Abstract
The mammalian Distal-less (Dlx) clusters (Dlx1-2, Dlx5-6, and Dlx3-7) have a nested expression pattern in developing visceral (branchial) arches. Genetic regulatory mechanisms controlling Dlx spatial expression within the visceral arches have not yet been defined. Here we show that an enhancer in the Dlx3-7 cluster can regulate the visceral arch specific expression pattern of the Dlx3 gene. We have used a 79-kb transgene construct containing the entire Dlx3-7 bigene cluster with a LacZ reporter inserted in frame in the first exon of the Dlx3 gene. Visceral arch expression is absent when a 4-kb element located within the Dlx3-7 intergenic region is deleted. A 245-bp element (I37-2) whose DNA sequence is highly conserved between human and mouse located within the 4kb-deleted region can drive visceral arch expression when fused to a hsp68-lacZ reporter transgene construct. Reporter expression is detected in 9.5 and 10.5 days postcoitum transgenic embryos in a manner consistent with the endogenous Dlx3 expression pattern in the mesenchyme of the first and second visceral arches. Thus the I37-2 element is both necessary and sufficient for Dlx3 expression. The I37-2 element contains several putative binding sites for several transcription factors including Dlx and other homeodomain proteins within the evolutionarily conserved region. Significantly, the I37-2 element shows a sequence-match including a Dlx binding site to a cis-element in the Dlx5-6 intermediate region designated mI56i [Zerucha, T., Stuhmer, T., Hatch, G., Park, B. K., Long, Q., Yu, G., Gambarotta, A., Schultz, J. R., Rubenstein, J. L. & Ekker, M. (2000) J. Neurosci. 20, 709–721], despite distant phylogenetic relationship between these clusters. Our results provide evidence for a concerted role for DLX auto- and cross-regulation in the establishment of a nested expression pattern for Dlx3-7 and Dlx5-6 clusters within the visceral arches.
Keywords: cis-element‖transgenic mouse‖APKA ectodermal ridge‖ homologous recombination
The Dlx (Distal-less) genes of mammals are arranged in three convergently transcribed bigene clusters. The Dlx genes regulate pattern formation in the lower cranial and anterior pharyngeal regions derived from the visceral (branchial) arches. The paired Dlx genes (Dlx1-2, Dlx5-6, and Dlx3-7) show highly overlapping expression patterns in the first and second visceral arches. Phylogenetic relationships indicate that the mammalian Dlx genes arose initially by a lateral gene duplication to form a primordial bigene cluster. Subsequently, three bigene clusters eventuated as a result of genome duplication and selected cluster loss (1). Sequence comparisons support this view, showing that the Dlx genes fall into two internally related clades, namely Dlx 1,6,7 and 2,5,6 each having diverged from the daughter genes of the primordial cluster. Each gene pair within a bigene cluster thus has a longer evolutionary history than the bigene clusters themselves. One might expect given this history that the expression patterns of genes within a bigene cluster would have diverged to a greater degree than that shown by the bigene clusters themselves. Actually, the opposite is true: the genes within a cluster show highly similar expression patterns. These patterns suggest the existence of a cluster regulatory mechanism that constrains the diverged expression of the genes within the clusters. A plausible explanation is that the genes within clusters share common enhancers possibly located within the common 3′ intergenic noncoding region separating the coding domains of the two genes.
Dlx clusters show a nested expression pattern in the visceral arches along the visceral arches proximal–distal axis. Dlx1-2 shows an early and broad expression in both the maxillary and mandibular arches. Dlx5-6 is expressed exclusively in the distal portion of the mandibular arch. Dlx3-7 is also is expressed in the mandibular arch, but more restricted distally and laterally. It has been suggested that the proximodistal patterning of the Dlx bigene clusters plays a critical role in jaw patterning of higher vertebrates (2). A genetic mechanism responsible for nested proximodistal patterning is unknown. However, it was shown in our previous studies that an intergenic region within the Dlx3-7 intermediate domain (I37-2) is a likely site of cis-enhancer sharing, and thus could also be a site for cross-cluster regulation among the Dlx bigene clusters. Functional Dlx binding sites have also been reported in the Dlx5-6 intergenic region (mI56i) (3). The mI56i site is responsive to the Dlx2 protein and governs Dlx5-6 expression in forebrain, limbs, and visceral arches.
