose levels. Gluconeogenesis begins 4–6 h after the onset of fasting and reaches maximal activity as hepatic glycogen stores are reduced. Fatty acid oxidation is also activated during fasting and provides ATP for the liver. The very rapid oxidation of fatty acids also leads to generation and export of ketone bodies, which provide an important alternative fuel source to glucose, especially for the central nervous system. Gluconeogenesis, β-oxidation of fatty acids, and ketogenesis all are suppressed by insulin and elevated in poorly controlled diabetes. Elevated gluconeogenesis is a major contributor to the hyperglycemia of diabetes.
Peroxisome proliferator-activated receptor (PPAR)γ coactivator-1α (PGC-1α) is a transcriptional coactivator that regulates a wide range of processes involved in energy production and utilization, namely adaptive thermogenesis, mitochondrial biogenesis, glucose uptake in muscle, and skeletal muscle fiber-type switching (1–4). PGC-1α is induced in the liver in fasting or diabetes and is a potent stimulator of the entire program of hepatic gluconeogenesis (5, 6). Primary murine hepatocytes and rats receiving recombinant PGC-1α through adenoviral delivery exhibit a dramatic rise in the expression of key gluconeogenic enzymes in the liver, such as phosphoenolpyruvate carboxykinase (PEPCK) and glucose-6-phosphatase (G6Pase), and in the subsequent production of glucose. The function of PGC-1α in other aspects of the hepatic fasting response is unknown, although PGC-1α has been shown to induce mitochondrial fatty acid oxidation in cardiac myocytes (7, 8).
As a multifaceted regulator of energy metabolism, PGC-1α demonstrates binding to several nuclear hormone receptors as well as other transcription factors. With regard to gluconeogenesis, preliminary analysis of the PEPCK promoter suggests that PGC-1α functions through several nuclear receptors: the glucocorticoid receptor, hepatocyte nuclear factor 4α (HNF4α), PPARα, and PPARγ (5, 9–12). The interaction of PGC-1α with glucocorticoid receptor and HNF4α requires the major LXXLL motif of this coactivator and utilizes the AF2 domain of each receptor. Because the analysis of PGC-1α action on gluconeogenic promoters to date has been entirely of the gain-of-function variety, a critical question is: Which of these interactions are required for the activation of gluconeogenesis? Given that this process occurs mainly in the liver and HNF4α is the only liver-enriched component implicated thus far with PGC-1α, we focused our studies on this protein.
A total knockout of HNF4α is embryonic-lethal, but mice lacking HNF4α selectively in the liver are viable for several weeks, although they suffer from defective lipid homeostasis and weight loss (13). By using primary hepatocytes isolated from control (HNF4α flox) and HNF4α liver-specific knockout mice (13), the ability of PGC-1α to activate components of the fasting response in these cells in the presence and absence of HNF4α was analyzed. These studies demonstrate that HNF4α is absolutely required for PGC-1α to induce the expression of the key gluconeogenic genes PEPCK and G6Pase. Furthermore, we show that PGC-1α is capable of inducing several genes of β-oxidation and ketogenesis in the liver, but these processes, unlike gluconeogenesis, do not depend on HNF4α.
Materials and Methods
Animal Experiments.
All animal experiments were performed in accordance with Association for Assessment and Accreditation of Laboratory Animal Care guidelines and approved by institutional committees at the Dana Farber Cancer Institute and the National Cancer Institute. For the fasting time-course experiment, 45-day-old mice were either fed ad libitum or subjected to 14-, 24-, or 48-h fasts that began during the dark period. Animals then were killed by CO2 asphyxiation, and their livers were harvested. Livers were snap-frozen in liquid nitrogen and stored at −80°C. RNA was collected from aliquots of frozen liver by using Trizol reagent (Invitrogen) and then analyzed by Northern blotting with specific cDNA probes.
Cell Culture.
