Abstract
The yeast CHA1 promoter is activated in the presence of serine or threonine. Activation requires the Cha4p activator, and it results in perturbation of a nucleosome that incorporates the TATA element under noninducing conditions. We show that in yeast lacking the amino terminus of histone H3, the promoter is constitutively active and the chromatin is concomitantly perturbed. This derepression occurs in the absence of elevated intracellular levels of serine or threonine and is not observed in cells lacking Rpd3p, Tup1p, or the amino terminus of histone H4. Furthermore, derepression in the absence of the H3 amino terminus requires the primary activator of this promoter, Cha4p, which we show by chromatin immunoprecipitation to be constitutively bound to the CHA1 promoter in WT yeast. Thus, the H3 amino terminus is required to prevent Cha4p from activating CHA1 in the absence of inducer. We also present results of a microarray experiment showing that the H3 amino terminus has a substantial repressive effect on a genome-wide scale.
The principal protein components of chromatin are the histones. The four core histones, H2A, H2B, H3, and H4, can be divided into central structured domains and relatively unstructured amino and, in some cases, carboxyl-terminal regions, often referred to as the histone tails (1, 2). These latter regions do not contribute to core nucleosome structure (3), but are subject to numerous posttranslational modifications that contribute to multiple aspects of chromatin function (4–6). Specific modifications have been correlated with transcriptional activation and others with repression, including the formation of stably repressed domains of heterochromatin (7).
One approach to studying the role of the histone tails in transcriptional regulation has been to focus on specific histone-modifying enzymes, or specific residues in the histone amino termini. An alternative, complementary approach has been to examine the effects of deleting individual histone amino termini. Such work has shown that in budding yeast (Saccharomyces cerevisiae), the individual histone amino termini are dispensable, but the H3 and H4 tails cannot be simultaneously removed without loss of viability, although they can be interchanged (8–11). Other experiments have shown that the H3 and H4 tails are involved in silencing at telomeres and the silent mating-type loci (12, 13), and have implicated the H3 and H4 amino termini in both positive and negative aspects of gene regulation (14–17).
In this work we demonstrate a role for the histone H3 amino terminus in the regulation of the CHA1 promoter in yeast. Our initial interest in the CHA1 promoter stems from its tightly regulated chromatin structure. In its uninduced state, the CHA1 TATA element is incorporated near the center of a positioned nucleosome, and this nucleosome is perturbed on activation of CHA1 by increased intracellular levels of serine or threonine (18). Nucleosome perturbation requires the principal CHA1 activator, Cha4p, and can be elicited under conditions in which recruitment of TATA box binding protein (TBP) and transcription are prevented (18). Furthermore, a CHA1 promoter containing a LexA binding site is not activated by artificial recruitment of TBP (as a LexATBP fusion protein), and its chromatin is not remodeled (19). These results implicate chromatin remodeling as a necessary, activator-mediated step in CHA1 activation, which may be separable from recruitment of the basal transcription machinery. However, CHA1 activation occurs independently of both the SWI/SNF complex and GCN5, and so it must depend on other chromatin remodeling and/or modification activities (18). Some chromatin remodeling complexes cannot use nucleosomes lacking histone tails as templates in vitro (2), and so it seemed possible that removal of individual histone tails might preclude remodeling and transcriptional activation. Instead, we found that the histone H3 amino terminus is required to prevent activation of the CHA1 promoter under noninducing conditions.
Materials and Methods
Plasmids.
The CHA1-LexA-MEL1 reporter plasmid has been described (19). PCR was used to convert the sequence GGTATATAAATAT containing the TATA element to GGTGTAATAT to create the tataΔ version of this plasmid. The LexA site was inserted for earlier experiments and does not affect CHA1 regulation (data not shown), and for simplicity the reporter is referred to here as CHA1-MEL1. Plasmids pNS329, pNS338, and pNS358 were made by subcloning the EcoRI–SalI fragments carrying HHT1-HHF1, hhf1-8(H4Δ2–26)-HHT1, and HHF1-hht1-2(H3Δ1–28), respectively, from pMS329, pMS338, and pMS358 (10) into pRS414 (20). Amino-terminally HA3-tagged Cha4p was expressed in cha4Δ yeast from the CHA4 promoter and carried on pRS414 (20). The HA3 tag sequence was amplified from the expression vector for HA3-tagged TBP (21) and ligated into the CHA4 coding sequence following the second codon as a PstI–BamHI fragment.
Yeast Strains and Growth Conditions.