In this report, we show that an evolutionarily conserved element in the intergenic region of the Dlx3-7 cluster (I37-2) is a visceral arches mesenchymal enhancer. This site shows sequence similarity to the previously reported element mI56i. Both sites possess elements capable of binding particular transcription factors as for example DLX and GATA. Our data reported here support the conclusions that the I37-2 site (i) is necessary and sufficient for Dlx3-7 expression in the visceral arches, and (ii) serves to coordinate the nested expression of the Dlx bigene clusters within the visceral arches.
Materials and Methods
Transgenic Construct.
The Dlx37-lacZ-Δ(I37-2,5) construct is a deleted version of Dlx37-lacZ 79 kb (1) from 52,847 to 57,017. A targeting construct (trp213-HMA-HMB) was prepared as follows. The recombinant termini required for yeast homologous recombination were obtained by PCR. For the left arm, termed HMA, we designed two primers: IgHM1 (5′-CAAGCGGCCGCACTGGGCTGGCCTGGAACTC-3′) and IgHM2 (5′-CAAGGATCCCCCATCCTTCCCTCCCTATC-3′). These primers amplify a region from 52,482 to 52,847. The PCR product was cloned into the trp213 plasmid carrying TRP marker gene, by using BamHI and NotI sites. For the left recombinant arm, called HMB, we used two primers designated IgHM3 (5′-CAAATCGATATCAGAAGCCCAATGTAATC-3′) and IgHM4 (5′-CAACTCGAGAGGAAGCCCATAAACAATAG-3′). The PCR product was digested with ClaI and XhoI and cloned into trp213-HMA. We digested resulting trp213-HMA-HMB with NotI and XhoI to produce a linear fragment required for yeast homologous recombination that has been described (1). Resulting positive clones were screened by digestion pattern of EcoRI and further validated by Southern blotting.
Small genomic fragments (I37-2 and I-37-2,8) were recovered from the P1 clone by PCR using PCR primers including SalI and HindIII sites, and cloned into a general promoter-reporter system vector termed HSF51, containing a hsp68 heat shock protein promoter and a lacZ reporter gene. The resulting plasmid was linearized by digestion with ScaI and HindIII, respectively.
Production of Transgenic Mice.
Linearized constructs were purified by ultra centrifugation on a 10–40% sucrose gradient. Concentration of DNA dialyzed against buffer (10 mM Tris/0.1 mM EDTA) was adjusted to 4 ng/μl and injected into pronuclei of one-cell mouse embryos as described (1). Injected embryos were transferred into the oviduct of pseudopregnant CD1 mice, dissected at appropriate stages, and stained for β-galactosidase activity. Two types of transgenic mice were studied: “transient” transgenic animals sampled after injection at various developmental stages, and “stable” transgenic animals obtained from stably transformed lines derived from independent founder transformants.
Results
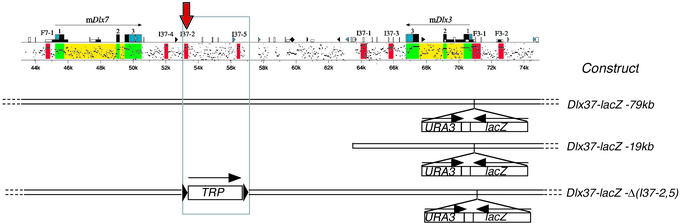
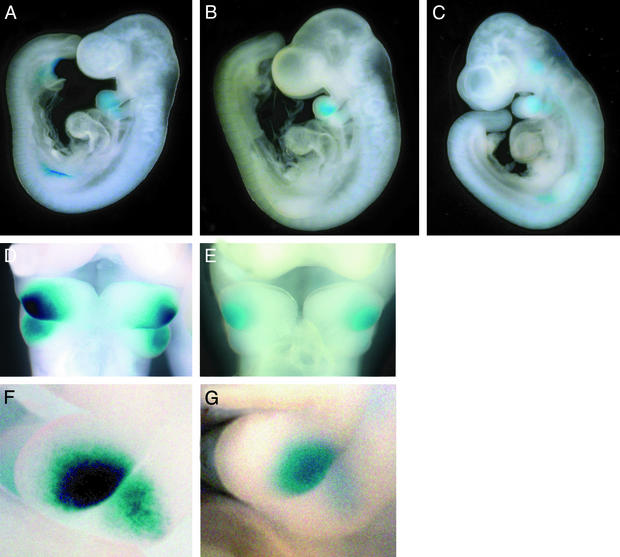
