Abstract
The relationship between mammalian basal metabolic rate (BMR, ml of O2 per h) and body mass (M, g) has been the subject of regular investigation for over a century. Typically, the relationship is expressed as an allometric equation of the form BMR = aMb. The scaling exponent (b) is a point of contention throughout this body of literature, within which arguments for and against geometric (b = 2/3) and quarter-power (b = 3/4) scaling are made and rebutted. Recently, interest in the topic has been revived by published explanations for quarter-power scaling based on fractal nutrient supply networks and four-dimensional biology. Here, a new analysis of the allometry of mammalian BMR that accounts for variation associated with body temperature, digestive state, and phylogeny finds no support for a metabolic scaling exponent of 3/4. Data encompassing five orders of magnitude variation in M and featuring 619 species from 19 mammalian orders show that BMR ∝ M2/3.
Pioneering work published by Max Rubner (1) in the 1880s reported that mammalian basal metabolic rate (BMR) was proportional to M2/3. In accordance with simple geometric and physical principles, it was therefore thought that an animal's rate of metabolic heat production was matched to the rate at which heat was dissipated through its body surface. However, Max Kleiber's influential monograph (2), published in 1932, concluded that basal metabolic rate scaled not in proportion with surface area, but with an exponent significantly greater than that of Rubner's surface law. Kleiber's work was later supported by Brody's (3) famous mouse-to-elephant curve, and an exponent of 3/4 (henceforth referred to as Kleiber's exponent) remains in widespread use. Quarter-power scaling is often regarded as ubiquitous in biology: metabolic rate has been reported as proportional to M3/4 in organisms ranging from simple unicells to plants and endothermic vertebrates (4, 5). Kleiber's exponent has become so widely accepted that metabolic scaling relationships that deviate from an exponent of 3/4 are often considered somehow flawed or are summarily dismissed. However, examination of the species compositions of early studies (2, 3) shows that they poorly reflect Mammalia. Most data points are derived from domestic species, which have been under artificial energetic constraints for many generations (6). Additionally, the order Artiodactyla is consistently over-represented; both Kleiber's (2) and Brody's (3) data sets include ≈20% artiodactyls, but only ≈5% of Recent mammals are artiodactyls (7). Being near the upper mass limit of the regressions, these animals exert a disproportionate influence on the scaling exponent. Their inclusion is problematic, because microbial fermentation of cellulose may delay or prohibit entrance into a postabsorptive state (8). This elevates metabolic rate above basal levels and, when coupled with a large body mass, artificially inflates the calculated scaling exponent. Examination of Brody's (3) data reveals the same problems (6). Because measurement of BMR must be obtained from inactive, postabsorptive, adult, nonreproductive, and thermoregulating animals in their inactive circadian phase and in a thermoneutral environment (8), measurements for large herbivores must be excluded from analyses of mammalian BMR, or included with caution.
The problematic inclusion of ruminants was also recognized by Kleiber (2), whose compilation included 13 data points derived from eight species (two steers, cow, man, woman, sheep, male dog, female dog, hen, pigeon, male rat, female rat, and ring dove). Kleiber addressed the problem by providing b values calculated for all 13 data points and for a subset of 9 data points with ruminants excluded. Using Kleiber's data (ref. 2; Table 1), exponents of 0.737 (r2 = 0.999) and 0.727 (r2 = 0.999) can be calculated for these groups, respectively. In this case, quarter-power scaling remained following the exclusion of ruminants, because of the influence of the four data points for male and female dogs and humans. The large b value can then be attributed to the high metabolic rate of domestic carnivores (6, 9, 10) and humans (180–200% of that predicted by the equations described below). Calculation of b from the remaining five data points yields a value of 0.667 (r2 = 0.999). The widespread use and acceptance of Kleiber's exponent can probably be attributed to a remarkably tight regression fit (r2). For Kleiber's thirteen data points, M alone explains 99.9% of the variation in BMR. To put this r2 in perspective, we randomly selected 250,000 groups of 13 species from a list of 391 species compiled by Heusner (11) (This compilation was selected because it includes data for domestic ruminants, as did Kleiber's.) Each group had a mass range of 3–4 orders of magnitude to match Kleiber's data, which spanned 3.7 orders of magnitude. Of the 250,000 least square regressions, only four had an r2 greater than 0.998 and none had an r2 greater than 0.999. The strength of Kleiber's exponent therefore seems to stem from an exceedingly fortuitous selection of data.