Primary mouse hepatocytes were isolated as described (14) and cultured in Williams' E medium containing 10% FBS and 2% DMSO at 37°C in 5% CO2. Murine hepatocytes transformed by simian virus 40 (SV40) large T antigen were cultured at 33°C in 5% CO2 in α-MEM with 2 mM glutamine/10 nM dexamethasone/10% FBS.
Adenovirus Infection.
Primary hepatocytes were infected 14 h after plating with adenoviruses expressing either GFP alone or GFP and PGC-1α at an multiplicity of infection of 50 (7). Twenty-four hours after infection, fresh growth medium containing 500 nM cAMP and 1 mM dexamethasone was added. Forty-eight hours after infection, cells were harvested for RNA isolation and Northern blotting with specific cDNA probes.
Plasmids.
PGC-1α plasmids (15) and the −1227/+57 and −180/+57 G6Pase luciferase reporter constructs have been described (16, 17). G6Pase constructs (−470/+57 and −298/+57) were made through restriction digest followed by Klenow fill-in reactions and blunt-end ligations. The region between −1227 and −470 was removed through a KpnI/BstX I double digest, and the region between −1227 and −298 was removed through a SacI/TthIII I double digest. Mutants A–D of the G6Pase promoter were made by using the QuikChange site-directed mutagenesis kit (Stratagene).
Transcriptional Activation Assays.
Transformed mouse hepatocytes were transiently transfected by using Superfect (Qiagen, Valencia, CA) at 60% confluence. After a 2- to 3-h incubation, fresh growth medium was added. Cells were lysed 24 h posttransfection, and aliquots were used to measure β-galactosidase (as a transfection control) and luciferase activities.
Gel-Shift Assays.
The sequences of each oligonucleotide corresponding to sites A–D are 5′-CCAGGAGGGCAGACCCTTGCACTGCCAAGAAGC-3′ (A), 5′-ATGTAGACTCTGTCCTGTGTCTCTGGCCTGGTA-3′ (B), 5′-GCCAAGAAGCATGCCAAAGTTAATCATTGGCCC-3′ (C), and 5′-GGCCCTGCTGAGTACATGGCCGATCAGGCTGTT-3′ (D). Each 5′ → 3′ oligonucleotide (Integrated DNA Technologies, Coralville, IA) was 32PO4-labeled by using T4 DNA polynucleotide kinase and then annealed to an excess of its unlabeled 3′ → 5′ complementary oligonucleotide. Radiolabeled duplex probes were spun through a G50 column and then incubated with in vitro-transcribed/translated proteins and antibodies for 20 min at 4°C in a buffer containing 12% glycerol, 12 mM Hepes (pH 7.9), 4 mM Tris (pH 7.9), 60 mM KCl, 1 mM EDTA, and 1 mM DTT. Complexes were resolved on a 4.5% acrylamide Tris-glycine gel. Antibodies against HNF4α and HNF3β (used as a nonspecific antibody) were from Santa Cruz Biotechnology.
Results and Discussion
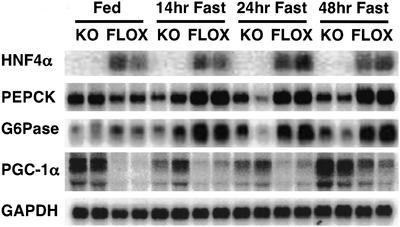
We first examined mice lacking liver HNF4α for the expression of gluconeogenic genes under conditions of feeding and fasting. Previous studies of mice lacking HNF4α in the liver revealed defects in lipid homeostasis, but gluconeogenesis was not investigated (13). As seen in Fig. 1 and as expected, fasting increases levels of mRNAs encoding the enzymes PEPCK and G6Pase in control mice (flox) at all time points. However, mice lacking HNF4α in the liver fail to induce PEPCK and G6Pase mRNA to any significant extent after the same periods of food deprivation. This shows that HNF4α is required for activating the genes of gluconeogenesis during fasting. Fig. 1 also shows, as expected, that PGC-1α is induced in the liver of the control (flox) mice under fasting conditions. Interestingly, the level of PGC-1α mRNA in the fed state and at all time points during the fast is greater in livers lacking HNF4α as compared with control livers. These data suggest an unexpected role for HNF4α as a regulator of PGC-1α as well as a target of this coactivator. Whether this effect of HNF4α is mediated through direct actions on the PGC-1α promoter or through more indirect actions is not known. In either case, the elevation in PGC-1α expression could represent part of a physiological system to compensate for the absence of HNF4α in these mice.