Five major conserved elements (I37-1 to -5) between human and mouse were found within the Dlx3-7 intergenic region (1). A full-length (79 kb) transgene construct drives expressions in limbs and mesenchyme of visceral arches, and a truncated transgene construct lacking the Dlx7 portion (Dlx37-lacZ-19kb, Fig. 1) show enhancer activity primarily in limbs, but not in visceral arches (1). This truncated construct retains two intergenic conserved motifs (I37-1 and -3) and lacks three motifs (I37-2, -4, and -5). We hypothesized that these conserved motifs in the intergenic region could be functional cis-elements responsible for the similar visceral arches expression patterns of the Dlx3 and Dlx7 genes. We made a targeted deletion construct lacking a 4-kb region including I37-2 and I37-5 [Dlx37-lacZ-Δ(I37-2,5), Fig. 1] to test whether those elements are necessary for enhancer activity in visceral arches. Transgenic animals with this construct show expression similar to that of Dlx37-lacZ-19kb. Limb expression was not affected by lack of the I37-2,5 region, but mesenchymal expression in visceral arches was fully abolished by this mutation (Fig. 2). This finding was highly consistent among all transgenic animals tested (Table 1). Our result suggests that a 4-kb region including I37-2 and I37-5 is indispensable for visceral arch expression. I37-2 is a prominently conserved element within this region showing high similarity (88% over 245 bp) between human and mouse (its location is shown by a red arrow in Fig. 1). We consider I37-2 as a prime candidate as a regulatory element governing mesenchymal visceral arch expression.
Figure 1.
Schematics of Dlx3-7 constructs. (Top) A percentage identity plot (PIP) that shows homology between human and mouse genomic sequences (1). A segment from Dlx3 to Dlx7 is shown. Actual sizes of the genomic constructs are 79 kb (Dlx3-lacZ-79kb), 15 kb (Dlx3-lacZ-19kb), and 75 kb [Dlx3-lacZ-Δ(I37-2,5)]. Dlx3-lacZ-79kb and Dlx3-lacZ-19kb were described (1). Dlx3-lacZ-Δ(I37-2,5) was generated from Dlx3-lacZ-79kb by replacing a 4-kb intergenic region from I37-2 to I37-5 with a TRP marker gene by yeast homologous recombination. Red boxes indicate evolutionarily conserved elements in the intergenic region. The red arrow indicates the position of the I37-2 evolutionarily conserved element. Green and yellow boxes represent exons and introns, respectively. Black triangles flanking the TRP insert represent loxP sites that were not used in this study.
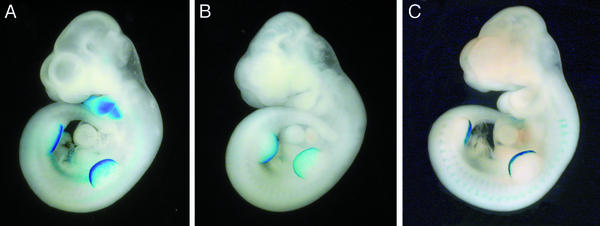
Figure 2.
Transgene expression in whole-mount mouse embryos. The developmental stage of the embryos is 10.5 dpc. (A) Full-length construct (Dlx3-lacZ-79kb) transgenic lacZ-stained embryo. First and second visceral (branchial) arches, AER of both fore and hind limbs are strongly stained. (B) Truncated Dlx3-lacZ-19-kb transgenic lacZ stained embryo. Expression in visceral arches was not detected, whereas AER expression was unaffected. (C) Targeted deletion of the I37-2,5 region [Dlx3-lacZ-Δ(I37-2,5)]; lacZ stained embryo. Deletion of I37-2,5 presents an expression pattern similar to that of Dlx3-lacZ-19kb at 10.5 dpc. Expression in the AER and lack of expression in visceral arches were highly consistent in all stable mouse lines.
Table 1.
