Abstract
Annual and perennial habit are two major strategies by which grasses adapt to seasonal environmental change, and these distinguish cultivated cereals from their wild relatives. Rhizomatousness, a key trait contributing to perenniality, was investigated by using an F2 population from a cross between cultivated rice (Oryza sativa) and its wild relative, Oryza longistaminata. Molecular mapping based on a complete simple sequence-repeat map revealed two dominant-complementary genes controlling rhizomatousness. Rhz3 was mapped to the interval between markers OSR16 [1.3 centimorgans (cM)] and OSR13 (8.1 cM) on rice chromosome 4 and Rhz2 located between RM119 (2.2 cM) and RM273 (7.4 cM) on chromosome 3. Comparative mapping indicated that each gene closely corresponds to major quantitative trait loci (QTLs) controlling rhizomatousness in Sorghum propinquum, a wild relative of cultivated sorghum. Correspondence of these genes in rice and sorghum, which diverged from a common ancestor ≈50 million years ago, suggests that the two genes may be key regulators of rhizome development in many Poaceae. Many additional QTLs affecting abundance of rhizomes in O. longistaminata were identified, most of which also corresponded to the locations of S. propinquum QTLs. Convergent evolution of independent mutations at, in some cases, corresponding genes may have been responsible for the evolution of annual cereals from perennial wild grasses. DNA markers closely linked to Rhz2 and Rhz3 will facilitate cloning of the genes, which may contribute significantly to our understanding of grass evolution, advance opportunities to develop perennial cereals, and offer insights into environmentally benign weed-control strategies.
The shift from perennial to annual habit was of great importance for the evolution of cereal crops from their wild progenitors (1). Perenniality in plants may result from several vegetative organs such as root pieces, rhizomes, stolons, or tubers. Among these traits, rhizomes are one of the key features that distinguish cultivated cereals from their wild relatives and also permit many grass species to survive in harsh environments. Rhizomes are a key means of propagation and persistence of many important weeds (2), but they may also be a useful attribute under some circumstances. For instance, strong rhizomes are certainly a desirable trait for many species of turf grasses. In many mountainous areas of Asia that depend on upland rice for subsistence, cultivation has resulted in serious soil erosion and damage to the fragile ecosystems in these areas. Perennial crop-production systems (3, 4) may provide an environmentally sound and economically viable alternative for use in such areas; however, perennial forms of high-yielding rice cultivars do not exist.
Among the 24 species comprising the genus Oryza are four perennials, Oryza longistaminata, Oryza officinalis, Oryza australiensis, and Oryza rhizomatis. O. longistaminata is the only rhizomatous species with the same AA genome type (2× = 24) as cultivated rice, Oryza sativa. Distributed widely throughout tropical Africa, O. longistaminata is an out-crossing species characterized by long anthers, self-incompatibility, allogamy, and strong rhizomes (5). It has been valuable as a source of resistance to biotic and abiotic stresses and other important traits in rice improvement (6–8). Because of the high level of molecular polymorphism between O. longistaminata and O. sativa, it was also used to construct the first high-density rice restriction fragment length polymorphism map (9).
Rice is widely recognized as an important model for other grasses, many of which have much larger and more complex genomes (10). Characterization of genes controlling rhizome formation and traits related to perenniality that distinguish O. longistaminata and O. sativa offers important insights into the evolution of cereal crops and provides a foundation for utilization of the rice sequence to study the molecular basis of perenniality and to identify potential targets for efficient weed control.
Materials and Methods
Genetic Materials and Rhizome Evaluation.
We mapped an F2 population derived from a cross between O. sativa cultivar RD23 (an indica cultivar from Thailand) and an unnamed O. longistaminata accession with long and strong rhizomes, originally collected from Niger and kindly provided by Hiroshi Hyakutake (Institute of Physical and Chemical Research, Saitama, Japan). The F1 plant of the RD23/O. longistaminata cross was obtained by direct hybridization followed by embryo rescue and had 32.5% pollen fertility, indehiscent anthers, rhizomes that were intermediate in size, and abundance between the parents. The F1 plant was grown in Hainan, China, and produced 381 F2 seeds by self-pollination. F2 seeds were germinated on Murashige and Skoog medium (3% sucrose plus 0.7% agar at pH 5.8) from which 268 seedlings were obtained and transplanted to an irrigated field. A subset of 228 randomly selected F2 plants, together with parents and F1, were multiplied by cuttings and planted in the field in a randomized complete block design with three replications on September 15, 2000. At the time of flowering, all plants were dug up, and the roots were washed free of soil, evaluated for the presence or absence of rhizomes, and measured for nine traits related to rhizome abundance/growth. These traits included tiller number (TN) per plant, rhizome number per plant, degree of primary branching, number of secondary branches, total length (in centimeters), internode length (in centimeters), internode number, and dry weight (in grams) of up to five main rhizomes per plant.
