Abstract
The transcription factor recombination signal sequence-binding protein Jκ (RBP-J) is a key downstream element in the signaling pathway of all four mammalian Notch receptors that are critically involved in the control of embryonic and adult development. RBP-J-deficient mice display complex defects and die around day 9.5 postcoitum. Here, we investigate the function of RBP-J in the development of mesodermal cell lineages by using the OP9 stroma coculture system. RBP-J-deficient embryonic stem (ES) cells gave rise to cardiomyocytes, endothelial cells, and primitive and definitive hematopoietic cells. Thus, RBP-J-mediated signals are not required for generation of these cell types. However, when compared with parental RBP-J-expressing ES cells, cardiomyogenesis derived from RBP-J-deficient ES cells was increased. Repression over the cardiogenic pathway was restored by expressing RBP-J in RBP-J-deficient ES cells. Our data indicate that Notch signaling via RBP-J plays an important role for the correct specification of myocardial cell fates.
Recombination signal sequence-binding protein Jκ (RBP-J), the mammalian homologue of the Drosophila Suppressor of Hairless protein, is a highly conserved transcription factor that is ubiquitously expressed from the stage of embryonic stem (ES) cells to adult (1). RBP-J plays a central role for the signal transduction of Notch receptors that are critically involved in the control of embryonic and adult development (1, 2). After activation by one of its cognate ligands of the Delta and Serrate/Jagged family, the Notch receptor transmembrane domain is proteolytically cleaved, releasing the Notch intracellular domain (NIC) from the membrane. The NIC then translocates to the nucleus, where it can modulate gene expression via association with RBP-J. Thus, sequence-specific DNA binding of RBP-J directs Notch transactivation to target genes specific for this pathway. In the absence of NIC, RBP-J acts as a transcriptional repressor, but on binding to NIC, RBP-J is converted to a transcriptional activator. In addition to mediating repression and Notch-dependent transcriptional activation, a Notch-independent, cell-type-specific autoactivation function has been described for Suppressor of Hairless in Drosophila (3).
Notch signaling mediates a wide array of cell fate decisions in many tissues (2). Expectedly, mutations in the components of the Notch signaling pathway have severe clinical consequences. For example, translocations of the human Notch1 locus (TAN1) result in leukemia (4), and mutations in the human Notch ligand Jagged1 are associated with Alagille syndrome, an autosomal dominant disorder involving multiple organs, including the heart (5, 6).
Notch and its cognate ligands are expressed in the heart region of vertebrate embryos before and after formation of a linear heart tube (7–11). In Xenopus, Notch/RBP-J signaling is involved in the specification of myocardial and nonmyocardial cell fates within the early heart field (12). Mice with a null mutation in either Notch1 or RBP-J exhibit, among other complex defects, pericardial edema and die early in embryonic development (13–15). Whether these malformations are caused by aberrant heart field patterning and/or result from alterations in cardiogenic development at an earlier stage of development remains to be addressed.
In the hematopoietic system, Notch signaling influences cell fate at several stages during differentiation of hematopoietic stem cells along the T and B cell and myeloid lineages (16–20). Nevertheless, the precise functions of this signaling pathway in hematopoiesis remain to be elucidated, mainly because of the complexity of the mammalian Notch signaling system (21): thus far, four different Notch receptors and five different ligands for Notch (Delta-like 1, 3, and 4 and Jagged 1 and 2) have been identified in mammals. To date, little is known about potential functional differences between the four Notch receptors or between the different ligands.
The central role of RBP-J in signaling of all four Notch receptors makes it an attractive target by which to study the biological importance of this regulatory system in mammals. However, the death of RBP-J-deficient mice early in development, exhibiting gross developmental abnormalities similar to but more severe than those of the different Notch null mutant mice (14), precludes investigation of developmental systems beyond day 10 of gestation.
To understand further the role of Notch/RBP-J signaling in the development of mesodermal cell lineages, we analyzed the differentiation of RBP-J-deficient ES cells, employing an in vitro culture system that allows the differentiation of ES cells into cardiac muscle, endothelial, and hematopoietic cells. In this culture system, ES cells are cocultured with the stromal cell line OP9, which was established from macrophage colony-stimulating factor (M-CSF)-deficient mice (22). The absence of M-CSF in the culture system prevents the dominant production of macrophages and, thus, allows the efficient generation of the other cell lineages. Recapitulating in vivo development, ES cells cultured on OP9 first differentiate into embryonic mesodermal cells and then, further, into endothelial cells and cardiomyocytes, as well as primitive nucleated erythrocytes and definitive hematopoietic cells including definitive erythrocytes and myeloid and B lymphoid cells (23, 24). The different stages of ES cell differentiation can be followed by the expression of stage-specific cell surface markers, and the respective populations can be purified by fluorescence-activated cell sorting (FACS; ref. 25). In this study, we have investigated the developmental potential of RBP-J-deficient ES cells in vitro when grown on OP9 stromal cells.