Yeast strain MSY202 [MATα ura3-52 lys2-Δ201 leu2-3, 112 trp1-289 HHT1-HHF1 Δ(HHT2-HHF2)] is derived from the copy II deletion strain MSY156 (22). The H3Δ1–28 strain MSY975 [MATα ura3-52 lys2-Δ201 leu2-3,-112 trp1-289 hht1-2 HHF1 Δ(HHT2 HHF2)] was constructed by integrating the hht1-2 amino-terminal deletion allele (10) at its chromosomal locus by one-step gene replacement as described (23). Disruption of the CHA4 gene in strains MSY202 and MSY975, and of RPD3 (from −12 to +742) in strain MSY202, was accomplished by one-step disruption using a LEU2 marker, and was verified by Southern blotting. Strains NSY429, NSY438, and NSY458 were derived from MX1-4C (MATα ura3-52 leu2-3,112 trp1-289 his3Δ1 Δ(hht1-hhf1)(hht2-hhf2) pMS329 [CEN ARS URA3 HHT1-HHF1]) (10) by transforming with pNS329, pNS338, and pNS358, respectively, carrying HHT1-HHF1, hhf1-8 (H4Δ2–26)-HHT1, and HHF1-hht1-2 (H3Δ1–28) on plasmid pRS414.
Yeast cells were grown at 30°C in complete synthetic dropout medium (Bio 101) containing 2% dextrose. The CHA1 promoter was induced by supplementing cultures with 1 mg/ml serine for a minimum of 4 h. Yeast transformation was done by using a standard lithium acetate protocol (24), and intracellular amino acid content was analyzed essentially as described (25).
Enzyme Assays and Analysis of Chromatin Structure.
Measurement of α-galactosidase activity was performed as described (26). At least three independent yeast clones were assayed for each reported value. Chromatin structure was analyzed as described (27, 28). All chromatin-mapping experiments were done at least in duplicate, and with a range of micrococcal nuclease (MNase) concentrations.
Chromatin Immunoprecipitation (ChIP).
ChIP was performed essentially as described (29), by using 20 μl (4 μg) of anti-hemagglutinin (HA) serum (Santa Cruz Biotechnology, SC7392). Crosslinked samples were sonicated by using three repetitions of six pulses at 90% duty cycle, 20% output with a sonifier (Model 250, Branson Ultrasonics, Danbury, CT), resulting in fragments ranging from 0.2 to 1.0 kb. In some experiments, immunoprecipitated (IP) samples were eluted by the addition of 0.1 mg of HA peptide (Roche Molecular Biochemicals), followed by 30-min incubation at room temperature, and results were similar to experiments using standard elution protocols. For analysis, typically 1–2 μl of IP DNA or 1 μl of a 1:1,000 dilution of input DNA was amplified by using primers spanning the CHA1 promoter, and, for a control, primers that were within the HIS4 promoter. PCR amplification was performed for 22 and 27 cycles, and aliquots were electrophoresed, Southern blotted, and hybridized to verify that amplification was in the linear range. Quantitation was performed by using a Molecular Dynamics PhosphorImager.
Microarrays and Northern Analysis.
RNA was prepared by hot phenol extraction (30) from yeast cultures grown in yeast extract/peptone/dextrose medium to mid-logarithmic phase. For microarray analysis, RNA was reverse-transcribed, and cDNA was prepared according to the supplier's protocol (Affymetrix, Santa Clara, CA). Three separate RNA preparations were analyzed for H3Δ1–28 (MSY975) yeast cells, and two for the corresponding WT yeast (MSY202); two preparations were analyzed for H4Δ2–26 (NSY438) and the corresponding WT (NSY429) yeast. See Tables 2 and 3, which are published as supporting information on the PNAS web site, www.pnas.org, and are also accessible on the web at www.wadsworth.org/resnres/bios/morse.htm.
Northern analyses were performed on at least three independent RNA preparations, including the same ones used for the microarray analysis, and were done as described (31). Probes for INO1, RNR3, BNA1, and PHO84 were prepared by PCR, and the BglII fragment from pGEM-PYK1, a gift from Joan Curcio (Wadsworth Center), was used to probe for PYK1. Message levels were quantitated by PhosphorImager and were normalized to PYK1 RNA.
Results
The CHA1 Promoter Is Constitutively Activated in the Absence of the Amino Terminus of Histone H3 but Not Histone H4.