Figure 1.
HNF4α is required for gluconeogenesis. Control (HNF4α flox) mice or mice lacking liver HNF4α were fed ad libitum or fasted for 14, 24, or 48 h. RNA taken from the livers of these mice was analyzed for HNF4α, PEPCK, G6Pase, and PGC-1α. GAPDH was blotted to verify equal loading.
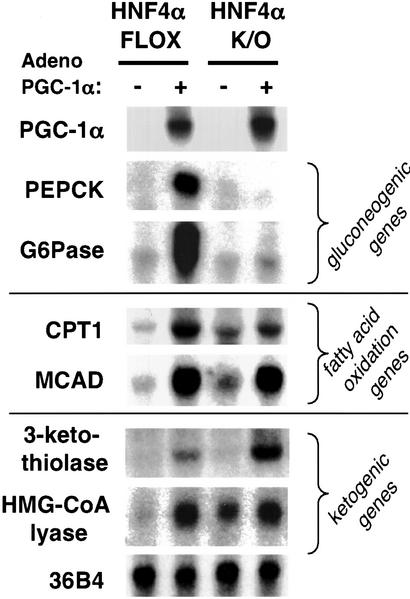
To investigate the requirement for HNF4α in PGC-1α function, primary hepatocytes were isolated from HNF4α liver-specific knockout mice and from control (HNF4α flox) mice, which then were infected with adenovirus encoding either GFP alone or GFP plus PGC-1α. Fig. 2 Top demonstrates that, as shown previously, expression of PGC-1α in control primary hepatocytes powerfully induces the expression of PEPCK and G6Pase mRNA (5). In marked contrast, infection of adenoviral PGC-1α into hepatocytes lacking HNF4α fails to elicit any detectable expression of either of these genes despite equivalent expression of PGC-1α mRNA. This indicates that PGC-1α's regulation of the genes encoding these gluconeogenic enzymes completely depends on HNF4α.
Figure 2.
PGC-1α requires HNF4α to induce the genes of gluconeogenesis. Primary hepatocytes taken from control and liver HNF4α knockout mice were infected with adenovirus encoding either GFP or PGC-1α as described in Materials and Methods. RNA was subsequently harvested and analyzed for the expression of gluconeogenic, β-oxidative, and ketogenic markers. 36B4 was blotted as an equal loading control. CPT1, carnitine palmitoyl transferase 1; MCAD, medium-chain acyl-CoA dehydrogenase; HMG-CoA lyase, 3-hydroxy-3-methylglutaryl-CoA lyase.
We next tested whether PGC-1α can regulate other aspects of the fasting response in the liver and whether these pathways also require HNF4α. Carnitine palmitoyl transferase 1 and medium-chain acyl-CoA dehydrogenase are two enzymes essential for the mitochondrial transport and β-oxidation of fatty acids. As seen in Fig. 2 Middle, the infection of primary hepatocytes with adenoviral PGC-1α strongly induces mRNAs for both of these genes. These inductions, unlike that of the gluconeogenic program, are observed in both the HNF4α flox hepatocytes and the HNF4α null hepatocytes, although there is a slight decrease in carnitine palmitoyl transferase 1 expression in the latter. Ketone body synthesis requires the expression of 3-ketothiolase, 3-hydroxy-3-methylglutaryl-CoA synthase, and 3-hydroxy-3-methylglutaryl-CoA lyase. These enzymes catalyze the conversion of acetyl CoA, the end product of β-oxidation, into ketone bodies. Fig. 2 Bottom demonstrates that expression of PGC-1α potently increases mRNA levels for 3-ketothiolase and 3-hydroxy-3-methylglutaryl-CoA lyase. This transcriptional coactivation does not depend on HNF4α, because the induction of these genes by PGC-1α is observed in both the control and knockout cells. These data indicate that PGC-1α can induce several key aspects of the liver's response to fasting, but only expression of the gluconeogenic genes depends on HNF4α.