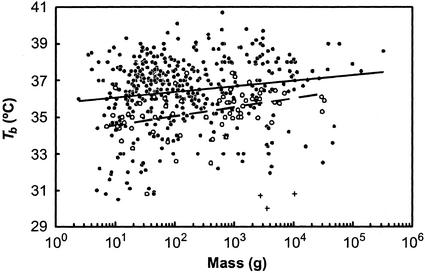
Another problem with previous analyses is that they all neglect differences in body temperature (Tb, °C) between species. This is important because Tb and M are primary determinants of metabolic rate (5) and Tb is significantly correlated with M for marsupials (ref. 12; Tb = 34.1 + 0.49 log M, n = 66; Fig. 1), eutherians (ANOVA F1,436 = 21.5, P = 0.01, Tb = 35.8 + 0.30 log M, n = 437; Fig. 1), and mammals in general (ANOVA F1,507 = 37.0, P < 0.001, Tb = 35.8 + 0.21 log M, n = 507). An accurate estimation of the relationship between BMR and M is therefore best obtained by normalizing the measured BMRs of all species to a common Tb.
Figure 1.
Relationship between body mass (M, g) and body temperature (Tb, °C) for eutherians (● and unbroken line; Tb = 35.8 + 0.30 log M, n = 437), marsupials (○ and broken line; Tb = 34.1 + 0.49 log M, n = 67), and monotremes (+, n = 4).
In the 70 years since Kleiber's monograph, a wealth of BMR and Tb data has accumulated. This report draws on the most comprehensive and representative database available, to analyze the relationship between BMR and body size. Although BMR is an artificial physiological construct that animals rarely show under natural conditions, it remains an established benchmark for comparing metabolic intensity between species. More importantly, if theoretical analyses are ever to explain the nonlinear relationship between metabolic rate and body size, it is essential to establish what that relationship actually is, without the confounding influences of Tb and digestive state.
Methods
Data for 619 species were compiled from the literature (see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Wherever possible, M, Tb, and BMR were sourced from the same paper. Where multiple values were available for a species, the arithmetic mean was calculated. BMR and Tb values were accepted only if the animals were resting, normothermic, postabsorptive, inactive, and conscious. Data that did not fulfill these criteria were disregarded. Adult body mass was obtained from multiple published sources when body mass was not provided in papers from which measurements were taken. The data were disregarded if no body mass could be found in the established literature. To allow for the overestimation of degrees of freedom problem inherent in comparative analyses of species data, a nested ANOVA was used to determine the appropriate taxonomic level at which averages should be calculated (13). Nested ANOVA showed that this order value captures 85% of the variation in M and 86% of the variation in BMR, and was therefore the appropriate level for analysis (9). Data were log transformed, and genera values were calculated as the average of species within genera, family values were calculated as averages of genera within families, and order values were calculated as the average of families within orders.
Least-square linear regressions of the form log(BMR) = log(a) + b log(M) were fitted to log–log transformed data. This enabled calculation of an allometric equation of the form BMR = aMb. When Tb was available for a species (n = 507), BMR data were normalized to a Tb equal to the mean Tb of all species (36.2°C). Order values were transformed using the average Tb of species within the order, using the same nested average calculation as used for BMR and M. Traditionally, correction for temperature differences is undertaken using Q10 principles (5, 14) such that
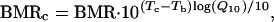 |
where BMRc is temperature-corrected BMR, Tc is the temperature to which all observations are corrected (36.2°C), and Q10 is the factorial increase in BMR associated with a temperature increase of 10°C. To select the appropriate Q10 for temperature correction in this analysis, results obtained with a series of values between 2 and 4 were compared. A Q10 of 3.0 was used because this value produced the highest r2 when log BMRc was regressed against log M, and therefore minimized the variation in BMRc. As an alternative to Q10 principles, Gillooly et al. (5) proposed a correction factor based on the universal temperature dependence (UTD) of biological processes, suggesting that correction using a single temperature-independent Q10 value could introduce an error as great as 15% over the range of biologically relevant temperatures (≈0–40°C). UTD correction considers metabolic rate to be the sum of many biological reactions, where each reaction rate is proportional to the product of the concentration of reactants, the fluxes of reactants and the kinetic energy of the system. Although the potential error introduced by Q10 correction is likely to be considerably less than 15% within the modest Tb range in the present analysis (≈30–40°C), both Q10 and UTD correction methods were used. Only the results obtained with Q10 correction are presented, however, because UTD correction accounted for marginally less of the residual variation and did not alter the conclusions. No attempt was made to distinguish between BMR values obtained in the active (α) or resting (ρ) phase of the day. This did not compromise the study, because the ≈33% elevation in BMR observed in the α phase (15) can be wholly accounted for with a Q10 of 3.0 and only a 2.4°C difference between α and ρ Tb, which is within the range of observed mammalian daily Tb variation (16). Assuming that BMR and Tb were measured in the same circadian phase, correction to a common Tb therefore accounts for circadian fluctuations in BMR.