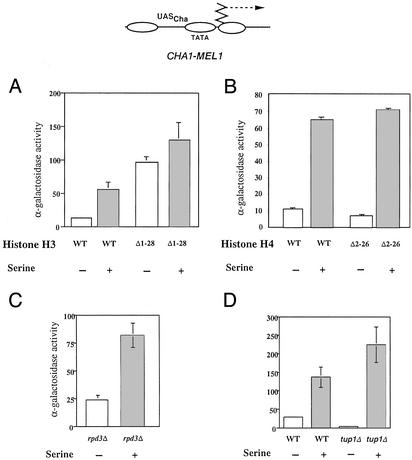
To gain possible new insight into aspects of chromatin-mediated regulation of CHA1, we examined activation of a CHA1-MEL1 reporter gene in WT yeast and congenic strains lacking the amino terminus of either histone H3 or H4. The MEL1 gene encodes α-galactosidase, which is easily measured in a colorimetric assay, and the fusion-gene promoter retains the chromatin structure and properties of the native CHA1 gene (19). The CHA1 promoter was strongly activated in the absence of serine in yeast expressing histone H3 lacking the amino-terminal 28 aa (H3Δ1–28 yeast), a region that includes most of the modifiable amino acids and that is genetically and structurally distinct from more interior portions (32, 33). MEL1 activity in H3Δ1–28 yeast in the absence of serine was greater than that seen under induced conditions in the corresponding WT strain (Fig. 1A). Induction by serine gave rise to a substantial increase in MEL1 activity in WT cells, as expected, and a modest increase in H3Δ1–28 yeast. Yeast expressing H3Δ1–28 from a CEN plasmid (isogenic to yeast expressing H4Δ2–26, below) behaved similarly to MSY975 (data not shown). In contrast, congenic yeast cells expressing from a CEN plasmid WT H3 and H4 lacking the amino-terminal residues 2–26 (H4Δ2–26), which includes most of the modifiable amino acids, and again, is genetically and structurally distinct from more interior regions (14, 32, 33), exhibited normal CHA1 regulation (Fig. 1B). Thus, the H3 amino terminus appears to have a specific role in preventing CHA1 activation in the absence of inducer.
Figure 1.
Derepression of CHA1 by deletion of the histone H3 amino terminus. α-Galactosidase expression of the CHA1-MEL1 reporter, schematized at the top, in the presence and absence of 1 mg/ml serine in WT (MSY202) and H3Δ1–28 (MSY975) yeast (A); WT (NSY429) and H4Δ2–26 (NSY438) yeast (B); rpd3Δ yeast (RMY16, derived from MSY202; C); and WT (FY23) and FY23 tup1Δ yeast (D). Standard deviations are shown.
The histone deacetylase Rpd3p and the Tup1p/Ssn6p(Cyc8p) complex are global repressors that act, at least in part, through the histone tails (34–36), and loss of histone H3 or H4 amino termini can cause derepression of genes that they regulate (refs 16 and 34; N.S. and R.H.M., unpublished results). We therefore asked whether the CHA1 promoter was activated in rpd3 or tup1 yeast. An rpd3Δ strain derived from MSY202 showed slightly increased CHA1 activation in the absence of serine compared with the WT control (Fig. 1C; compare with WT in Fig. 1A), whereas a tup1Δ strain showed decreased CHA1-MEL1 expression in the absence of serine, and increased expression in the presence of serine, compared with WT yeast (Fig. 1D). We conclude that the histone H3 amino terminus regulates the CHA1 promoter through a mechanism independent of Rpd3p or Tup1p/Ssn6p.
The CHA1 promoter is normally induced by high intracellular levels of serine or threonine. As such elevated levels could occur by means of indirect effects (25), we measured intracellular levels of amino acids in H3Δ1–28 yeast and the corresponding WT strain. We found no change in threonine or serine levels (Table 1), indicating that activation of the CHA1 promoter by removal of the H3 amino terminus most likely occurs through a direct effect at the CHA1 promoter.
Table 1.
Intracellular amino acid levels in WT and H3Δ1–28 yeast cells
| Strain | Amino acid level*
|
||||
|---|---|---|---|---|---|
| Asp | Ser | Thr | Ile | Ala | |
| WT | 55 | 36 | 25 | 46 | 100 |
| H3Δ1–28 | 59 | 40 | 27 | 46 | 104 |
Averages from two independent determinations were normalized to alanine concentration in WT cells, which was arbitrarily set to 100. The values from the two experiments were all within 20% of the average value (most were within 5%).
The CHA1 Promoter Adopts a Constitutively Active Chromatin Configuration in the Absence of the Histone H3 Amino Terminus.
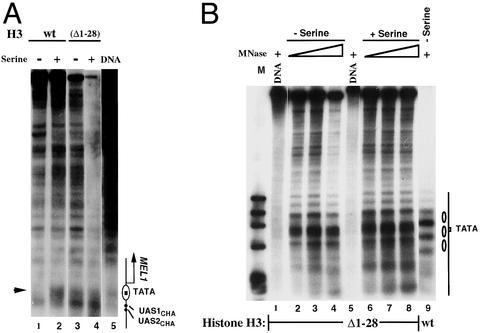
We next sought to determine whether the constitutive activation of the CHA1 promoter in H3Δ1–28 yeast was accompanied by a constitutively active chromatin configuration. We therefore mapped the chromatin structure of the CHA1 promoter in both the CHA1-MEL1 reporter and the endogenous CHA1 locus by indirect end-label-mapping of MNase cleavage sites. In both the CHA1-MEL1 reporter and the endogenous locus, the TATA element is protected against MNase digestion in chromatin from uninduced cells expressing WT histones (Fig. 2 A, lane 1, and B, lane 9), consistent with earlier work, indicating that a nucleosome incorporates the TATA element under uninduced conditions (18, 19). On induction by addition of serine, the TATA elements of the CHA1-MEL1 reporter and the endogenous CHA1 promoter become accessible to MNase, indicating disruption of the TATA-containing nucleosome (Fig. 2A, lane 2, and data not shown; ref. 18). In contrast, in H3Δ1–28 yeast, the TATA element is constitutively accessible to MNase, and additional changes in the MNase cleavage pattern in the endogenous CHA1 promoter characteristic of the induced state are also apparent (Fig. 2 A, lanes 3 and 4, and B, lanes 2–4 and 6–8). Thus, the constitutive activation of the CHA1 promoter in H3Δ1–28 yeast is reflected in its chromatin structure.