Materials and Methods
Plasmid Constructions.
Reading frames for murine RBP-J and VP16-RBP-J derived from the plasmids pCMX-mRBP-J and pCMX-VP16-mRBP-J (26) and for a histone 2B-GFP fusion protein derived from the plasmid pBOS-H2B-GFP (PharMingen) were cloned into the expression vector pCAG-IP (27). CMV-RBP-J-GFP and CMV-VP16-RBP-J-GFP were generated by cloning the same reading frames into CMV-Exp3-GFP (28).
In Vitro Differentiation of ES Cells.
ES cell lines were maintained on gelatin-coated culture dishes in the presence of leukemia inhibitory factor. To generate mesodermal cells, ES cells were allowed to differentiate on collagen type IV-coated plates for 4 days (25). Lateral plate mesodermal cells then were isolated by FACS of Flk1+ cells with an average purity of 85%. To generate endothelial–hematopoietic precursor cells, ES cells were differentiated on OP9 stroma (22). After 5 days, platelet-endothelial cell adhesion molecule (PECAM) 1+ cells were sorted and then cultured on OP9 for further differentiation or used in methylcellulose colony assays. To assess the development of endothelial colonies, Flk1+ cells were cultured on OP9 stroma and PECAM-1+ colonies were counted after 6 days. To induce the development of cardiomyogenic colonies, Flk1+ cells generated from ES cells were cultured on OP9 stroma in the presence of stem cell factor. Six days later, cultures were counted for spontaneously contracting colonies and assayed for Nkx-2.5, GATA4, and ventricular myosin expression. Induction of hematopoiesis was performed by culturing PECAM-1+ cells on OP9 in differentiation medium containing 100 units/ml stem cell factor, 60 units/ml IL-7, and 50 units/ml Flt3-ligand (R & D Systems) for 12 days. Half of the culture medium was exchanged every 4 days, and the hematopoietic cells that had arisen were analyzed by flow cytometry. To determine the frequency of hematopoietic precursors, PECAM-1+ cells were cultured in methylcellulose colony assays in the presence of 100 units/ml stem cell factor, 200 units/ml mouse IL-3, 100 units/ml granulocyte CSF, and 2 units/ml erythropoietin. In some assays, Flt3-ligand, IL-6, thrombopoietin, and granulocyte CSF/M-CSF, each at 10 ng/ml, were also added. Colonies were scored after 8 days. Mixed colonies consisted of erythroid cells and at least two other cell lineages. Pure erythroid colonies on day 8 were attributed to burst-forming unit E, and colonies containing granulocytes and/or macrophages were attributed to granulocyte/macrophage. Cytospin preparations were stained by using Hemacolor staining reagents.
Transient Transfections, Luciferase Assays, and Reporter Plasmids.
ES cells were transfected by lipofection according to manufacturer instructions (GIBCO). One day after transfection, preparation of protein extracts and measurement of luciferase activities were performed by using the Dual-Luciferase Kit (Promega) according to manufacturer instructions. To confirm the absence of functional RBP-J and transient rescue of RBP-J in RBP-J−/− ES clones, 5 × 105 ES cells were transfected with the following plasmids: 1 μg of (RBP-J RE)12-Luc [pGa981-6, firefly luciferase reading frame under the control of a minimal promoter and 12 RBP-J binding sites (29)], 0.1 μg of phRL-CMV (constitutive expression of Renilla luciferase for transfection efficiency control), 2 μg of CMV-RBP-J-GFP or CMV-Exp-GFP, and 1 μg of CMV-NICFL-GFP (16) or CMV-Exp-GFP.
To confirm expression of functional RBP-J in RBP-J rescues, 5 × 105 cells were transfected with the following plasmids: 1 μg of (RBP-J RE)12-Luc, 0.1 μg of phRL-CMV, and 1 μg of CMV-NICFL-GFP or CMV-Exp-GFP. To confirm expression of functional VP16-RBP-J in VP16-RBP-J rescues, 5 × 105 cells were transfected with the following plasmids: 1 μg of (RBP-J RE)12-Luc or (minimal promoter)-Luc [pGa50-7, firefly luciferase reading frame under the control of a minimal promoter (29)], 0.1 μg of phRL-CMV, and 1 μg of Bluescript.
Stable Transductions and Selection Procedure.
ES cells were stably transduced by electroporation using standard procedures and selection in 1 μg/ml Puromycin (Sigma).
Antibodies.