Numbers of reporter gene expression-positive transgenic animals in visceral arches and AER
| Construct | Total expression- positive animals
|
Expression area
|
||||
|---|---|---|---|---|---|---|
| VA
|
AER
|
|||||
| T | S | T | S | T | S | |
| Dlx37-lacZ-79kb- (I37-2,5) | 1 | 3 | 0 | 0 | 1 | 3 |
| I37-2-lacZ-A | 2 | – | 2 | – | 0 | – |
| I37-2-lacZ-B | 3 | 2 | 3 | 2 | 0 | 0 |
| I37-2,8-lacZ | 7 | 1 | 6 | 1 | 0 | 0 |
Numbers of lacZ expression-positive animals are listed separately for transient embryos (T) and stable mouse lines (S). VA, visceral arch.
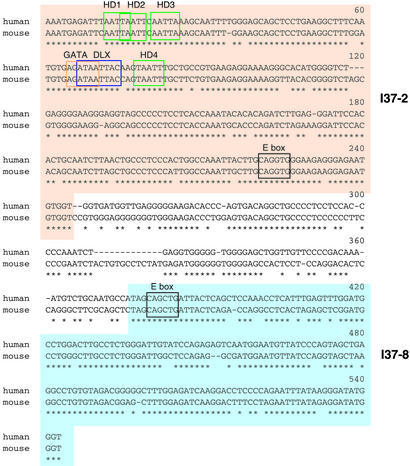
An alignment of the I37-2 site and its flanking region is shown in Fig. 3. A 245-bp I37-2 segment shows 88% identity between human and mouse. A flanking 167 bp I37-8 segment also shows high human–mouse identity (85%). There are several putative binding sites for transcription factors within the conserved segments. Several homeodomain binding core sites (TAATT) are found, including a perfectly matched site to the Dlx3 binding motif. Interestingly, this putative Dlx motif is immediately followed by another homeodomain binding motif. There is another region rich in homeodomain binding sites 5′ to the Dlx binding motif. In this area, putative binding sites for the Q50-PRD-like family (4) is found. A GATA binding site is also found in a position that overlaps the Dlx binding site. There are E-boxes in the conserved area that is putative target sites for basic helix–loop–helix factors.
Figure 3.
Evolutionarily conserved element I37-2 and the adjacent element I37-8. Conserved elements were identified by using BLAST search as described (1). Note that these elements are based on an homology score and do not necessarily signify functional units. Asterisks indicate identical nucleotides between human and mouse nucleotides. Gaps are shown by hyphens. Green boxes show putative homeodomain binding core sites (TAATT). The blue box designates a putative DLX binding site [(A/C/G)TAATT(G/A)(C/G), ref. 8]. The red box identifies a putative GATA binding site (AGATAA). Black boxes mark putative E-box (basic helix-loop-helix) binding sites.
We tested the enhancer activity of I37-2 and its flanking area by fusing it upstream of heat shock protein promoter hsp68 and a lacZ reporter in both orientations (Fig. 4). The result was highly reproducible among transgenic animals we tested (Table 1). The transgenic animals showed expression pattern in first and second visceral arches as early as 9.5 days postcoitum (dpc), similar to endogenous expression timing (Fig. 5B). At this time lacZ expression is found mostly in the distal part of the first arch, and weak expression is also found in the distal part of the second arch. Expression was restricted to the visceral arches. Expression is similar to the endogenous pattern at 10.5 dpc (Fig. 5 F and G), but in most transgenic lines signal intensity is weaker than that of the longer 79-kb construct (Fig. 5A). Only one transgenic line showed very strong reporter expression in the visceral arches with some additional expression in the head, limb, and trunk, patterns that are absent in other transgenic lines. No expression was detected in several typical Dlx3 expression sites such as apical ectodermal ridge (AER) or the ventral side of tail ectoderm.
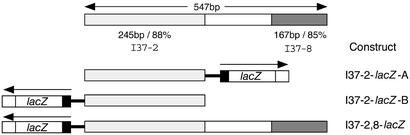
Figure 4.