Figure 2.
Indirect end-label mapping of the chromatin structure of the CHA1-MEL1 reporter gene and the endogenous CHA1 promoter. (A) MNase cleavage sites were mapped for naked DNA and for chromatin from WT (MSY202) and H3Δ1–28 (MSY975) yeast, in the presence and absence of serine, as indicated. Cleavage sites were mapped relative to the SalI site 623 bp 5′ of the TATA element. Samples were digested with MNase at 20 units/ml (lane 1), 50 units/ml (lane 2), 5 units/ml (lanes 3 and 4), and 4 units/ml (lane 5). The arrowhead indicates a cleavage site close to the TATA element that is protected in the inactive promoter. The diagram on the right shows the location of the nucleosomal TATA element and the binding sites for the Cha4p activator (UAS1CHA and UAS2CHA). UAS, upstream activating sequence. (B) MNase cleavage sites were mapped from the BamHI site from chromatin prepared from WT (MSY202) and H3Δ1–28 yeast (MSY975), in the presence or absence of serine, as indicated. Lane M contains phage φX174 DNA cut with HaeIII. MNase was used at 4 units/ml (lanes 1 and 5), 5 units/ml (lanes 2 and 6), 20 units/ml (lanes 3, 7, and 9), and 50 units/ml (lanes 4 and 8). The structure of the uninduced promoter region is indicated on the right, with the ovals representing positioned nucleosomes.
The Histone H3 Amino Terminus Is Required to Prevent the Constitutively Bound Activator Cha4p from Activating the CHA1 Promoter in the Absence of Inducing Signal.
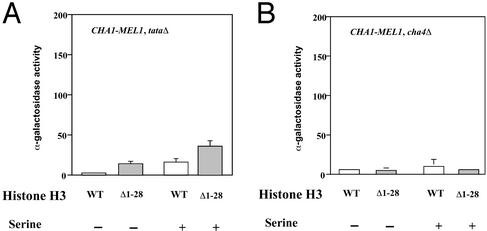
The change in chromatin structure of the uninduced CHA1 promoter seen in H3Δ1–28 cells (Fig. 2) could be a cause or consequence of the observed transcriptional derepression. If alteration of chromatin structure caused by the histone H3 tail deletion led directly to derepression (for instance, by allowing access by RNA polymerase II), the normal requirements for an activator or TATA element might be bypassed. To test this possibility, we examined CHA1-MEL1 transcription of a reporter gene lacking a TATA element, and in cha4Δ yeast cells, which lack the principal activator for CHA1 (Fig. 3). The results show that expression is reduced 3- to 5-fold by deletion of the TATA element in both WT and H3Δ1–28 yeast cells, and is essentially eliminated by loss of the Cha4p activator protein. These results suggest that the activation of the CHA1 promoter in the absence of inducing signal in H3Δ1–28 cells occurs via the normal transcriptional activation pathway, as it depends on both a TATA element and the activator Cha4p.
Figure 3.
Derepression of the CHA1 promoter in H3Δ1–28 yeast requires the TATA element and the Cha4p activator. α-Galactosidase expression of the CHA1-MEL1 reporter with mutated TATA element (tataΔ) in WT (MSY202) and H3Δ1–28 (MSY975) yeast (A), and in WT (MSY202) and H3Δ1–28 (MSY975), cha4Δ yeast (B), in the presence and absence of 1 mg/ml serine. Standard deviations are indicated.