FITC-conjugated mAb against Ly9.1, Gr-1, Ter119, and Flk1 and phycoerythrin-conjugated mAb against CD19 and PECAM-1 were used for FACS (PharMingen). Mac-1, B220, c-kit, and VE-cadherin mAbs used for FACS were all conjugated to allophycocyanin; the latter two were done in our laboratory. Unconjugated PECAM-1 mAb (PharMingen) was used as a marker for endothelial colonies and was detected by goat anti-rat Ig (H+L; The Jackson Laboratory). Unconjugated ventricular myosin mAb (Alexis, Lausen, Switzerland) was used for immunostaining and was detected by using goat anti-mouse Ig conjugated to horseradish peroxidase (BioSource International, Camarillo, CA). Unconjugated antiventricular myosin also was detected by goat anti-mouse Ig conjugated to Alexa Fluor 488. Immunofluorescence was performed by using unconjugated goat anti-GATA4 or rabbit anti-Nkx-2.5 (A-16, Santa Cruz Biotechnology). These were detected by donkey anti-goat Ig (H+L) or goat anti-rabbit Ig (H+L) conjugated to Alexa Fluor 594 (Molecular Probes).
Fluorescent Cell Sorting and Analysis.
Fluorescent cell sorting and analysis of cell cultures was performed as described (25).
Immunostaining.
Cultures were fixed with 2% paraformaldehyde in PBS for 10 min, washed with PBS, and blocked with PBS containing 1% skim milk powder and Triton X-100. To stain endothelial colonies, cultures were incubated with a 1:100 dilution of unconjugated anti-mouse PECAM-1 mAb (PharMingen) in PBS containing 1% skim milk powder and Triton X-100. The following day, the cultures were washed with PBS containing Triton X-100 (PBST). Primary antibody was detected by goat anti-rat Ig conjugated to horseradish peroxidase used at a 1:100 dilution in PBST. After overnight incubation with secondary antibody, cultures were washed with PBST and horseradish peroxidase was visualized by using NBT/BCIP color/substrate (Roche, Gipf-Oberfrick, Switzerland). To stain cardiac muscle colonies, cultures were incubated with antiventricular myosin mAb overnight and washed, and antibody binding was detected by using goat anti-rat Ig conjugated to horseradish peroxidase.
For double-immunostaining, the cultures were fixed, blocked as described, and incubated overnight with mouse antiventricular myosin (1:10 dilution) and with either rabbit anti-GATA4 polyclonal antibody or goat anti-Nkx-2.5, both at a 1:20 dilution in PBS containing 1% skim milk powder and Triton X-100. Primary antibodies were detected after washes with PBST by using anti-mouse Ig Alexa Fluor 488 and either anti-goat Ig or anti-rabbit Ig conjugated to Alexa Fluor 594. All secondary antibodies were used at a 1:100 dilution in PBST. To prevent cross-reaction of the secondary steps, rabbit anti-goat Ig-Alexa Fluor 594 was added and unbound antibody was washed before the addition of goat anti-mouse-Alexa Fluor 488. Slides then were washed and mounted in ProLong antifade mountant (Molecular Probes). Micrographic pictures were taken with a Leica TCS SP2.
Western Blot Analysis.
Western blot analysis was performed with 40 μg of protein extract per lane and a mAb against murine RBP-J (30) by using standard procedures and the enhanced chemiluminescence kit (Amersham Pharmacia) for visualization.
Results and Discussion
Expression of RBP-J Is Rescued in RBP-J-Deficient ES Cells.
To analyze whether and how RBP-J deficiency affects cardiogenic, endothelial, and hematopoietic development, we examined RBP-J-deficient (14) and RBP-J-expressing ES cell clones for their in vitro differentiation capacity by using the OP9 stroma coculture system. For this study, three RBP-J−/− clones (clones 24, 49, and 68), two RBP-J+/− cell clones (3B4 and 3B5), the wild-type RBP-J+/+ D3 cell line (14), six RBP-J−/− cell clones expressing RBP-J from a transgene, and six mock-transfected RBP-J−/− cell clones (see below) were chosen.
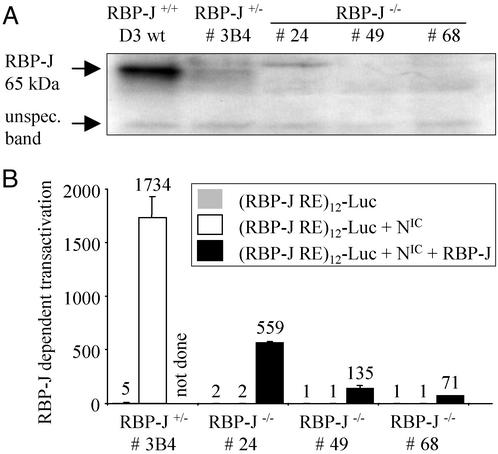
None of the RBP-J−/− clones expressed RBP-J protein, and RBP-J+/− ES cell clones had reduced levels of RBP-J compared with wild-type ES cells (Fig. 1). When tested in a functional assay by transiently expressing a RBP-J-dependent reporter construct and activated Notch, RBP-J−/− clones did not show RBP-J-dependent transactivation unless RBP-J was exogenously expressed in the cells (Fig. 1).
Figure 1.