Genotyping and Data Analyses.
A total of 181 well distributed simple sequence-repeat markers developed by Cornell University (New York) and the International Rice Microsatellite Initiative (Ithaca, NY) were used to genotype the parents, F1 plant, and F2 population following standard protocols (11, 12). The computer program MAPMAKER/EXP 3.0b (13) was used to construct the linkage map of the F2 population. SAS PROC FREQ (14) was used to test associations of rhizome phenotypes with markers and to test the fit of predicted genetic models with the observed data. Composite interval mapping (15) was used to identify and map quantitative trait loci (QTLs) by using the threshold of logarithm of odds = 2.50. Comparative mapping to identify corresponding genomic regions across the rice and sorghum genomes were conducted by using common anchor markers described by Ware et al. (16) and Paterson et al. (17).
Results
Construction of the Linkage Map and Genomic Composition of the F2 Population.
The linkage map (Fig. 1) was comprised of 181 codominant simple sequence-repeat markers spanning 1,758.6 centimorgans (cM) and covering all 12 chromosomes at an average distance of 9.7 ± 6.5 cM between adjacent markers. Marker orders and locations were nearly identical to the previously published arrangement of these simple sequence repeats (12) except for five small inversions on chromosomes 3 (between OSR16 and OSR18), 5 (between RM159 and RM122and between RM291 and RM161), and 7 (between RM47 and RM18 and between RM118 and RM134). Segregation distortion of allelic frequencies from the expected 1:1 ratio was observed at 73 markers (40%) in 18 genomic regions, including 40 markers on chromosomes 1–6, 9, and 11 favoring the O. sativa allele and 33 on chromosomes 1, 3, 8, 9, and 12 favoring the O. longistaminata allele, respectively (Fig. 1). Overall, O. longistaminata alleles accounted for ≈45.5% of the genome in the F2 population.
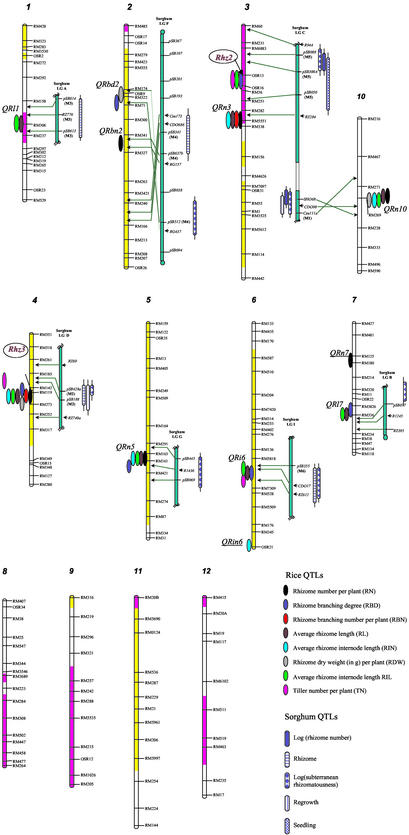
Figure 1.
Comparative mapping of genes/QTLs affecting rhizome expression and abundance in O. longistaminata and S. halepense. DNA markers in normal Italian were used in gene/QTL mapping; those indicated by arrows are locations inferred relative to closely linked restriction fragment length polymorphism markers from rice (9) and sorghum (2) by using the comparative mapping search tool from the Gramene database (16), where M1, M2, and M5 represent maize chromosomes 1, 2, and 5, respectively. The yellow portions of the chromosomes show the genomic regions of distorted segregation favoring the RD23 (O. sativa) allele, and the pink portions show the genomic regions of distorted segregation favoring the O. longistaminata allele. The two underlined rice QTLs were not matched with corresponding sorghum QTLs.
Identification and Mapping of a Pair of Dominant-Complementary Genes for Rhizome Formation in O. longistaminata.