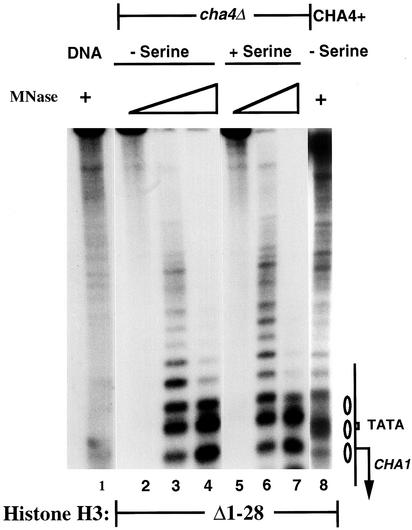
To determine whether Cha4p is also required for the altered, constitutively active chromatin configuration at the CHA1 promoter in H3Δ1–28 yeast, we examined CHA1 chromatin structure by MNase digestion and indirect end-label analysis. The results, shown in Fig. 4, show that the CHA1 promoter in H3Δ1–28 yeast lacking the Cha4p activator adopts the inactive configuration under both induced and uninduced conditions, as did the CHA1-MEL1 reporter (data not shown). Thus, the activated chromatin structure seen at the CHA1 promoter in H3Δ1–28 yeast under uninduced conditions is not a direct consequence of the absent H3 amino terminus, but instead it requires Cha4p.
Figure 4.
Chromatin structure perturbation of the CHA1 promoter in H3Δ1–28 yeast requires the Cha4p activator. MNase cleavage sites in the endogenous CHA1 promoter were mapped from the BamHI site in cha4Δ or CHA4+ H3Δ1–28 (MSY975) yeast, in the presence or absence of serine, as indicated. Samples were digested with MNase at 10 units/ml (lane 1), 0 units/ml (lanes 2 and 5), 0.5 units/ml (lanes 3 and 6), 2 units/ml (lanes 4 and 7), or 20 units/ml (lane 8). The sample in lane 8 was run on a separate gel. The CHA1 promoter is schematized on the right, with the ovals representing positioned nucleosomes present in the repressed promoter.
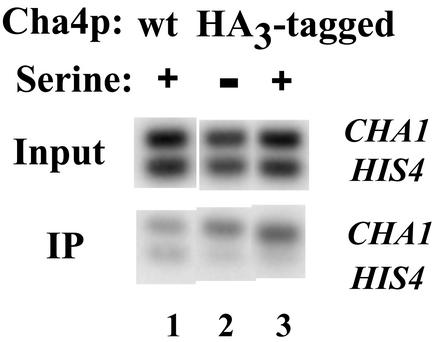
The results of Figs. 3 and 4 indicate that the histone H3 amino terminus is normally required to prevent activation of the CHA1 promoter, with concomitant alteration in chromatin structure, by Cha4p. To test whether Cha4p binds constitutively to the CHA1 promoter in cells expressing WT histones, as has been reported (18), we performed ChIP experiments. We placed a triple HA tag two amino acids from the amino terminus of Cha4p; this resulted in a very modest (2-fold) derepression of CHA1-MEL1 under uninduced conditions, and slightly decreased activation, with expression increasing by 3- to 4-fold on induction. We then used anti-HA antibodies in ChIP experiments using cells expressing amino-terminally HA-tagged Cha4p and control cells lacking any HA tag (Fig. 5). We compared the amount of IP CHA1 promoter sequence to input DNA for each sample, and normalized to IP HIS4 promoter fragment as a nonspecific control. In five independent experiments with two distinct yeast clones expressing HA-tagged Cha4p, we found that CHA1 was preferentially IP in cells expressing HA-tagged Cha4p relative to control cells by 1.3- to 3.4-fold. Most significantly, we observed no difference between induced and uninduced cells (ratios of IP CHA1 DNA in induced to uninduced varied from 0.9 to 1.3). These results indicate that Cha4p is indeed constitutively bound to the CHA1 promoter. Taken together, our results indicate that the H3 amino terminus is needed to prevent promoter-bound Cha4p from activating CHA1 under noninducing conditions.
Figure 5.
ChIP analysis of Cha4p binding to the CHA1 promoter. ChIP was performed by using anti-HA antibody, using cells expressing WT Cha4p or HA3-tagged Cha4p, and in the presence or absence of serine, as indicated. Input and IP DNA was amplified by using primers for the CHA1 and HIS4 promoters simultaneously, and the amplified DNAs were then electrophoresed, blotted, and hybridized with the labeled PCR products.
The Histone H3 Amino Terminus Is Repressive on a Genome-Wide Scale in Yeast.