Confirmation of the absence of functional RBP-J in RBP-J−/− ES cell lines. (A) Absence of RBP-J protein in RBP-J−/− ES cells. Western blot analysis of RBP-J+/+, RBP-J+/−, and RBP-J−/− ES cells is shown. The unspecific band shows equal loading of all lanes. (B) Absence of functional, RBP-J-dependent Notch signaling in RBP-J−/− ES cells. Transient reporter transfections were done by using a luciferase reporter under the control of 12 RBP-J-binding sites [(RBP-J RE)12-Luc]. Transient cotransfection of RBP-J restored RBP-J-dependent signaling of activated Notch (NIC).
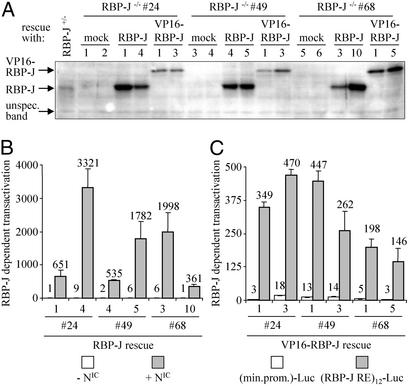
To confirm that observed effects depend on Notch/RBP-J signaling, RBP-J−/− ES cell clones were generated that expressed RBP-J or VP16-RBP-J, a transcriptionally active derivative of RBP-J, from a transgene. For our study, we needed to select a vector system that was capable of expression in primitive ES cells as well as all progenitor cells and intermediate and mature cell types produced during cardiogenic, hematopoietic, and endothelial differentiation. Because the pCAG-IP expression vector was shown to drive gene expression in undifferentiated and in differentiated ES cells (27, 31), we chose to use this vector. Expression of transgene(s) throughout ES cell differentiation along the hematopoietic, endothelial, and cardiomyogenic lineages from the pCAG-IP expression vector was confirmed by using GFP as a marker gene (see Fig. 6 and Movie 1, which are published as supporting information on the PNAS web site, www.pnas.org). RBP-J−/− ES cell clones 24, 49, and 68 were transfected with the pCAG-IP expression vector driving expression of RBP-J or VP16-RBP-J, respectively, and stable cell lines were generated. As shown by Western blotting, expression of RBP-J was rescued in the transfected RBP-J−/− ES cell clones (Fig. 2). Functionality of RBP-J and VP16-RBP-J then was tested in a reporter assay (Fig. 2). In all of 46 clones analyzed, the transgenes were functional (Fig. 2 and data not shown). For each original RBP-J−/− ES clone (24, 49, and 68), two subclones showing functional expression of RBP-J and VP16-RBP-J from the transgene, respectively, and two mock-transfected control clones then were chosen for further analysis.
Figure 2.
Generation of RBP-J−/−-derived ES cell clones expressing RBP-J or VP16-RBP-J. (A) Expression of the RBP-J and VP16-RBP-J proteins in rescued RBP-J−/− ES clones. The unspecific band shows equal loading of all lanes. (B) Detection of functional RBP-J in RBP-J rescued clones. Rescue of RBP-J-dependent signaling of activated Notch (NIC) was confirmed by using a luciferase reporter under the control of 12 RBP-J-binding sites. (C) Detection of functional VP16-RBP-J in VP16-RBP-J rescued clones. Constitutive activation of RBP-J-dependent signaling was detected by using luciferase reporters under the control of a minimal promoter with or without additional 12 RBP-J-binding sites.
For differentiation of ES cells, two techniques that allow the generation of hematopoietic, cardiomyogenic, and endothelial cells from ES cells were used. ES cells were induced to differentiate on collagen type IV-coated dishes for 4 days in the absence of leukemia inhibitory factor. Subsequently, Flk1+ cells were purified by FACS and cocultured with OP9 stromal cells in the presence of hematopoietic cytokines. Alternatively, ES cells first were differentiated into PECAM-1+ cells by coculture with OP9 stroma for 5 days in the absence of leukemia inhibitory factor. Purified PECAM-1+ cells then were cocultured further on OP9. These techniques are thought to reflect the differentiation from lateral plate mesodermal (Flk1+) and endothelial–hematopoietic (PECAM-1+) precursors.
In Vitro Cardiomyogenesis Is Increased in the Absence of RBP-J.
Mice with a null mutation in RBP-J exhibit, among other complex defects, pericardial edema and die early in embryonic development (14). To ask whether these malformations result from alterations in cardiogenic development, wild-type, RBP-J+/−, RBP-J−/−, and RBP-J−/− ES cell clones expressing RBP-J or VP16-RBP-J from the transgene were differentiated into mesodermal (Flk1+) cells, which then were isolated and cultured on OP9 cells for cardiogenic differentiation. As in all experiments, at least two clones of each genotype were analyzed.