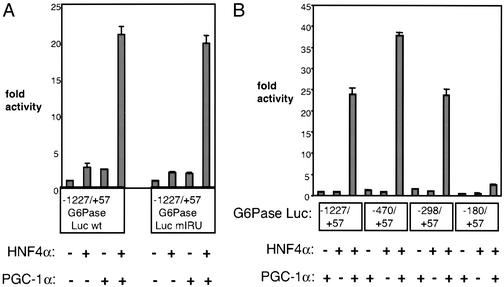
PGC-1α interacts with HNF4α through a well defined response element on the PEPCK promoter (5, 10). However, the role of HNF4α in the hormonal and fasting induction of G6Pase through its promoter remains unclear. Based on the data presented above, we sought to determine how PGC-1α and HNF4α cooperate to regulate the expression of this gluconeogenic gene. By using a reporter plasmid containing 1.2 kb of the human G6Pase promoter placed upstream of the luciferase gene (−1227/+57 G6Pase Luc), transient transfection assays were conducted in hepatocytes transformed with SV40 large T antigen (18). Transfection of a plasmid expressing either PGC-1α or HNF4α activates this promoter very poorly (Fig. 3A). However, cotransfection of PGC-1α and HNF4α together causes a dramatic activation of this promoter by at least 20-fold. These data suggest that although HNF4α cannot activate the G6Pase promoter by itself, it can do so quite effectively in combination with PGC-1α. This function of HNF4α, in principle, could be through direct binding to the promoter or through the regulation of other factors known to bind the G6Pase promoter.
Figure 3.
PGC-1α strongly coactivates HNF4α on the G6Pase promoter. (A) Coactivation is independent of the IRU. SV40-transformed hepatocytes were transfected with HNF4α ± PGC-1α. Transcriptional activity was measured by using a reporter construct containing the G6Pase promoter upstream of luciferase. A wild-type version of the reporter (−1227/+57 G6Pase Luc wt) was compared with one lacking an intact IRU (−1227/+57 G6Pase Luc mIRU). (B) PGC-1α coactivates HNF4α on a region of the promoter between nucleotides −298 and −180. Reporter constructs representing various truncations of the promoter were cotransfected with HNF4α ± PGC-1α. These graphs are representative of at least three independent trials.
FOXO1, an important regulator of G6Pase expression, is a member of the forkhead group of transcription factors that has been shown to be a key target for insulin suppression of gluconeogenesis (17, 19, 20). The insulin response unit (IRU) between −196 and −156 in the human G6Pase promoter contains three well characterized FOXO1-binding sites (17, 21). Because FOXO1 interacts with many nuclear hormone receptors including glucocorticoid receptor, thyroid receptor, and estrogen receptor (22), we first considered the possibility that HNF4α may regulate G6Pase expression by forming a complex with FOXO1 and PGC-1α on the IRU. However, HNF4α and PGC-1α are able to coactivate a mutant version of the reporter lacking FOXO1-binding sites (mIRU) as well as they coactivate the wild-type reporter (Fig. 3A). In contrast, the mutant IRU promoter is completely unresponsive to FOXO1 (ref. 17 and data not shown).
To investigate more systematically sites on the G6Pase promoter responsive to coactivation of HNF4α by PGC-1α, a series of deletion mutants from the parent (−1227/+57) G6Pase promoter–luciferase vehicle was constructed. Cotransfected HNF4α and PGC-1α robustly activate promoters originating at nucleotides −470 and −298 (Fig. 3B). However, there is a 90% decrease in coactivation of HNF4α when the region between −298 and −180 is removed, suggesting that this short segment of DNA contains sites through which HNF4α functions with PGC-1α.