Of the 228 F2 individuals evaluated, 122 had rhizomes and 106 did not. This fits a 9:7 ratio (χ2 = 0.70 and P = 0.40) and excludes other candidate ratios (for a 3:1 ratio, χ2 = 56.17 and P = 0), suggesting that two dominant-complementary genes may largely control rhizomatousness in O. longistaminata. Two genomic regions were strongly associated with the rhizome phenotype, one near OSR16 on chromosome 3 (χ2 = 40.3 and P < 0.0001) and the other near RM119 on chromosome 4 (χ2 = 64.4 and P < 0.0001). Both regions showed significant segregation distortion with the O. longistaminata allele favored at OSR16 on chromosome 3 and the O. sativa allele favored at RM119 on chromosome 4. To test whether the genes at the two regions fit the model of two dominant-complementary genes that we inferred based on the segregation data, the predicted rhizome phenotypes of individual F2 plants were generated based on the marker genotypes at OSR16 and RM119 and subjected to an association analysis. The χ2 value from the two-gene model was 113.2 (P < 0.0001), much greater than that obtained from either single-marker model. The goodness-of-fit test also indicated that the predicted rhizome phenotypes of the F2 individuals based on the genotypes at the two markers were consistent with the observed rhizome expression (χ2 = 1.29 and P = 0.26). However, there were 35 individuals that did not show the predicted rhizome phenotypes based on their genotypes at the two loci. These were all heterozygous at both loci and had a greater than average proportion of O. sativa chromatin across the remainder of the genome, particularly at QTLs affecting rhizome abundance discussed below, suggesting that the dominance of the rhizome genes may be modified by interactions with other genes that are beyond the resolution of our data.
To determine the exact positions of the two genes, we examined those F2 individuals having crossovers between the markers flanking OSR16 and RM119 and the changes (presence or absence) in the rhizome phenotype resulting from the crossovers. For OSR16, we found that crossovers between OSR13 and OSR16 resulted in changes in rhizome phenotype but not between OSR16 and RM36. In this way, the first locus was mapped to chromosome 3, 1.3 cM from OSR16 and 8.1 cM from OSR13. Similarly, changes in rhizome phenotype resulted from crossovers between RM119 and RM273 but not between RM142 and RM119. Thus, the second locus was mapped to chromosome 4, 2.2 cM from RM119 and 7.4 cM from RM273 (Fig. 1). We tentatively designated this pair of interacting genes Rhz2 and Rhz3.
QTLs Affecting Abundance of Rhizomes and Related Growth Traits.
The F2 individuals having rhizomes showed considerable variation for traits related to rhizome abundance and growth. Composite interval mapping revealed 16 QTLs on 8 of the 12 rice chromosomes that affected the nine rhizome traits. The O. longistaminata alleles at all of these QTLs increased the values of the rhizome-related traits (Fig. 1 and Table 1). As expected, the O. longistaminata allele at Rhz3 increased values of all nine measured rhizome traits. However, the O. longistaminata allele at Rhz2 was associated only with increased degree of primary branching, internode length, and TN. Six QTLs appeared to be more important, and each affected more than one measured rhizome trait (Table 1). Of these, QRn3, QRn5, and QRn10 appeared to control the number and branching of rhizomes, and QRl1, QRl6, and QRl7 influenced primarily the length of rhizomes. Eight additional QTLs had relatively small effects, and each affected only one of the rhizome traits (Table 1).
Table 1.