A conservative approach was then adopted where lineages for which the conditions required for BMR measurement were suspected to be difficult or impossible to achieve were excluded. The lineages excluded were Artiodactyla, Macropodidae (Diprotodontia), Lagomorpha, and Soricidae (Insectivora). Exclusion of artiodactyls was considered necessary because the length of time for which they were fasted (2–3 days) was probably insufficient to produce a postabsorptive state [which requires 2–7 days to achieve in domestic ruminants (17), but may be in fact unachievable (8)]. Similarly, macropod marsupials are large herbivores with a complex voluminous stomach that is a major site for microbial fermentation (18). Lagomorphs were excluded because their hindgut is a major site for microbial fermentation (18) and they have high metabolic rates relative to other eutherians (6), possibly associated with microbial fermentation. Shrews (Soricidae) were excluded because they may become hyperactive when postabsorptive, hence postabsorptive and inactive conditions are mutually exclusive (19). Although some other lineages (e.g., Cetacea and Proboscidae) are not present in the data set, their absence stems solely from a lack of measurements that satisfy the basic requirements for BMR.
Results
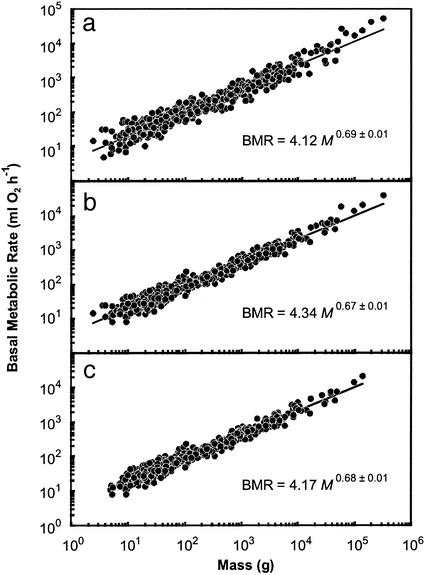
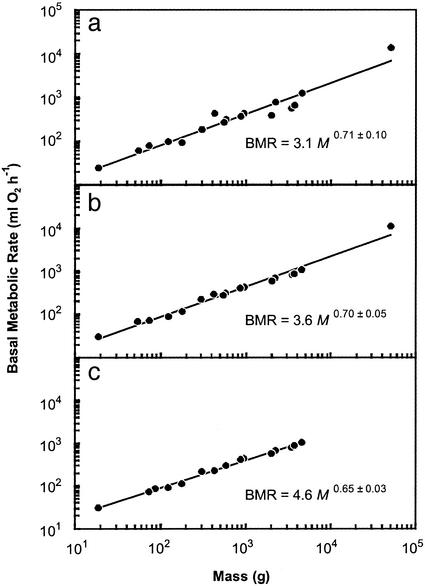
Both interspecific and interordinal analyses were made. For the 619 species for which BMR data have been published (Table 1), M alone accounted for 94% of the interspecific variation in BMR, but the 95% confidence intervals of the allometric exponent (0.69) do not include 3/4 or 2/3 (Fig. 2a). However, this finding may by misleading, because species values do not represent statistically independent data on which to base a comparison (13). This leads to overestimation of degrees of freedom, which artificially narrows confidence intervals and can result in the false rejection of null hypotheses. The use of an average value calculated for some higher taxonomic level reduces degrees of freedom and addresses the nonindependence problem inherent in nonphylogenetically informed analyses (13). As has been previously demonstrated (9), the order level was identified as that which captures a large proportion of the variation in M and BMR, but does not unnecessarily reduce sample size (see Methods). For the 17 mammalian orders represented by at least three species, M also accounts for 94% of the variation in BMR, but the allometric exponent is not significantly different from 3/4 or 2/3 (Fig. 3a). Additionally, the variation not accounted for by M (the BMR residuals) is significantly positively correlated with Tb for both the interspecific (BMR residual = 0.05 Tb − 1.8; n = 507, r2 = 0.32, P < 0.001) and interordinal (BMR residual = 0.07 Tb − 2.4; n = 17, r2 = 0.76, P < 0.001) analyses. When BMR values are normalized to a Tb of 36.2°C by using Q10 principles, both the interspecific and interordinal allometric exponents decreased and neither was found to be significantly different from 2/3, whereas only the interspecific exponent was significantly different from 3/4 (Figs. 2b and 3b). Finally, exclusion of Artiodactyla, Macropodidae (Diprotodontia), Lagomorpha, and Soricidae (Insectivora) further refined the predictions such that M and Tb accounted for 96% of the interspecific variation in BMR and 99% of the interordinal variation in BMR (Figs. 2c and 3c). Both interspecific (0.68) and interordinal (0.65) allometric exponents were significantly different from 3/4 and were not significantly different from 2/3 (Figs. 2c and 3c).