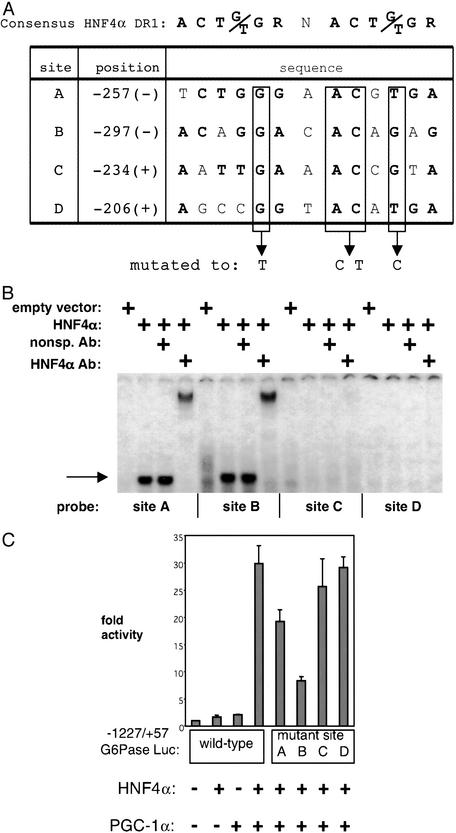
This region of the G6Pase promoter (−298/−180) was analyzed by NUBISCAN 1.0PB, a computer algorithm that predicts nuclear receptor response elements by using a weighted matrix of single hexamer half-sites (23). A search for potential HNF4α-binding sites (DR1) in this region of the G6Pase promoter was conducted, and the four highest-scoring sites are listed in Fig. 4A. To determine whether each of these sites bind HNF4α, we performed electrophoretic mobility-shift assays. Radiolabeled duplex probes were designed to encompass each of the 13-bp sites, identified above, surrounded by 10-bp flanking sequences. After the addition of in vitro-transcribed/translated HNF4α to each probe, a specific complex is clearly visible on sites A and B (Fig. 4B). This complex is supershifted by the addition of an antibody raised against HNF4α. No specific HNF4α complex is detected on sites C and D. The functional significance of these HNF4α-binding sites was determined by mutating each one within the context of the parent G6Pase promoter. Fig. 4A shows the nucleotides that were subjected to point mutation in sites A–D. The ability of PGC-1α to coactivate HNF4α decreases 30% and 70% when sites A and B, respectively, are mutated (Fig. 4C). There is little or no effect on the activity of HNF4α in combination with PGC-1α when sites C and D are mutated. These data, in total, suggest that HNF4α by itself is a poor activator of the G6Pase promoter, but two distinct HNF4α sites can function to activate this promoter when PGC-1α is present.
Figure 4.
Identification of HNF4α-binding sites in the G6Pase promoter. (A) Computer algorithm search for HNF4α-binding sites. The four highest-scoring potential HNF4α-binding sites (DR1s) between −298 and −180 of the G6Pase promoter are depicted with their position and sequence. The nucleotides in bold correspond to the consensus HNF4α-binding site. Boxed nucleotides were mutated within the context of the parent promoter construct. (B) HNF4α protein binds in vitro to sites A and B. Electrophoretic mobility-shift assays were conducted in which in vitro-translated HNF4α was incubated with radiolabeled probes corresponding to the sites depicted in A. The single arrow marks the level of the HNF4α-specific complex. A slower migrating complex is visible after the addition of an antibody against HNF4α. Free probe was in excess and is not shown. (C) Mutation of site B, and to a lesser extent of site A, impairs PGC-1α coactivation of HNF4α on the G6Pase promoter. Sites A–D on the G6Pase promoter were mutated as depicted in A. SV40-transformed hepatocytes were cotransfected with HNF4α and PGC-1α in the presence of these mutant reporters. The graph is representative of three independent trials.