Schematic of the I37-2 and the adjacent I37-8 elements. Dotted box represents the I37-2 evolutionarily conserved 245-bp element. Shaded box represents another evolutionarily conserved 167-bp element (I37-8) situated adjacent to the I37-2 element. Black box 5′ of the lacZ gene is the heat shock protein promoter hsp68, and the white box 3′ of the lacZ gene is a simian virus 40 (SV40) poly(A) signal. Together, these elements form a minimal reporter. The I37-2 element was tested by its insertion upstream of the minimal reporter in both direction (I37-2-lacZ-A and -B). An extended genomic fragment including the I37-8 element is fused to the minimal reporter system in the same direction as the I37-2-lacZ-B construct (I37-2,8-lacZ).
Figure 5.
Reporter gene expression in I37-2 and I37-2,8 transgenic mice embryos. (A–C) lacZ-stained transgenic embryos. Embryo stages are 9.5 dpc. (A) Dlx37-lacZ-79kb construct transgene embryo. (B) lacZ-stained I37-2-lacZ construct transgene embryo. Expression was first detected strongly at 9.5 dpc in the first visceral arch and less strongly in the second arch. No expression is found in the AER in both fore and hind limbs. (C) lacZ-stained I37-2,8-lacZ construct trangene embryo. I37-2,8 construct also shows expression pattern and timing similar to that of I-37-2 except for additional expression in head and trunk. (D–G) Frontal (D and E) and lateral (F and G) view of lacZ-stained 10.5 dpc embryos. (D and F) Dlx37-lacZ-79kb construct transgene embryos. (E and G) lacZ-stained I37-2-lacZ construct transgene embryos. I37-2 embryos showed expression pattern in mesenchyme of the first and second arches similar to that of Dlx37-lacZ-79kb embryos, but with reduced signal intensity.
We also tested the enhancer activity of a 547-bp fragment that included both I37-2 and the adjacent site I37-8, another small evolutionarily conserved element adjacent to I37-2 (Fig. 3). Transgene expression with this slightly longer construct (I37-2, 8) showed a similar expression pattern to that of the I37-2 construct. Expression initiated at 9.5 dpc (Fig. 5C), and is found primarily in the first and second arches. This pattern is similar to that of I37-2 (Fig. 5B), with some additional expression in the trunk and head (Fig. 5C).
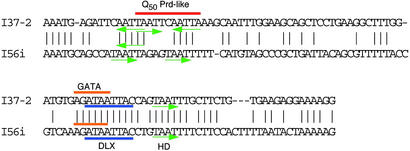
I37-2 contains several putative homeodomain binding sites and a putative DLX binding site (Fig. 3). Those sites are highly conserved between human and mouse. Interestingly, these motifs are also found in a functional regulatory element in the Dlx5-6 intergenic sites (mI56i, ref. 2; Fig. 6). The DLX binding site shows a perfect match between the two bigene clusters, and furthermore, the mI56i site has been identified previously as an active DLX binding site (3). The putative binding sites are not only similar with respect to sequence, but also in regard to their relative spatial positions.
Figure 6.
Sequence and motif comparisons between I37-2 and I56i. Identical nucleotides are indicated by a line. Putative homeodomain binding sites (TAATT) are indicated by green arrows. A putative DLX binding site and an overlapping putative GATA site are shown by blue and red underline, respectively. Note that putative binding sites are highly conserved between the Dlx3-7 element (I37-2) and the Dlx5-6 element (I56i). Spacings between putative binding sites are also highly similar.
Discussion
I37-2 Is Necessary and Sufficient to Drive Early Expression in Visceral Arches Mesenchyme.
Dlx3 is first expressed in the first and second visceral (branchial) arches at 9.5 dpc in both epithelium and mesenchyme (5). Previous studies by Morasso and coworkers showed that mesenchymal expression in the visceral arches is not supported by a 5′ segment of the Xenopus Dlx3 driving a reporter construct, whereas other endogenous expression sites such as limb, whisker follicle, and genital tubercle are supported (6). We have shown previously that a 79-kb mouse Dlx3-7 reporter construct with a LacZ reporter in frame in Dlx3 can recapitulate mesenchymal expression in the visceral arches, but when truncated in the intergenic region between Dlx3 and Dlx7, mesenchymal expression is lost (1). These experiments suggest that essential cis-element(s) necessary for mesenchymal expression in the visceral arches are located 3′ of the Dlx3 gene, possibly within the intergenic region. Further, our data presented here indicates that the evolutionarily conserved element I37-2 serves specifically to initiate visceral arch expression. In addition, when I37-2 is deleted from the 79-kb genomic construct, visceral arch expression is lost. These experiments strongly support the view that the I37-2 element is an indispensable cis-regulatory core element.