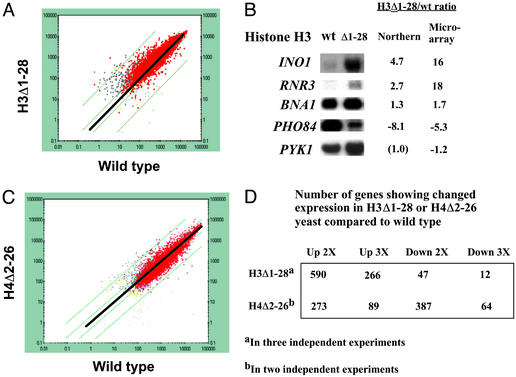
To determine the scope of genes that might be repressed by similar, or by distinct, mechanisms involving the H3 amino terminus, we conducted a microarray analysis of transcription in H3Δ1–28 yeast (MSY975) and the corresponding WT strain (MSY202) grown in rich medium (yeast extract/peptone/dextrose), conditions in which many genes are not fully active. Results from one such experiment are depicted in a scattergram in Fig. 6A, in which each point represents the expression of a particular gene in WT and H3Δ1–28 cells. Results for several genes were verified by Northern analysis (Fig. 6B and data not shown). The skewed distribution in the scattergram, with more points above than below the diagonal that represents equal expression in WT and H3Δ1–28 cells, indicates that the H3 amino terminus exerts a substantial repressive influence on a genome-wide scale. This result was confirmed in three independent comparisons of H3Δ1–28 yeast to WT yeast (Fig. 6D); furthermore, a Student's t test on the same three data sets indicated that 366 genes increased in expression with P < 0.05, whereas only 101 decreased with P < 0.05.
Figure 6.
Effect of the H3Δ1–28 deletion on gene expression. (A) Gene-expression data from microarray analysis of H3Δ1–28 (MSY975) yeast are plotted vs. data from WT (MSY202) yeast. Each point represents expression data for an individual gene. Red indicates high confidence for the value from each data set; gray points represent data for which confidence is high for one set but low for the other. For points above the thicker diagonal, confidence is lower for the value from WT yeast, and for those below the diagonal, confidence is lower for the value from H3Δ1–28 yeast. The thick diagonal represents no change in expression between WT and H3Δ1–28 yeast, and the thin diagonals represent increase or decrease in expression by 2- and 30-fold, respectively. (B) Northern analysis of selected genes in WT vs. H3Δ1–28 yeast. Ratios are averages from two experiments for Northern analysis data, and from three experiments for microarray data. Negative values indicate a decrease in H3Δ1–28 compared with WT yeast by the value shown. (C) Gene-expression data from microarray analysis of H4Δ2–26 (NSY438) yeast are plotted vs. data from WT (NSY429) yeast as in A, but the thin diagonal lines represent increase or decrease by 2- or 10-fold, respectively. (D) Summary of gene-expression data from three independent comparisons of H3Δ1–28 or H4Δ2–26 yeast to WT yeast.
We also wished to determine whether the histone H4 amino terminus exerted a generally repressive influence on a genome-wide scale. Results of two independent microarray experiments indicate that the H4 amino terminus exerts a substantial regulatory role; however, in contrast to the H3 amino terminus, its overall effect is nearly balanced between positively and negatively influencing transcription, with a possible small bias toward an activating effect (Fig. 6 C and D). We note that the strains used in this analysis carry the histone H3 and H4 genes on a CEN plasmid, unlike MSY202 and MSY975. Microarray experiments comparing H3Δ1–28 yeast to WT in a congenic strain expressing H3Δ1–28 from a CEN plasmid (NSY458) yielded similar results to those using MSY975 and MSY202 (data not shown).
Inspection of Fig. 6A reveals that genes increasing in expression in H3Δ1–28 cells were mostly expressed at low-to-moderate levels in WT cells. Examination of the gene expression data did not reveal substantial overrepresentation of any one class of gene (e.g., transcription factors, cell-cycle components, DNA repair, etc.), although 10 of 55 genes examined that are involved in meiosis or sporulation were derepressed by 3-fold or more. This finding makes sense, as many such genes are regulated by Tup1p, which interacts physically and genetically with the H3 and H4 tails, and that recruits histone deacetylases as part of its repressive mechanism (34, 37, 38). In fact, of 119 genes that increased in expression by 3-fold or more in a tup1Δ compared with a WT strain (39), 30 also increase their expression by that amount in H3Δ1–28 cells. Another gene-regulatory protein that interacts with the histone amino termini is Rpd3p, a histone deacetylase that also functions as a corepressor. The putative principal targets of Rpd3p are the histone H3 and H4 amino termini (35, 36, 40), and, in support of this idea, we found that of 258 genes that increased in expression by 3-fold or more in rpd3Δ compared with WT yeast (data not shown), 76 are also up-regulated by that amount in H3Δ1–28 yeast. This finding contrasts somewhat with the data of Bernstein et al. (41), although the trend is similar: of 117 genes that increase >2-fold in rpd3Δ cells in two independent determinations (41), 24 increase >3-fold in our H3Δ1–28 yeast (in three independent determinations). Of 43 genes that increase >2-fold in hda1 yeast compared with WT (41), 13 increase in H3Δ1–28 yeast. Thus, many genes repressed by histone deacetylases require the histone H3 amino terminus for full repression. However, many others, including CHA1, are evidently repressed by other mechanisms involving the H3 amino terminus.