Gluconeogenesis is a major contributing factor in the hyperglycemia of both type I and type II diabetes. It therefore is critical that we develop a better understanding of the molecular mechanisms driving this process to facilitate the development of improved therapies. We have shown previously that PGC-1α is increased in the liver during fasting and in multiple animal models of diabetes (5). Expression of this coactivator in cultured liver cells or in livers of live rats at physiological levels is sufficient to activate the entire program of gluconeogenesis. Although PGC-1α can interact with and coactivate the glucocorticoid receptor, HNF4α, and the PPARs on promoters of genes involved in gluconeogenesis, no data are available concerning the relative requirement for any of these transcription factors. Using hepatocytes deficient in HNF4α, we show that PGC-1α completely loses its ability to activate genes of gluconeogenesis, namely PEPCK and G6Pase, in the absence of this liver-enriched nuclear receptor. Analysis of the G6Pase promoter indicates that PGC-1α functions, in large measure, through HNF4α-binding sites that have detectable activity only in the presence of both HNF4α and PGC-1α. Mutation of these sites significantly impairs PGC-1α coactivation but does not completely abrogate it. Because HNF4α is known to induce HNF1α in hepatocytes (24, 25), it is possible that some degree of PGC-1α coactivation may occur through an HNF1α-binding site in the G6Pase promoter (26–28). It is also possible that PGC-1α is acting in part through other HNF4α-binding sites outside the region of the G6Pase promoter studied here; indeed, a functional site for HNF4α distinct from those described here has been found recently at −76 of the mouse G6Pase promoter (29).
Although HNF4α is absolutely required for PGC-1α-mediated induction of PEPCK and G6Pase mRNA, it is very likely that this coactivator also requires other transcription factors known to bind the complex promoters of these genes. Recent data suggest that PGC-1α also binds to and coactivates FOXO1 through the IRU of the G6Pase promoter (P.P., unpublished data). The PEPCK promoter is also activated by an array of accessory factors and receptors that require precise positioning in order for maximal induction to occur (30). For example, the binding of HNF4α on this promoter has been shown to facilitate the binding of the glucocorticoid receptor on nonconsensus glucocorticoid response elements (31). It is likely that PGC-1α plays a critical and dynamic role in modulating these interactions and multiprotein assembly.
The role of the liver in metabolic adaptation to the fasted state extends beyond gluconeogenesis to β-oxidation of fatty acids and ketogenesis. We show here that PGC-1α can induce several key genes of both of these processes in primary hepatocytes. Hence, PGC-1α may be viewed as a more general mediator of the fasted state in the liver. Interestingly, although the absence of HNF4α completely abrogates PGC-1α's induction of the gluconeogenic program, it has little or no effect on PGC-1α's regulation of these other hepatic pathways. This is despite previous studies showing that the promoters of the medium-chain acyl-CoA dehydrogenase and carnitine palmitoyl transferase 1 genes have binding sites for HNF4α (32, 33). These data, combined with results shown here, indicate that not every binding site for HNF4α is a target for coactivation by PGC-1α in vivo. The induction of the β-oxidation genes by PGC-1α in the absence of HNF4α likely occurs through other transcription factors such as PPARα and the estrogen-related receptor α (8, 34–38). Likewise, hepatic thiolase has been identified as a target of PPARα transactivation (39). Similar genetic analysis of these pathways to determine the factors through which PGC-1α is working must be performed.
The data presented here may have important therapeutic implications for both type I and type II diabetes, where inappropriately activated gluconeogenesis contributes greatly to hyperglycemia. The antidiabetic effects of the drug metformin illustrate that suppression of gluconeogenesis can have therapeutic value in this disease (40, 41). The data presented here, in combination with previous findings (5), strongly suggest that chemical inhibition of the docking of PGC-1α on HNF4α might have important clinical effects. Although development of an inhibitor working directly at the protein–protein interface between these factors is possible, it seems far more likely that a ligand antagonist could be developed for HNF4α that may preferentially inhibit the docking of PGC-1α versus other coactivator proteins.