Genes/QTLs affecting rhizome traits in the O. sativa (RD23)/O. longistaminata F2 population
| Loci | Ch. | Marker interval | Trait | LOD | A | PA | D | D/A |
|---|---|---|---|---|---|---|---|---|
| QRl1 | 1 | RM306–RM237 | RL | 3.10 | −1.17 | 0.0024 | 1.28 | −1.09 |
| RIL | 3.40 | −0.16 | 0.0013 | 0.17 | −1.06 | |||
| Rhz2 | 3 | OSR13–OSR16 | RBD | 12.71 | −0.83 | 0 | — | — |
| RIL | 10.91 | −0.53 | 0 | — | — | |||
| TN | 6.64 | −21.7 | 0 | — | — | |||
| QRn3 | 3 | RM282–RM5551 | RN | 7.11 | −1.58 | 0 | — | — |
| RBN | 8.51 | −0.51 | 0 | — | — | |||
| RL | 7.78 | −3.15 | 0 | — | — | |||
| RIN | 12.38 | −1.01 | 0 | — | — | |||
| Rhz3 | 4 | RM119–RM273 | RN | 3.85 | −0.60 | 0.0975 | 1.28 | −2.13 |
| RBD | 14.39 | −0.89 | 0 | — | — | |||
| RBN | 8.11 | −0.41 | 0.0001 | 0.34 | −0.83 | |||
| RL | 13.12 | −1.93 | 0 | 2.31 | −1.20 | |||
| RIL | 15.58 | −0.31 | 0 | 0.37 | −1.19 | |||
| RIN | 7.46 | −0.66 | 0.0002 | 0.58 | −0.88 | |||
| TN | 8.82 | −23.5 | 0 | 11.8 | −0.50 | |||
| SDW | 13.33 | −31.0 | 0 | 18.7 | −0.60 | |||
| RDW | 2.31 | −0.25 | 0.4181 | 1.15 | −4.6 | |||
| QRn5 | 5 | RM161–RM274 | RN | 2.06 | −1.03 | 0.0022 | — | — |
| RL | 2.85 | −1.22 | 0.003 | — | — | |||
| RIL | 3.38 | −0.24 | 0.0001 | — | — | |||
| RIN | 3.64 | −0.71 | 0.0001 | — | — | |||
| RBD | 3.20 | −0.33 | 0.0017 | −0.25 | 0.76 | |||
| QRl6 | 6 | RM30–RM7309 | RBD | 2.92 | −0.48 | 0.0013 | — | — |
| RL | 2.86 | −1.47 | 0.0004 | — | — | |||
| RIL | 3.15 | −0.25 | 0.0002 | — | — | |||
| TN | 4.13 | −17.3 | 0.0001 | — | — | |||
| QRl7 | 7 | RM336–RM234 | RBD | 2.77 | −0.31 | 0.0004 | — | — |
| RL | 3.51 | −1.46 | 0.0002 | — | — | |||
| RIL | 3.74 | −0.23 | 0.0001 | — | — | |||
| QRn10 | 10 | RM271–RM269 | RN | 3.74 | −1.20 | 0.0001 | −0.65 | 0.54 |
| RL | 4.34 | −1.59 | 0 | −0.80 | 0.50 | |||
| RIL | 5.22 | −0.24 | 0 | −0.14 | 0.58 | |||
| RIN | 3.81 | −0.61 | 0 | — | — | |||
| RDW | 2.12 | −0.92 | 0.0035 | — | — | |||
| QRbd2 | 2 | RM71–RM300 | RBD | 4.21 | −0.39 | 0 | — | — |
| RDW | 3.66 | −1.30 | 0.0001 | −0.75 | 0.58 | |||
| QRn6 | 6 | RM345–OSR21 | RN | 2.50 | −1.11 | 0.0018 | — | — |
| RIN | 5.51 | −0.86 | 0 | −0.61 | 0.71 | |||
| QRn2 | 2 | RM341–RM327 | RN | 4.09 | −1.55 | 0 | −1.00 | 0.65 |
| QRn7 | 7 | RM125–RM180 | RN | 3.46 | −1.03 | 0.0006 | 0.75 | −0.73 |
Ch., chromosome; RN, rhizome number per plant; RBD, rhizome branching degree; RBN, rhizome branching number per plant; RL, average rhizome length (in cm); RIL, average rhizome internode length (in cm); RIN, average rhizome internode number; RDW, rhizome dry weight (in g) per plant; TN, tiller number per plant; A and D, QTL additive and dominance effects, respectively; PA, probability associated with QTL additive effects. The presented dominance effects are all significant at P < 0.05, and − represents insignificant effects.
Comparative Mapping.