First, we tested the generation of mesodermal (Flk1+) cells from ES cells. No difference in the generation of Flk1+ cells could be detected between RBP-J-expressing or RBP-J-deficient ES cell clones. However, ES cell clones selected for stable VP16-RBP-J expression retained an undifferentiated morphology and hardly gave rise to Flk1+ cells or PECAM-1+ precursor cells when cultured on OP9 cells (data not shown). The observed incapacity of ES cell clones expressing the transcriptionally active form of RBP-J, VP16-RBP-J, to differentiate suggests that dysregulated expression of transcriptionally active RBP-J may block differentiation of ES cells into lateral plate mesodermal (Flk1+) and endothelial–hematopoietic progenitor (PECAM-1+) cells. However, during selection for stable RBP-J−/− ES cell clones expressing VP16-RBP-J, we observed increased cell death and differentiation in the transfected cultures (data not shown). Thus, it is possible that ES cells carrying mutations that render them incapable of differentiating were selected during the establishment of stable VP-16-RBP-J-expressing cell lines. The increased cell death during selection may be due to the relatively toxic effects of the VP16-RBP-J fusion protein, which carries the strong viral transactivation domain, VP16. Whether the increased differentiation occurring initially in the VP16-RBP-J-transfected ES cell cultures results from the VP16-RBP-J fusion protein or from the transcriptionally active RBP-J remains to be addressed. Because the capacity of VP16-RBP-J-expressing ES cells to differentiate into mesodermal precursor cells was impaired, conditional systems will be necessary to define the consequences of Notch/RBP-J signaling on differentiation beyond lateral plate mesodermal precursors.
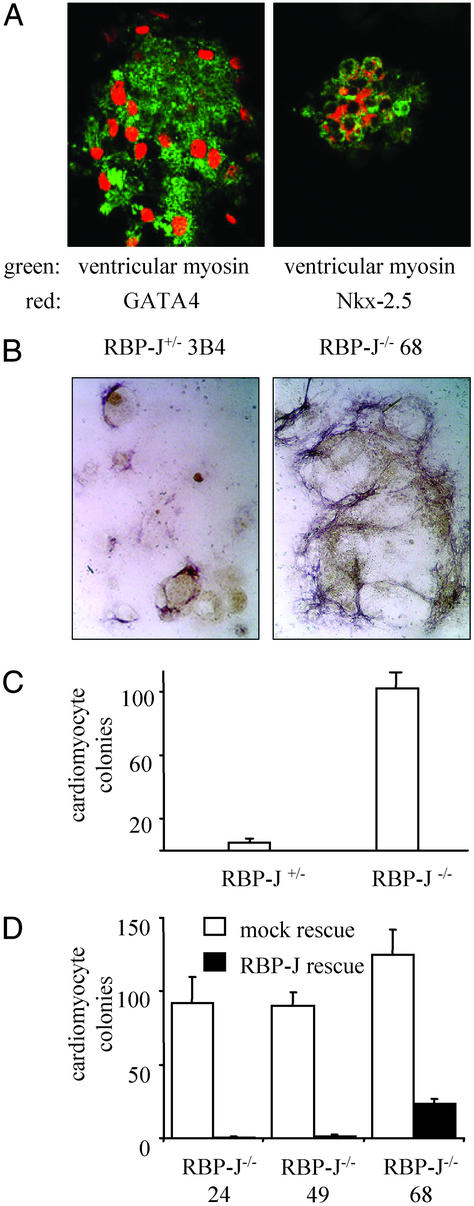
Next, we analyzed whether the lack of RBP-J influences the development of cardiac muscle cells from mesodermal (Flk1+) cells. To do this, Flk1+ cells derived from ES cells were cultured further on OP9, and the number of spontaneously contracting colonies generated after 6 days was determined. To ensure that the beating cells within the colonies were cardiomyocytes and not skeletal or smooth muscle cells, the colonies were stained for expression of cardiac-specific ventricular myosin, a contractile protein considered a marker of myocardial differentiation, and for GATA4 and Nkx-2.5, two markers of the early heart field (32–34). As shown by double-immunofluorescence staining, the cells within the beating colonies expressed both cardiac-specific ventricular myosin and GATA4 or Nkx-2.5, respectively (Fig. 3A). In the absence of RBP-J, the number of cardiac muscle colonies generated from Flk1+ cells on OP9 cells was increased considerably (Fig. 3 B and C). In RBP-J−/− ES cells expressing RBP-J from the transgene, repression over the cardiogenic pathway was restored (Fig. 3D). Thus, these results demonstrate that RBP-J is critically involved in the regulation of cardiomyogenic differentiation in mammals. In addition to a RBP-J-mediated block of skeletal muscle differentiation by activated Notch (26), RBP-J-independent signaling from Notch has been described (35–37). Whether such a RBP-J-independent pathway also functions during cardiogenesis remains to be addressed. Our observation that the absence of RBP-J leads to an increase in cardiac muscle development is consistent with recent experiments in Xenopus (12), showing that Notch/RBP-J signaling is necessary to specify cell fates within the heart field by suppressing cardiomyogenic differentiation. Cardiac defects are common in patients with Alagille syndrome, a disease caused by autosomal-dominant mutations in the gene coding for the Notch receptor Jagged1 (38). How these cardiac defects arise, however, is unknown. The in vitro model for mammalian cardiomyogenic development described here offers a valuable tool with which to dissect the role of Notch signaling in cardiac defects, such as in Alagille syndrome.