Discussion
The two major findings in this work are that the histone H3 amino terminus is required to prevent activation of the CHA1 promoter by the constitutively bound activator Cha4p under noninducing conditions; and that the histone H3 amino terminus plays a generally repressive role on a genome-wide basis. Previous work showed hyperactivation of Gal4p-regulated genes on loss of the H3 amino terminus, but no activation was seen under repressed conditions (14, 15). The mechanism by which the H3 amino terminus regulates CHA1 appears, to our knowledge, to be novel, and remains to be explored in detail. First, it is likely that the H3 amino terminus exerts its effect directly at the CHA1 promoter, as intracellular levels of serine and threonine remain normal in H3Δ1–28 yeast (Table 1). Second, it is independent of the global repressors Rpd3p or Tup1p, which are known to act at least in part through the histone amino termini (Fig. 1). Third, although the CHA1 promoter is also derepressed in yeast lacking Sir4p or a functional RSC complex (18, 42), this derepression does not require Cha4p, so it must occur by a mechanism distinct from that which is reported here. We speculate that the H3 amino terminus, possibly in combination with other proteins present at the CHA1 promoter, interacts with a protein or proteins that block activation by Cha4p in the absence of inducing signal. Alternatively, the H3 amino terminus may block activation by stabilizing higher-order chromatin structure (2).
Microarray experiments revealed that the histone H3 amino terminus exerts a substantial repressive effect on a genome-wide scale. Some of the genes showing decreased expression in H3Δ1–28 yeast are regulated by Tup1p and some by Rpd3p, consistent with findings that these global repressors interact with the histone amino termini (34–36, 40). The chromatin remodeler ISW2 also interacts with the histone amino termini (although likely predominantly with the histone H4 tail; ref 43) and represses a number of genes, especially those involved in meiosis (44, 45). However, although some genes that are derepressed in isw2 yeast, such as INO1 and SGA1, are also derepressed in H3Δ1–28 yeast, others such as REC104 are not (45). Thus, although there is overlap among the genes affected by loss of the H3 amino terminus and genes regulated by enzymes that interact with the histone tails, the correlation is not complete.
Some genes are doubtless derepressed in H3Δ1–28 yeast through indirect effects. For example, the a1 mating factor (Mfa1) is expressed from the MFA1 locus, which is regulated by Tup1p and is derepressed in H3Δ1–28 cells; thus, genes that are positively regulated by Mfa1 (such as SGA1) should also show greater expression in H3Δ1–28 cells. Nevertheless, it seems likely that a significant number of genes are repressed by direct effects involving the H3 amino terminus, as appears to be true for CHA1. Detailed examination of the mechanisms by which the H3 amino terminus contributes to repression of specific genes will be required to reveal the full spectrum of means by which this chromatin constituent contributes to gene regulation in yeast.
Finally, although histone-modifying enzymes that act as coactivators or corepressors in yeast are generally conserved in higher eukaryotes (6, 7), little is known of the overall contributions of the histone tails to transcriptional regulation in multicellular organisms. Because the histone genes exist in multiple copies in these organisms, it is not practical to do gene replacements, as has been done in yeast. However, dominant phenotypes might be observed in the presence of histone mutants such as those used here. This approach has recently been used to examine effects of histone methylation in Neurospora crassa (46) and may also prove fruitful in examining contributions of the histone amino termini to gene regulation in other organisms. We speculate that such studies might reveal a repressive contribution of the histone H3 amino terminus that has been conserved through much of the eukaryotic kingdom.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of Tim Moran of the Wadsworth Center Molecular Genetics Core Facility. We thank Grace Stafford for critically reading the manuscript; our colleagues at the Wadsworth Center for helpful discussions; Chuck Lowry for yeast strains; David Stillman, Laurent Kuras, and Kevin Struhl for plasmids; and Mike Ryan and Robin Pietropaolo of the Wadsworth Center Microarray Facility, the Wadsworth Center Molecular Genetics Core, and the Wadsworth Center Protein Sequencing and Amino Acid Analysis Core. This work was supported by National Institutes of Health Grants GM51993 (to R.H.M.) and GM28920 (to M.M.S.) and National Science Foundation Grant MCB-0133399 (to R.H.M.).