Using the comparative genome-mapping tool and the Gramene database (12, 16) together with our published data (2, 17), we were able to evaluate the degree of correspondence between O. longistaminata rhizome genes and QTLs to those reported from Sorghum propinquum (Fig. 1). For instance, Rhz3 appeared to correspond closely to a major QTL flanked by csu40 and pSB188 on sorghum linkage-group D, which affects subterranean rhizomatousness, regrowth, and TN in S. propinquum. Rhz2, QRn3, and QRn10 corresponded closely to three linked QTLs affecting subterranean rhizomatousness, rhizome number and length, and TN on sorghum linkage-group C. QRl1 and QRl7 mapped to the corresponding regions harboring two QTLs influencing regrowth and subterranean rhizomatousness on sorghum linkage-groups A and B. QRbd2 and QRbn2 mapped to regions harboring two linked QTLs affecting regrowth and subterranean rhizomatousness on sorghum linkage-group F; QRi6 corresponded to a QTL associated with regrowth and subterranean rhizomatousness on sorghum linkage-group I; and QRn5 corresponded to a QTL associated with subterranean rhizomatousness on sorghum linkage-group G (2). We did not find sufficient evidence to suggest the corresponding locations of QRn7 and QRin6 on sorghum linkage groups, nor did we know clearly the rice genomic locations corresponding to sorghum linkage-groups H and J, at which two additional rhizome QTLs locate (2). To evaluate the likelihood of this degree of correspondence, specifically of 10 of 16 rice QTLs (62.5%) to 9 of 11 sorghum QTLs (81.8%), we used published methods (17) based on somewhat conservative assumptions that we could identify correspondence for ≈90% (1,700 cM) of the present rice map to the sorghum map and that the size of interval that we could confidently assign QTLs to was ≈30 cM. The likelihood of the observed number of matches occurring by chance was 0.00004, indicating that QTLs controlling rhizomatousness in O. longistaminata and S. propinquum fall in largely corresponding genomic locations.
Discussion
Our results have several important implications for understanding the genetics and evolution of perenniality in grasses. Annual habit represents a widespread strategy by which grasses adapt to seasonal environmental changes (temperature, water, solar intensity, and associated factors). The sympatric distribution of annual and perennial species within many grass genera and the general association of perenniality with wild species and annual habit with cultigens suggest that perenniality is ancestral to annual habit. This notion was supported by the dominant nature of the Rhz2 and Rhz3 loci discovered in this study that control rhizome expression in O. longistaminata. The strong interaction between Rhz2 and Rhz3 and their pleiotropic effects on many rhizome traits indicate that Rhz2 and Rhz3 are likely to be regulatory genes. Both also contributed strongly to TN, a trait associated strongly with regrowth (ratooning) and persistence of perennial grasses. The dominant nature of both genes implies that a single recessive, null, or loss-of-function mutation at either Rhz3 or Rhz2 may shut off rhizome expression. Furthermore, the presence of Rhz3 mutants in more O. sativa accessions (18) suggests that the first mutation leading to the switch from perenniality and rhizomatousness to the current annual habit of O. sativa was more likely to have occurred at Rhz3. Rhz3 accounted for greater phenotypic variance and affected more rhizome-related traits than did Rhz2, implying that Rhz3 may be more “upstream” in the rhizomatousness pathway. The pleiotropic effects of Rhz2 on degree of primary branching, internode length, and TN, on the other hand, suggest its influence on the growth of axillary buds and/or intercalary meristems. Similarly, it is not surprising that all rhizome-enhancing alleles at the identified QTLs were from O. longistaminata, because mutations at these QTLs affecting the abundance of rhizomes in the ancestral species would have become neutral and accumulated at much faster rates after the rhizome pathway was shut off by the mutation at Rhz3. It is conceivable that mutations at these QTLs were less likely to be associated with other fitness traits. Otherwise, individuals with these mutations would have been eliminated from the O. sativa gene pool long ago.
The close correspondence between most rhizome genes/QTLs in rice and sorghum suggests that some of the same genes may influence rhizome expression and abundance in these distantly related grass species, lending support to the notion of “grasses as a single genetic system” (19). Our results regarding rhizomatousness clearly fit a model developed by Paterson et al. (17) for other traits, suggesting that in many cases convergent evolution of independent mutations at corresponding loci contributed substantially to the domestication of grasses.
The strong interaction between Rhz3 and Rhz2 and their pleiotropic effects on many rhizome traits indicated that they are additional examples of mutations at key loci leading to dramatic evolutionary changes such as have been reported in several plants, including the tbl locus that governs the fate of the axillary meristems in maize and distinguishes cultivated maize, Zea mays L. ssp. mays, from its probable wild progenitor, Z. mays ssp. Parviglumis (20). Our results suggest that similar to tb1, Rhz3 and Rhz2 may be regulatory genes that have special importance in plant evolution as proposed first by Britten and Davidson (21) and elucidated further by Doebley and Lukens (22). With the availability of the complete sequence of the rice genome (23, 24), determining the locations of Rhz2 and Rhz3 represents a key step toward cloning and molecular characterization of these two important genes, which will contribute significantly to our understanding of the evolutionary history of the grass family, create opportunities to develop perennial cereals, and offer insights into environmentally benign weed-control strategies.