Figure 3.
Increased cardiomyogenesis from RBP-J−/− ES cells. Flk1+ cells derived from ES cells were cultured on OP9 cells for 6 days and analyzed for spontaneously contracting colonies and expression of cardiac-specific markers. (A) ES cells can be differentiated into cardiomyocytes coexpressing the cardiac-specific markers ventricular myosin and GATA4 or Nkx-2.5. The double-positive colonies showed the typical morphology of colonies spontaneously contracting in living cultures. Magnification is ×630. (B) Increased generation of cells expressing the cardiac marker ventricular myosin in the absence of RBP-J. Expression of ventricular myosin is visualized by a dark-purple staining. Unstained compact ES cell-derived colonies show a light-brown color. Three independent RBP-J+/− and RBP-J−/− clones were analyzed in triplicate or quadruplicate cultures. Photomicrographs of one representative RBP-J+/− clone and one representative RBP-J−/− clone are shown. Magnification is ×25. (C) Increased generation of cardiomyocyte colonies in the absence of RBP-J. Three independent RBP-J+/− and RBP-J−/− clones were analyzed in triplicate or quadruplicate cultures. Mean values ± SEM of all replicates of the three clones are shown as one bar, respectively. Synchronously beating colonies were counted as one cardiomyocyte colony. The experiment was repeated six times with virtually identical results. One representative experiment is shown. The increase in cardiomyocyte colonies derived from RBP-J-deficient ES cells is statistically significant (P < 0.001). (D) The increased cardiomyogenesis of RBP-J−/− ES cells can be reverted by exogenous RBP-J expression. Two independent mock- and RBP-J-rescued clones (for the three original RBP-J−/−clones) were analyzed in quadruplicate cultures. The experiment was repeated four times with virtually identical results. One representative experiment is shown. The decrease in cardiomyocyte colonies derived from RBP-J-rescued ES cells is statistically significant (P < 0.001).
RBP-J Is Not Essential for the Generation of Endothelial and Hematopoietic Cells.
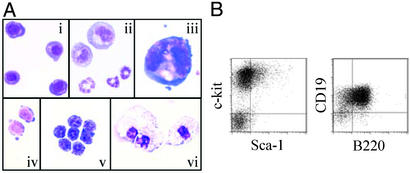
To investigate the role of RBP-J in hematopoietic development, Flk1+ cells derived from RBP-J+/+, RBP-J+/−, or RBP-J−/− ES cells were analyzed for their ability to differentiate into hematopoietic cells during coculture with OP9 cells. Primitive and definitive erythroid cells, granulocytes, macrophages, megakaryocytes, and B lymphoid cells were identified in cultures of RBP-J−/− cells (Fig. 4) as in control cultures of cells expressing RBP-J (data not shown). Thus, the data presented here show, in line with previous reports that used mice carrying an induced deletion of Notch or RBP-J in adult bone marrow cells (19, 39), that RBP-J is not essential for the development of hematopoietic cells. Furthermore, our work suggests that the Notch/RBP-J-signaling pathway also is not required for hematopoietic development during embryogenesis. Next, we asked whether embryonic hematopoietic–endothelial development from RBP-J−/− ES cells was altered. Although the morphology of the hematopoietic colonies revealed a normal appearance and cell type distribution, the number of colonies generated from endothelial–hematopoietic (PECAM-1+) precursors was reduced in RBP-J−/− clones (Fig. 5). The low hematopoietic colony formation, however, was maintained when RBP-J−/− ES cells expressing RBP-J from the transgene were differentiated on OP9 (Fig. 5). This suggests that the differences in colony formation reflect clonal variances and that hematopoietic development is largely unaffected by the absence of RBP-J.
Figure 4.
Hematopoietic development from RBP-J−/− ES cells in vitro. Flk1+ cells derived from three independent RBP-J+/− and RBP-J−/− clones were cultured on OP9 cells for 10 days. The experiment was repeated six times with virtually identical results. (A) Photomicrographs of hematopoietic cells derived from RBP-J−/− ES cells. Cytospin preparations were stained with May–Grünwald–Giemsa. Magnification is ×1,000. (i) Primitive erythroid cells. (ii) Neutrophil granulocytic cells. (iii) Megakaryocyte. (iv and v) Definitive erythroid cells. (vi) Macrophages. (B) Detection of hematopoietic progenitors and B lymphoid cells derived from RBP-J−/− ES cells by expression of cell surface markers. Expression of surface markers on cultured cells derived from RBP-J−/− J ES cells was analyzed by FACS on days 8 and 10 of differentiation on OP9 stroma. Results of one representative clone are shown.