Acknowledgments
We gratefully acknowledge members of the Spiegelman laboratory for helpful discussions on the project. We thank Dr. Dieter Schmoll (Ernst-Moritz-Arndt University, Greifswald, Germany) for providing us with G6Pase reporter constructs and Dr. Grant Mitchell (University of Montreal, Quebec) for the 3-hydroxy-3-methylglutaryl-CoA lyase cDNA. pcDNA3-Flag-FOXO1 was a gift from Dr. William Sellers (Dana–Farber Cancer Institute, Boston). SV40-transformed hepatocytes were generously provided by Dr. Domenico Accili (Columbia University, New York). P.P. was supported by a Lee Career Award. This work was supported by National Institutes of Health Grant DK 61562 (to B.M.S.).
Abbreviations
- PPAR
peroxisome proliferator-activated receptor
- PGC-1α
PPARγ coactivator-1α
- PEPCK
phosphoenolpyruvate carboxykinase
- G6Pase
glucose-6-phosphatase
- HNF4α
hepatocyte nuclear factor 4α
- SV40
simian virus 40
- IRU
insulin response unit
References
- 1.Puigserver P, Wu Z, Park C W, Graves R, Wright M, Spiegelman B M. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 2.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla R C, Spiegelman B M. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 3.Michael L F, Wu Z, Cheatham R B, Puigserver P, Adelmant G, Lehman J J, Kelly D P, Spiegelman B M. Proc Natl Acad Sci USA. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin J, Wu H, Tarr P T, Zhang C Y, Wu Z, Boss O, Michael L F, Puigserver P, Isotani E, Olson E N, et al. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 5.Yoon J C, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn C R, Granner D K, et al. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 6.Herzig S, Long F, Jhala U S, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, et al. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 7.Lehman J J, Barger P M, Kovacs A, Saffitz J E, Medeiros D M, Kelly D P. J Clin Invest. 2000;106:847–856. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vega R B, Huss J M, Kelly D P. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell J, Noisin E, Hall R, O'Brien R, Imai E, Granner D. Mol Endocrinol. 1994;8:585–594. doi: 10.1210/mend.8.5.8058068. [DOI] [PubMed] [Google Scholar]
- 10.Hall R K, Sladek F M, Granner D K. Proc Natl Acad Sci USA. 1995;92:412–416. doi: 10.1073/pnas.92.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson R W, Reshef L. Annu Rev Biochem. 1997;66:581–611. doi: 10.1146/annurev.biochem.66.1.581. [DOI] [PubMed] [Google Scholar]
- 12.Tontonoz P, Hu E, Devine J, Beale E G, Spiegelman B M. Mol Cell Biol. 1995;15:351–357. doi: 10.1128/mcb.15.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayhurst G P, Lee Y H, Lambert G, Ward J M, Gonzalez F J. Mol Cell Biol. 2001;21:1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaher H, Fernandez-Salguero P M, Letterio J, Sheikh M S, Fornace A J, Jr, Roberts A B, Gonzalez F J. Mol Pharmacol. 1998;54:313–321. doi: 10.1124/mol.54.2.313. [DOI] [PubMed] [Google Scholar]
- 15.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O'Malley B, Spiegelman B M. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 16.Schmoll D, Wasner C, Hinds C J, Allan B B, Walther R, Burchell A. Biochem J. 1999;338:457–463. [PMC free article] [PubMed] [Google Scholar]
- 17.Schmoll D, Walker K S, Alessi D R, Grempler R, Burchell A, Guo S, Walther R, Unterman T G. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 18.Rother K I, Imai Y, Caruso M, Beguinot F, Formisano P, Accili D. J Biol Chem. 1998;273:17491–17497. doi: 10.1074/jbc.273.28.17491. [DOI] [PubMed] [Google Scholar]