Abbreviations
- HA
hemagglutinin
- IP
immunoprecipitated
- MNase
micrococcal nuclease
- ChIP
chromatin immunoprecipitation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Arents G, Burlingame R W, Wang B C, Love W E, Moudrianakis E N. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen J C, Tse C, Wolffe A P. Biochemistry. 1998;37:17637–17641. doi: 10.1021/bi982409v. [DOI] [PubMed] [Google Scholar]
- 3.Bohm L, Crane-Robinson C. Biosci Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- 4.Cheung P, Allis C D, Sassone-Corsi P. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 5.Strahl B D, Allis C D. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Reinberg D. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 7.Rice J C, Allis C D. Curr Opin Cell Biol. 2001;13:263–273. doi: 10.1016/s0955-0674(00)00208-8. [DOI] [PubMed] [Google Scholar]
- 8.Kayne P S, Kim U J, Han M, Mullen J R, Yoshizaki F, Grunstein M. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- 9.Grunstein M. Annu Rev Cell Biol. 1990;6:643–678. doi: 10.1146/annurev.cb.06.110190.003235. [DOI] [PubMed] [Google Scholar]
- 10.Morgan B A, Mittman B A, Smith M M. Mol Cell Biol. 1991;11:4111–4120. doi: 10.1128/mcb.11.8.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ling X, Harkness T A, Schultz M C, Fisher-Adams G, Grunstein M. Genes Dev. 1996;10:686–699. doi: 10.1101/gad.10.6.686. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J S, Ling X, Grunstein M. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- 13.Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- 14.Durrin L K, Mann R K, Kayne P S, Grunstein M. Cell. 1991;65:1023–1031. doi: 10.1016/0092-8674(91)90554-c. [DOI] [PubMed] [Google Scholar]
- 15.Mann R K, Grunstein M. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roth S Y, Shimizu M, Johnson L, Grunstein M, Simpson R T. Genes Dev. 1992;6:411–425. doi: 10.1101/gad.6.3.411. [DOI] [PubMed] [Google Scholar]
- 17.Zhang W, Bone J R, Edmondson D G, Turner B M, Roth S Y. EMBO J. 1998;17:3155–3167. doi: 10.1093/emboj/17.11.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreira J M, Holmberg S. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan M P, Stafford G A, Yu L, Morse R H. Mol Cell Biol. 2000;20:5847–5857. doi: 10.1128/mcb.20.16.5847-5857.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christianson T W, Sikorski R S, Dante M, Shero J H, Hieter P. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- 21.Kuras L, Struhl K. Nature. 1999;399:609–613. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 22.Smith M M, Stirling V B. J Cell Biol. 1988;106:557–566. doi: 10.1083/jcb.106.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Megee P C, Morgan B A, Smith M M. Genes Dev. 1995;9:1716–1727. doi: 10.1101/gad.9.14.1716. [DOI] [PubMed] [Google Scholar]
- 24.Hill J, Donald K A, Griffiths D E, Donald G. Nucleic Acids Res. 1991;19:5791. doi: 10.1093/nar/19.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pedersen J O, Rodriguez M A, Praetorius-Ibba M, Nilsson-Tillgren T, Calderon I L, Holmberg S. Mol Gen Genet. 1997;255:561–569. doi: 10.1007/s004380050529. [DOI] [PubMed] [Google Scholar]
- 26.Ryan M P, Jones R, Morse R H. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent N A, Bird L E, Mellor J. Nucleic Acids Res. 1993;21:4653–4654. doi: 10.1093/nar/21.19.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stafford G A, Morse R H. J Biol Chem. 1997;272:11526–11534. doi: 10.1074/jbc.272.17.11526. [DOI] [PubMed] [Google Scholar]
- 29.Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt M E, Brown T A, Trumpower B L. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang S S, Yin X, Guzzo-Arkuran C, Jones V S, Davison A J. BioTechniques. 1993;14:380–381. [PubMed] [Google Scholar]
- 32.Lenfant F, Mann R K, Thomsen B, Ling X, Grunstein M. EMBO J. 1996;15:3974–3985. [PMC free article] [PubMed] [Google Scholar]
- 33.Luger K, Richmond T J. Curr Opin Genet Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 34.Edmondson D G, Smith M M, Roth S Y. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 35.Rundlett S E, Carmen A A, Kobayashi R, Bavykin S, Turner B M, Grunstein M. Proc Natl Acad Sci USA. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kadosh D, Struhl K. Mol Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watson A D, Edmondson D G, Bone J R, Mukai Y, Yu Y, Du W, Stillman D J, Roth S Y. Genes Dev. 2000;14:2737–2744. doi: 10.1101/gad.829100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Suka N, Carlson M, Grunstein M. Mol Cell. 2001;7:117–126. doi: 10.1016/s1097-2765(01)00160-5. [DOI] [PubMed] [Google Scholar]
- 39.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 40.Rundlett S E, Carmen A A, Suka N, Turner B M, Grunstein M. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein B E, Tong J K, Schreiber S L. Proc Natl Acad Sci USA. 2000;97:13708–13713. doi: 10.1073/pnas.250477697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moreira J M, Holmberg S. EMBO J. 1999;18:2836–2844. doi: 10.1093/emboj/18.10.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clapier C R, Langst G, Corona D F, Becker P B, Nightingale K P. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgel P T, Tsukiyama T, Wu C. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldmark J P, Fazzio T G, Estep P W, Church G M, Tsukiyama T. Cell. 2000;103:423–433. doi: 10.1016/s0092-8674(00)00134-3. [DOI] [PubMed] [Google Scholar]
- 46.Tamaru H, Selker E U. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.