In contrast to mesenchymal expression, epithelial expression in visceral arches at an early stage (9.5–10.5 dpc) is not supported by the 79-kb transgene. It is possible that mesenchymal and epithelial expression are regulated by separate elements, and that the cis-element necessary for epithelial expression is not located within the 79-kb transgene, but is located in a distal 5′ region, because the 79-kb genomic fragment extends only 7 kb 5′ of the Dlx3 coding region. Interestingly, a similar situation was reported for the paralogous Dlx2 cluster (7). In that study, Dlx2 ectodermal expression in the visceral arch was reconstituted by using a Dlx2 3.8 kb 5′ upstream fragment, and Dlx2 mesenchymal expression was not detected (7). Thomas and coworkers suggested that at least some of the regions responsible for first arch mesenchymal expression appear to lie in the intergenic region. Their data complements our own, indicating that cis-elements responsible for epithelial and mesenchymal expression are independent and located separately, 5′ for the ectodermal and intergenic for the mesenchymal expressions.
I37-2 Element Is Primarily an Early Visceral Arch Enhancer.
A proper pattern of mesenchymal expression in the arches is supported by the 250-bp I37-2 element at both 9.5 and 10.5 dpc. However, the reporter signal in arches is almost undetectable after 12.5 dpc, whereas reporter expression is very active in mice bearing the 79-kb construct at that time. A longer fragment containing the I37-2 region plus the I37-8 conserved region (I37-28-lacZ) supports a slight increase in signal intensity but mostly it is similar to I37-2. Obviously this result suggests that other cis-elements are required to maintain visceral arch expression after 12.5 dpc. We view I37-2 as a necessary core element whose expression control may be modified by other control elements in the intergenic domain. As we will discuss below, we also regard I37-2 as a target of Dlx2 and/or Dlx5/6 signaling. Thus, the proper expression of Dlx3-7 may be initiated through I37-2 and subsequently modified by additional control motifs within the intergenic domain. The specific control relationships can be further elucidated by combinatorial mutations of the multiple control elements.
I37-2 Contains DLX and Homeodomain Binding Sites That Are Highly Conserved Between Human and Mouse.
Sequence analysis shows a striking overall similarity between human and mouse I37-2 elements. Several homeodomain-binding motifs are present within the I37-2 conserved region. One shows a perfect match to a biochemically defined Dlx3 consensus binding motif (8). Because all DLX proteins possess highly conserved homeodomains, and are thus considered to share similar binding nature, the Dlx3 binding site is potentially important as both an autoregulation and a cross-regulation site responsive to other Dlx genes. Moreover, it is known that many homeobox genes are expressed in the arches, such as Hox, Dlx, Msx, and Gsc (5). MSX proteins can potentially bind to Dlx binding sites. There is also a GATA binding site (AGATAA, refs. 9 and 10) that overlaps the Dlx motif. GATA-3 shows temporal coexpression in the arches with respect to Dlx genes (11) so that a regulatory interaction between Dlx3 and GATA-3 is a distinct possibility. Another putative binding motif is arista-less PRD-like (Q50) factors. Prx1, Prx2, Alx3, Alx4, and Cart1 are members of this class and can bind to the 5′-TAATNNNATTA-3′ motif (4). Interestingly, these arista-less related factors are expressed adjacent to the more laterally positioned DLX transcription factors in a proximodistal arrangement. It has been shown that the aristaless and Dlx factors share complementary functions within their expression domains (12). An interesting possibility is that the aristaless factors may serve to antagonize Dlx3 expression to sculp proper Dlx3-specific expression.
Paralogous Dlx Clusters Share Similar DLX Homeodomain Binding Motifs.
As described above, a functional cis-element (mI56i) has been detected in the intergenic region of the Dlx5-6 cluster that drives expression in the forebrain, visceral arches, and/or limbs (3). Because of its location in the intergenic region and similarities in expression pattern, we compared the mouse I37-2 element to that of mI56i. Significantly, it can be shown that the putative homeodomain binding sites found in I37-2 are also present in the mI56i element. The most striking result is that the Dlx binding elements are perfectly conserved. This DLX binding site has been shown to be a functional binding site in Dlx5-6, and most probably DLX2 binds to this element (3). Actually, both Dlx5-6 and Dlx3-7 are expressed within the Dlx2 expression domain, and later temporally than Dlx2 (5, 13). It is possible that both Dlx3-7 and Dlx5-6 are regulated by DLX2 through the I37-2 and mI56i elements. DLX5 and DLX6 are also candidates as binding factors to the I37-2 element, because it has been shown that Dlx3 is clearly regulated by Dlx5-6 factors (3).