- 19.Nakae J, Park B C, Accili D. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 20.Nakae J, Kitamura T, Silver D L, Accili D. J Clin Invest. 2001;108:1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streeper R S, Svitek C A, Chapman S, Greenbaum L E, Taub R, O'Brien R M. J Biol Chem. 1997;272:11698–11701. doi: 10.1074/jbc.272.18.11698. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H H, Herrera R E, Coronado-Heinsohn E, Yang M C, Ludes-Meyers J H, Seybold-Tilson K J, Nawaz Z, Yee D, Barr F G, Diab S G, et al. J Biol Chem. 2001;276:27907–27912. doi: 10.1074/jbc.M104278200. [DOI] [PubMed] [Google Scholar]
- 23.Podvinec M, Kaufmann M R, Handschin C, Meyer U A. Mol Endocrinol. 2002;16:1269–1279. doi: 10.1210/mend.16.6.0851. [DOI] [PubMed] [Google Scholar]
- 24.Tian J M, Schibler U. Genes Dev. 1991;5:2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- 25.Kuo C J, Conley P B, Chen L, Sladek F M, Darnell J E, Jr, Crabtree G R. Nature. 1992;355:457–461. doi: 10.1038/355457a0. [DOI] [PubMed] [Google Scholar]
- 26.Lin B, Morris D W, Chou J Y. Biochemistry. 1997;36:14096–14106. doi: 10.1021/bi9703249. [DOI] [PubMed] [Google Scholar]
- 27.Lin B, Morris D W, Chou J Y. DNA Cell Biol. 1998;17:967–974. doi: 10.1089/dna.1998.17.967. [DOI] [PubMed] [Google Scholar]
- 28.Streeper R S, Eaton E M, Ebert D H, Chapman S C, Svitek C A, O'Brien R M. Proc Natl Acad Sci USA. 1998;95:9208–9213. doi: 10.1073/pnas.95.16.9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boustead J N, Stadelmaier B T, Eeds A M, Wiebe P O, Svitek C A, Oeser J K, O'Brien R, M. Biochem J. 2003;369:17–22. doi: 10.1042/BJ20021382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J C, Stromstedt P E, Sugiyama T, Granner D K. Mol Endocrinol. 1999;13:604–618. doi: 10.1210/mend.13.4.0269. [DOI] [PubMed] [Google Scholar]
- 31.Stafford J M, Wilkinson J C, Beechem J M, Granner D K. J Biol Chem. 2001;276:39885–39891. doi: 10.1074/jbc.M105370200. [DOI] [PubMed] [Google Scholar]
- 32.Carter M E, Gulick T, Raisher B D, Caira T, Ladias J A, Moore D D, Kelly D P. J Biol Chem. 1993;268:13805–13810. [PubMed] [Google Scholar]
- 33.Louet J F, Hayhurst G, Gonzalez F J, Girard J, Decaux J F. J Biol Chem. 2002;277:37991–38000. doi: 10.1074/jbc.M205087200. [DOI] [PubMed] [Google Scholar]
- 34.Gulick T, Cresci S, Caira T, Moore D D, Kelly D P. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huss J M, Kopp R P, Kelly D P. J Biol Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 36.Mascaro C, Acosta E, Ortiz J A, Marrero P F, Hegardt F G, Haro D. J Biol Chem. 1998;273:8560–8563. doi: 10.1074/jbc.273.15.8560. [DOI] [PubMed] [Google Scholar]
- 37.Sladek R, Bader J A, Giguere V. Mol Cell Biol. 1997;17:5400–5409. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vega R B, Kelly D P. J Biol Chem. 1997;272:31693–31699. doi: 10.1074/jbc.272.50.31693. [DOI] [PubMed] [Google Scholar]
- 39.Kroetz D L, Yook P, Costet P, Bianchi P, Pineau T. J Biol Chem. 1998;273:31581–31589. doi: 10.1074/jbc.273.47.31581. [DOI] [PubMed] [Google Scholar]
- 40.Wiernsperger N F, Bailey C J. Drugs. 1999;58:31–39. doi: 10.2165/00003495-199958001-00009. [DOI] [PubMed] [Google Scholar]
- 41.Wiernsperger N F, Bailey C J. Drugs. 1999;58:75–82. doi: 10.2165/00003495-199958001-00009. [DOI] [PubMed] [Google Scholar]