Figure 2.
Relationship between mammalian body mass (M, g) and basal metabolic rate (BMR, ml of O2 per h) for all data (a; n = 619, r2 = 0.94), data corrected to a common body temperature (36.2°C) by using a Q10 of 3.0 (b; n = 507, r2 = 0.96), and data corrected to 36.2°C for all species excluding Artiodactyla, Lagomorpha, Soricidae (Insectivora), and Macropodidae (Diprotodontia) (c; n = 469, r2 = 0.96). Exponents are shown with 95% confidence intervals.
Figure 3.
Relationship between body mass (M, g) and basal metabolic rate (BMR, ml of O2 per h) for mammalian orders (see Methods for details) for all data (a; n = 17, r2 = 0.94), data corrected to a common body temperature (36.2°C) by using a Q10 of 3.0 (b; n = 17, r2 = 0.98), and data corrected to 36.2°C for all species excluding Artiodactyla, Lagomorpha, Soricidae (Insectivora), and Macropodidae (Diprotodontia) (c; n = 15, r2 = 0.99). Exponents are shown with 95% confidence intervals.
Discussion
This study finds that the BMR of mammals is proportional to M2/3, as is the case for birds (20–23). The relationships presented here fail to account for only 4% of the interspecific and 1% of the interordinal variation in mammalian BMR. Many factors have been suggested as proximal causes for the residual differences in mammalian BMR, and investigation of these factors is likely to continue to be a fruitful area of investigation in the future. Factors that have been implicated so far include phylogeny (6, 9), diet (10), geography (24), aridity (24), habitat productivity (24, 25), and relative organ masses (26). In many cases, separation of these influences is difficult, particularly when they are correlated or confounded (27).
In addition to the statistical analyses presented here, the validity of a BMR scaling exponent of 2/3 can be investigated by using this relationship to predict allometric exponents for complimentary variables (e.g., home range) that can reasonably be thought to be related to BMR. Such comparisons have previously been approached from the invalid assumption that BMR is proportional to M3/4. For example, a recent analysis of home range scaling (28) used a BMR exponent of 0.75 and predicted home range scaling exponents of 0.83, 1.33, and 1.5 for terrestrial mammalian herbivores, terrestrial mammalian carnivores, and terrestrial avian carnivores, respectively (ref. 28; Fig. 3). These predictions differed from the observed exponents (0.83, 1.21, and 1.37) by an average of 0.09. Recalculation of the predicted home range scaling exponents by using a BMR scaling exponent of 0.67 yields predictions of 0.75, 1.25, and 1.42, which differ from the observed exponent by only 0.002. This strengthens the case for a 2/3 exponent by linking BMR with home range size, a variable that integrates behavior, physiology, and population density (28).
The finding that BMR is proportional to M2/3 challenges a 70-year-old paradigm and suggests that a common cause underlies the influence of M on BMR for endothermic homeotherms. An exponent of 2/3 questions recent explanations for quarter power scaling (29–32), and indicates that other explanations need to be sought. Because the present analysis is concerned only with a description of the allometric relationship between BMR and M, any speculation regarding what factors might account for it has been avoided.
Supplementary Material
Acknowledgments
We thank Russell Baudinette for reviewing a draft version of this manuscript.