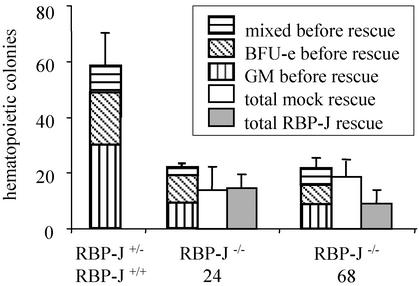
Figure 5.
Reduced but not RBP-J-rescued generation of hematopoietic colony-forming cells from RBP-J−/− endothelial–hematopoietic progenitors. ES cells differentiated into PECAM-1+ cells on OP9 stromal cells were cloned in methylcellulose in the presence of hematopoietic cytokines in triplicate. One RBP-J+/−, one RBP-J+/+, two different RBP-J−/−, and two independent mock- and RBP-J-rescued subclones (for two original RBP-J−/− clones) were analyzed in triplicate cultures. Mean values ± SEM of the colony number generated from the different genotypes after 8 days are shown as one bar. Mixed, colonies consisting of erythroid and myeloid cells; BFU-e, erythroid bursts; GM, colonies consisting of granulocytes and/or macrophages. The experiment was repeated twice with virtually identical results. One representative experiment is shown. No obvious differences in the colony types were apparent between RBP-J+/− and RBP-J−/− cultures. The decrease in hematopoietic colonies derived from RBP-J−/− ES cells is statistically significant (P < 0.001), but the numbers of hematopoietic colonies derived from mock- and RBP-J-rescued clones are not significantly different (P > 0.2).
Endothelial colonies derived from endothelial–hematopoietic (PECAM-1+) progenitor cells were similar in number in RBP-J-expressing and RBP-J-deficient clones (RBP-J−/− ES: 28 ± 5; RBP-J+/− ES: 24 ± 5; four experiments with two independent RBP-J+/− and three independent RBP-J−/− clones; P > 0.2; data not shown), suggesting that the generation of endothelial cells from PECAM-1+ progenitors is not disturbed by the absence of RBP-J. The defects in vascular development displayed by mice with targeted mutations in genes required for Notch signal transduction (40–42) therefore may result from alterations in vascular morphogenesis rather than from an influence of Notch/RBP-J signaling on the generation of endothelial cells.
Taken together, our data suggest a role for Notch/RBP-J signaling at various stages during mesodermal development, before and at the specification of mesodermal progenitors. A more precise temporal and dosage control of Notch/RBP-J activity achieved by induced deletion and/or activation of RBP-J in the relevant cell types in vivo will clarify further the involvement of RBP-J-mediated Notch signals in mesodermal development.
Supplementary Material
Acknowledgments
We thank Dr. Hitoshi Niwa for providing the pCAG-IP construct and Cornelia Kuklik-Roos, Hanna Eilken, and Heike Knetsch for expert technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (Regulatory Protein Networks Research Group to U.J. and G.W.B.) and Embryonic and Somatic Stem Cells–Regenerative Systems for Cell and Tissue Repair Priority Programme (to U.J.) and Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science, and Technology of the Japanese Government (Grants 1221 9209, 1267 0301, and 1221 5071 to M.O. and S.-I.N.). T.S. is a recipient of predoctoral fellowships from the Boehringer Ingelheim Fonds and the Deutscher Akademischer Austausch-dienst (gemeinsames Hochschulsonderprogramm III von Bund und Ländern). S.T.F. is a recipient of a postdoctoral fellowship from the Japan Society for the Promotion of Science.