Flanking bases 5′ of the DLX binding sites are also well conserved. Interestingly, this exactly corresponds to the putative GATA binding sites (AGATAA) that overlaps the DLX motif (ATAATTAC). Our data suggest that GATA factors may be modulaters of the expression of the Dlx gene family during development and possibly in later life. There is another highly conserved homeodomain binding site (TAATT) 3′ to the DLX binding site in both Dlx3-7 and Dlx5-6. The spacing between these sites is identical. Several putative homeodomain binding sites form clusters in both I37-2 and I56i elements about 50 bp 5′ to the Dlx site. Interestingly, only I37-2 has a Q50 PRD-like motif here. This finding is consistent with the difference between the Dlx3-7 and Dlx5-6 expression patterns. Dlx3-7 has a laterally restricted expression pattern, whereas Dlx5-6 has a broader pattern. Possibly, aristaless factors can repress Dlx3-7 expression in the more medial regions within the visceral arches. More experimental data will be necessary to confirm this hypothesis and other issues raised by our findings.
Acknowledgments
We thank Gemma deMartino for technical assistance. This research was supported by U.S. Department of Energy Grant DE-FG02-01ER63274, National Institutes of Health Grant GM 09966, and National Science Foundation Grant IBN-9905403 (to F.H.R.); and a Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad (to K.S.).
Abbreviations
- dpc
days postcoitum
- AER
apical ectodermal ridge
References
- 1.Sumiyama K, Irvine S Q, Stock D W, Weiss K M, Kawasaki K, Shimizu N, Shashikant C S, Miller W, Ruddle F H. Proc Natl Acad Sci USA. 2002;99:780–785. doi: 10.1073/pnas.012584999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depew M J, Lufkin T, Rubenstein J L. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- 3.Zerucha T, Stuhmer T, Hatch G, Park B K, Long Q, Yu G, Gambarotta A, Schultz J R, Rubenstein J L, Ekker M. J Neurosci. 2000;20:709–721. doi: 10.1523/JNEUROSCI.20-02-00709.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson D, Sheng G, Lecuit T, Dostatni N, Desplan C. Genes Dev. 1993;7:2120–2134. doi: 10.1101/gad.7.11.2120. [DOI] [PubMed] [Google Scholar]
- 5.Robinson G W, Mahon K A. Mech Dev. 1994;48:199–215. doi: 10.1016/0925-4773(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 6.Morasso M I, Mahon K A, Sargent T D. Proc Natl Acad Sci USA. 1995;92:3968–3972. doi: 10.1073/pnas.92.9.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas B L, Liu J K, Rubenstein J L, Sharpe P T. Development (Cambridge, UK) 2000;127:217–224. doi: 10.1242/dev.127.2.217. [DOI] [PubMed] [Google Scholar]
- 8.Feledy J A, Morasso M I, Jang S I, Sargent T D. Nucleic Acids Res. 1999;27:764–770. doi: 10.1093/nar/27.3.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merika M, Orkin S H. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko L J, Engel J D. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieuw K H, Li G, Zhou Y, Grosveld F, Engel J D. Dev Biol. 1997;188:1–16. doi: 10.1006/dbio.1997.8575. [DOI] [PubMed] [Google Scholar]
- 12.ten Berge D, Brouwer A, el Bahi S, Guenet J L, Robert B, Meijlink F. Dev Biol. 1998;199:11–25. doi: 10.1006/dbio.1998.8921. [DOI] [PubMed] [Google Scholar]
- 13.Qiu M, Bulfone A, Ghattas I, Meneses J J, Christensen L, Sharpe P T, Presley R, Pedersen R A, Rubenstein J L. Dev Biol. 1997;185:165–184. doi: 10.1006/dbio.1997.8556. [DOI] [PubMed] [Google Scholar]