Abbreviations
- BMR
basal metabolic rate
- UTD
universal temperature dependence
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rubner M. Zeitschrift fur Biologie. 1883;19:536–562. [Google Scholar]
- 2.Kleiber M. Hilgardia. 1932;6:315–353. [Google Scholar]
- 3.Brody S. Bioenergetics and Growth. New York: Reinhold; 1945. [Google Scholar]
- 4.Hemmingsen A M. Rep Steno Mem Hosp Nord Insulinlab. 1960;9:1–110. [Google Scholar]
- 5.Gillooly J F, Brown J H, West G B, Savage V M, Charnov E L. Science. 2001;293:2248–2251. doi: 10.1126/science.1061967. [DOI] [PubMed] [Google Scholar]
- 6.Hayssen V, Lacy R C. Comp Biochem Physiol A Physiol. 1985;81:741–754. doi: 10.1016/0300-9629(85)90904-1. [DOI] [PubMed] [Google Scholar]
- 7.Nowak R M. Walker's Mammals of the World. Baltimore: Johns Hopkins Univ. Press; 1999. [Google Scholar]
- 8.McNab B K. Physiol Zool. 1997;70:718–720. doi: 10.1086/515881. [DOI] [PubMed] [Google Scholar]
- 9.Elgar M A, Harvey P H. Funct Ecol. 1987;1:25–36. [Google Scholar]
- 10.McNab B K. Quart Rev Biol. 1988;63:25–54. doi: 10.1086/415715. [DOI] [PubMed] [Google Scholar]
- 11.Heusner A A. J Exp Biol. 1991;160:25–54. doi: 10.1242/jeb.160.1.25. [DOI] [PubMed] [Google Scholar]
- 12.Withers P C, Thompson G G, Seymour R S. Aust J Zool. 2000;48:241–258. [Google Scholar]
- 13.Harvey P H, Pagel M D. The Comparative Method in Evolutionary Biology. New York: Oxford Univ. Press; 1991. [Google Scholar]
- 14.Guppy M, Withers P C. Biol Rev. 1999;74:1–40. doi: 10.1017/s0006323198005258. [DOI] [PubMed] [Google Scholar]
- 15.Kenagy G J, Vleck D. In: Vertebrate Circadian Systems: Structure and Physiology. Aschoff J, Daan S, Groos G A, editors. Berlin: Springer; 1982. pp. 322–338. [Google Scholar]
- 16.Aschoff J. In: A Companion to Animal Physiology. Taylor C R, Johansen K, Bolis L, editors. Cambridge, U.K.: Cambridge Univ. Press; 1982. pp. 173–186. [Google Scholar]
- 17.Blaxter K L. The Energy Metabolism of Ruminants. London: Hutchinson; 1962. [Google Scholar]
- 18.Stevens C E, Hume I D. Comparative Physiology of the Vertebrate Digestive System. Cambridge, U.K.: Cambridge Univ. Press; 1995. [Google Scholar]
- 19.Speakman J R, McDevitt R M, Cole K R. Physiol Zool. 1993;66:1045–1049. [Google Scholar]
- 20.Bennett P M, Harvey P H. J Zool. 1987;213:327–363. [Google Scholar]
- 21.Reynolds P S, Lee R M., III Am Nat. 1996;147:735–759. [Google Scholar]
- 22.Tieleman B I, Williams J B. Physiol Biochem Zool. 2000;73:461–479. doi: 10.1086/317740. [DOI] [PubMed] [Google Scholar]
- 23.Frappell P B, Hinds D S, Boggs D F. Physiol Biochem Zool. 2001;74:75–89. doi: 10.1086/319300. [DOI] [PubMed] [Google Scholar]
- 24.Lovegrove B G. Am Nat. 2000;156:201–219. doi: 10.1086/303383. [DOI] [PubMed] [Google Scholar]
- 25.Mueller P, Diamond J. Proc Natl Acad Sci USA. 2001;98:12551–12554. [Google Scholar]
- 26.Konarzewski M, Diamond J. Evolution (Lawrence, Kans) 1995;49:1239–1248. doi: 10.1111/j.1558-5646.1995.tb04450.x. [DOI] [PubMed] [Google Scholar]
- 27.Cruz-Neto A P, Garland T, Jr, Abe A S. Zoology. 2001;104:49–58. doi: 10.1078/0944-2006-00006. [DOI] [PubMed] [Google Scholar]
- 28.Haskell J P, Ritchie M E, Olff H. Nature. 2002;418:527–530. doi: 10.1038/nature00840. [DOI] [PubMed] [Google Scholar]
- 29.West G B, Brown J H, Enquist B J. Science. 1997;276:122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 30.West G B, Brown J H, Enquist B J. Science. 1999;284:1677–1679. doi: 10.1126/science.284.5420.1677. [DOI] [PubMed] [Google Scholar]
- 31.Banavar J R, Damuth J, Maritan A, Rinaldo A. Proc Natl Acad Sci USA. 2002;99:10506–10509. doi: 10.1073/pnas.162216899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Darveau C A, Suarez R K, Andrews R D, Hochachka P W. Nature. 2002;417:166–170. doi: 10.1038/417166a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.