Abbreviations
- RBP-J
recombination signal sequence-binding protein Jκ
- ES
embryonic stem
- NIC
notch intracellular domain
- FACS
fluorescence-activated cell sorting
- PECAM
platelet-endothelial cell adhesion molecule
References
- 1.Honjo T. Genes Cells. 1996;1:1–9. doi: 10.1046/j.1365-2443.1996.10010.x. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 3.Barolo S, Walker R G, Polyanovsky A D, Freschi G, Keil T, Posakony J W. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- 4.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 5.Oda T, Elkahloun A G, Pike B L, Okajima K, Krantz I D, Genin A, Piccoli D A, Meltzer P S, Spinner N B, Collins F S, et al. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 6.Li L, Krantz I D, Deng Y, Genin A, Banta A B, Collins C C, Qi M, Trask B J, Kuo W L, Cochran J, et al. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 7.Franco del Amo F, Smith D, Swiatek P J, Gendron-Maguire M, Greenspan R J, McMahon A P, Gridley T. Development (Cambridge, UK) 1992;115:737–744. doi: 10.1242/dev.115.3.737. [DOI] [PubMed] [Google Scholar]
- 8.Reaume A G, Conlon R A, Zirngibl R, Yamaguchi T P, Rossant J. Dev Biol. 1992;154:377–387. doi: 10.1016/0012-1606(92)90076-s. [DOI] [PubMed] [Google Scholar]
- 9.Williams R, Lendahl U, Lardelli M. Mech Dev. 1995;53:357–368. doi: 10.1016/0925-4773(95)00451-3. [DOI] [PubMed] [Google Scholar]
- 10.Myat A, Henrique D, Ish-Horowicz D, Lewis J. Dev Biol. 1996;174:233–247. doi: 10.1006/dbio.1996.0069. [DOI] [PubMed] [Google Scholar]
- 11.Westin J, Lardelli M. Dev Genes Evol. 1997;207:51–63. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- 12.Rones M S, McLaughlin K A, Raffin M, Mercola M. Development (Cambridge, UK) 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 13.Conlon R A, Reaume A G, Rossant J. Development (Cambridge, UK) 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 14.Oka C, Nakano T, Wakeham A, de la Pompa J L, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak T W, et al. Development (Cambridge, UK) 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 15.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 16.Schroeder T, Just U. EMBO J. 2000;19:2558–2568. doi: 10.1093/emboj/19.11.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osborne B, Miele L. Immunity. 1999;11:653–663. doi: 10.1016/s1074-7613(00)80140-5. [DOI] [PubMed] [Google Scholar]
- 18.Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, Kuroda K, Suzuki A, Nakano T, Honjo T. Nat Immunol. 2002;3:443–450. doi: 10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 19.Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 20.Deftos M L, Bevan M J. Curr Opin Immunol. 2000;12:166–172. doi: 10.1016/s0952-7915(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 21.Kadesch T. Exp Cell Res. 2000;260:1–8. doi: 10.1006/excr.2000.4921. [DOI] [PubMed] [Google Scholar]
- 22.Kodama H, Nose M, Niida S, Nishikawa S. Exp Hematol (Charlottesville, Va) 1994;22:979–984. [PubMed] [Google Scholar]
- 23.Nakano T, Kodama H, Honjo T. Science. 1994;265:1098–1101. doi: 10.1126/science.8066449. [DOI] [PubMed] [Google Scholar]
- 24.Nakano T, Kodama H, Honjo T. Science. 1996;272:722–724. doi: 10.1126/science.272.5262.722. [DOI] [PubMed] [Google Scholar]
- 25.Hirashima M, Kataoka H, Nishikawa S, Matsuyoshi N. Blood. 1999;93:1253–1263. [PubMed] [Google Scholar]
- 26.Kuroda K, Tani S, Tamura K, Minoguchi S, Kurooka H, Honjo T. J Biol Chem. 1999;274:7238–7244. doi: 10.1074/jbc.274.11.7238. [DOI] [PubMed] [Google Scholar]
- 27.Niwa H, Masui S, Chambers I, Smith A G, Miyazaki J. Mol Cell Biol. 2002;22:1526–1536. doi: 10.1128/mcb.22.5.1526-1536.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIvor Z J, Heyworth C M, Johnson B A, Pearson S, Fiegler H, Hampson L, Dexter T M, Cross M A. Br J Haematol. 2000;110:674–681. doi: 10.1046/j.1365-2141.2000.02214.x. [DOI] [PubMed] [Google Scholar]
- 29.Strobl L J, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm G W, Zimber-Strobl U. Immunobiology. 1997;198:299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 30.Sakai T, Furukawa T, Iwanari H, Oka C, Nakano T, Kawaichi M, Honjo T. J Biochem (Tokyo) 1995;118:621–628. doi: 10.1093/oxfordjournals.jbchem.a124955. [DOI] [PubMed] [Google Scholar]
- 31.Era T, Witte O N. Proc Natl Acad Sci USA. 2000;97:1737–1742. doi: 10.1073/pnas.97.4.1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komuro I, Izumo S. Proc Natl Acad Sci USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heikinheimo M, Scandrett J M, Wilson D B. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 34.Lints T J, Parsons L M, Hartley L, Lyons I, Harvey R P. Development (Cambridge, UK) 1993;119:415–431. doi: 10.1242/dev.119.3.969. [DOI] [PubMed] [Google Scholar]
- 35.Wilson-Rawls J, Molkentin J D, Black B L, Olson E N. Mol Cell Biol. 1999;19:2853–2862. doi: 10.1128/mcb.19.4.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Development (Cambridge, UK) 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 37.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Development (Cambridge, UK) 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 38.Joutel A, Tournier-Lasserve E. Semin Cell Dev Biol. 1998;9:619–625. doi: 10.1006/scdb.1998.0261. [DOI] [PubMed] [Google Scholar]
- 39.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald H R, Aguet M. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 40.Hrabe de Angelis M, McIntyre J, II, Gossler A. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 41.Krebs L T, Xue Y, Norton C R, Shutter J R, Maguire M, Sundberg J P, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 42.Xue Y, Gao X, Lindsell C E, Norton C R, Chang B, Hicks C, Gendron-Maguire M, Rand E B, Weinmaster G, Gridley T. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.