Acknowledgments
We thank M. Gale and S. Cox for valuable comments and suggestions on the early version of the manuscript. This study was supported by grants from the National Natural Science Foundation of China, the Federal Ministry for Economical Cooperation and Development of German Government (to the International Rice Research Institute), the U.S. National Science Foundation and U.S. Department of Agriculture (to University of Georgia), and the Agricultural Department of Yunman Province, China.
Abbreviations
- TN
tiller number
- QTL
quantitative trait locus
- cM
centimorgan
References
- 1.Briggs F N, Knowles P F. Introduction to Plant Breeding. New York: Reinhold; 1977. [Google Scholar]
- 2.Paterson A H, Schertz K, Lin Y R, Liu S C, Chang Y L. Proc Natl Acad Sci USA. 1995;92:6127–6131. doi: 10.1073/pnas.92.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wagoner P. Crit Rev Plant Sci. 1990;9:381–408. [Google Scholar]
- 4.Cox T S, Bender M, Picone C, Van Tassel D L, Holland J B, Brummer C E, Zoeller B E, Paterson A H, Jackson W. Crit Rev Plant Sci. 2002;21:59–91. [Google Scholar]
- 5.Ghesquiere A. Proceedings of the Third International Rice Genetics Symposium. Manila, Philippines: Int. Rice Res. Inst.; 1985. pp. 15–27. [Google Scholar]
- 6.Brar D S, Khush G S. Plant Mol Biol. 1997;35:35–47. [PubMed] [Google Scholar]
- 7.Khush G S, Bacalango E, Ogawa T. Rice Genet Newsl. 1990;7:121–122. [Google Scholar]
- 8.Maekawa M. In: Proceedings of the First International Rice Genetics Symposium. Khush G S, editor. Manila, Philippines: Int. Rice Res. Inst.; 1996. pp. 428–433. [Google Scholar]
- 9.Causse M A, Fulton T M, Cho Y G, Ahn S N, Chunwongse J, Wu K, Xiao J, Yu Z, Ronald P C, Harrington S E, et al. Genetics. 1994;138:1251–1274. doi: 10.1093/genetics/138.4.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gale M D, Devos K M. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temnykh S, Genevieve D, Angelika L, Leonard L, Samuel C, McCouch S R. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCouch S R, Teytelman L, Xu Y, Lobos K B, Clare K, Walton M, Fu B, Maghirang R, Li Z, Xing Y, et al. DNA Res. 2002;9:199–207. doi: 10.1093/dnares/9.6.199. [DOI] [PubMed] [Google Scholar]
- 13.Lander E, Green P. Proc Natl Acad Sci USA. 1987;84:2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.SAS Institute. SAS/STAT. Cary, NC: SAS Inst.; 1996. [Google Scholar]
- 15.Zeng Z-B. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ware D, Jaiswal P, Ni J, Pan X, Chang K, Clark K, Teytelman L, Schmidt S, Zhao W, Cartinhour S, et al. Nucleic Acids Res. 2002;30:103–105. doi: 10.1093/nar/30.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paterson A H, Lin Y R, Li Z K, Schertz K F, Doebley J F, Pinson S R M, Liu S C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 18.Maekawa M, Inukai T, Rikiishi K, Matsuura T, Govindaraj K G. SABRAO J Breed Genet. 1998;30:69–72. [Google Scholar]
- 19.Freeling M. Plant Physiol. 2001;125:1191–1197. doi: 10.1104/pp.125.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doebley J, Stec A, Gustus C. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Britten R, Davidson E. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 22.Doebley J, Lukens L. Plant Cell. 2001;10:1075–1082. doi: 10.1105/tpc.10.7.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu J, Hu S, Wang J, Wong G K-S, Li S, Liu J B, Deng Y, Dai L, Zhou Y, Zhang X, et al. Science. 2002;296:79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 24.Goff S A, Ricke D, Lan T-H, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, et al. Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]