Abstract
The expression of activation-induced cytidine deaminase (AID) is prerequisite to a “trifecta” of key molecular events in B cells: class-switch recombination and somatic hypermutation in humans and mice and gene conversion in chickens. Although this critically important enzyme shares common sequence motifs with apolipoprotein B mRNA-editing enzyme, and exhibits deaminase activity on free deoxycytidine in solution, it has not been shown to act on either RNA or DNA. Recent mutagenesis data in Escherichia coli suggest that AID may deaminate dC on DNA, but its putative biochemical activities on either DNA or RNA remained a mystery. Here, we show that AID catalyzes deamination of dC residues on single-stranded DNA in vitro but not on double-stranded DNA, RNA–DNA hybrids, or RNA. Remarkably, it has no measurable deaminase activity on single-stranded DNA unless pretreated with RNase to remove inhibitory RNA bound to AID. AID catalyzes dC → dU deamination activity most avidly on double-stranded DNA substrates containing a small “transcription-like” single-stranded DNA bubble, suggesting a targeting mechanism for this enigmatic enzyme during somatic hypermutation.
The breakthrough discovery of activation-induced cytidine (CR) deaminase (AID) (1, 2) has opened a way to identify the biochemical reactions responsible for generating high-affinity antibodies. Although AID is normally expressed in germinal center B cells, its forced expression in B cells at the wrong stage of differentiation (3), in non-B cells (4, 5), and in Escherichia coli (6, 7) results in enhanced mutagenesis and mutational spectra mimicking somatic hypermutation (SHM). This suggests that AID can operate effectively out of its natural milieu and does not require either B cell-specific V-gene targeting elements or other B cell-specific proteins (8). Based on its similarity to apolipoprotein B mRNA-editing enzyme, which catalyzes the deamination of C on mRNA (9), it has been suggested that conversion of dC → dU on DNA, followed by the generation of an abasic site, might account for at least a subset of mutations targeted to SHM hot-spot sequences (6, 8, 10). Recent data showing that AID- and apolipoprotein B mRNA-editing enzyme-generated hypermutation is enhanced in uracil-N-glycosylase-deficient strains of E. coli imply that both AID and apolipoprotein B mRNA-editing enzyme can act on DNA (7).
Because previous studies have failed to detect AID activity on either DNA or RNA, the biologically relevant substrate for AID remains an important open question. In this article we identify and characterize AID activity on nucleic acid substrates and provide a surprising explanation for why its activity has been elusive.
Materials and Methods
Materials.
Ultrapure dNTP, 2′,3′-dideoxynucleoside triphosphate, and T4 polynucleotide kinase were purchased from Amersham Pharmacia; RNaseA, CR, deoxycytidine (CdR), uridine, and deoxyuridine were from Sigma; T7 sequenase (version 2.0) and RNase inhibitor were from United States Biochemical; and uracil DNA glycosylase (UDG) and apurinic endonuclease (APE) were generous gifts from D. Mosbaugh (Oregon State University, Corvallis). DNA oligonucleotides containing normal cytosine or 5-methylcytosine were synthesized on an Applied Biosystems 392 DNA/RNA synthesizer. DNA oligonucleotides RR-1 (5′-AGA-ATT-AAG-TTA-AGC-TAG-TTA-AGT-TAT-3′), RR-2 (5′-AGA-ATT-AAG-TTA-AGC-TAG-CTA-AGT-TAT-3′), RR-3 (5′-AGA-ATT-AAG-TTA-ATC-TAG-ACA-AGT-TAT-3′), and DDR-2 (5′-AAA-GGG-GAA-AGC-AAA-GAG-GAA-AGG-TGA-GGA-GGT-3′) were used as single-stranded (ss)DNA substrates for deamination reactions. An RNA oligonucleotide rDR1 (5′-AAA-GGG-GAA-AGC-AAA-GAG-GAA-AGG-UGA-GGA-GGU-3′) was used as an ssRNA substrate. Double-stranded (ds)DNA, DNA–DNA bubble substrates, and RNA–DNA hybrids were formed by annealing the oligos with their DNA or RNA complementary strands.
AID Purification.
AID cDNA was amplified from Ramos B cell mRNA and cloned into pAcG2T vector (PharMingen). AID was expressed as a GST fusion protein in Sf9 insect cells by using a baculovirus expression system. GST-AID (≈50 kDa) was batch-purified by using glutathione-Sepharose (Amersham Pharmacia) and had a purity of >75% chromatographically. Eluted samples were dialyzed with 20 mM Tris, pH 7.5/10 mM NaCl/0.1 mM DTT/20% glycerol and stored at −80°C.
Deamination Reactions.
In a typical deamination experiment (10-μl volume), AID (200 ng), RNaseA (1 μg), and DNA substrate (100 nM) were incubated in the reaction buffer (10 mM Tris⋅HCl, pH 8.0/1 mM EDTA/1 mM DTT) at 37°C for 5 min or as indicated. RNase inhibitor (200 units) was added when present. Reactions were terminated by extracting twice with phenol/chloroform/isoamyl alcohol (25:24:1).
Analysis of Deamination Products by UDG and APE (Assay 1).
Oligonucleotides were 5′-end-labeled with 32P by T4 kinase before use in deamination reactions. For ssDNA substrates, after incubation with AID, a complementary DNA strand was annealed to the ssDNA substrate followed by incubation with 3 units of UDG and 12.5 nM APE in buffer containing 50 mM Hepes, pH 7.5, 10 mM MgCl2, 20 mM NaCl, 2 mM DTT, and 0.5 mM ATP. This procedure allows the visualization of a putative AID-catalyzed dC → dU conversion by the presence of a 32P-labeled DNA oligomer corresponding in length to a nick at an original dC site (see, e.g., Fig. 1a Left). After 30 min of incubation at 37°C, the reactions were terminated by adding an equal volume of 95% formamide/20 mM EDTA. Reaction products were resolved and visualized by 19% denaturing PAGE and phosphorimaging. To examine AID activity on RNA–DNA hybrids, RNase was added in the presence of AID to hydrolyze the RNA portion of the RNA–DNA hybrid, and deamination products then were analyzed as described for ssDNA. One unit of UDG catalyzes the release of 1 nmol of uracil per hour.
Figure 1.
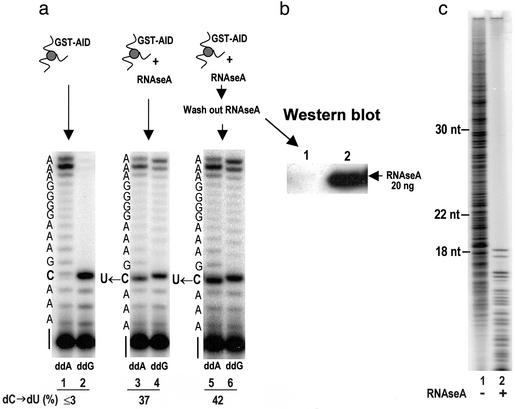
AID-catalyzed dC → dU conversion on ssDNA potentiated by RNase. (a) Assay 1 detects dC deamination by using UDG and APE. An AID-catalyzed 14-nt deamination product occurs with ssDNA but only in the presence of RNaseA (lane 1). (b) dC → dU conversion in SHM hot-spot and non-hot-spot sequence contexts using assay 1. The arrows indicate positions of deamination products after UDG and APE treatment. (c) Assay 2 detects dC deamination by using primer elongation–dideoxynucleotide termination. Deamination of dC is detected by the presence of an intense termination band (in lane 1, ddA) located opposite the original template C site (U ← C, see sequence to the left of gel) and a reduction in the intensity of the termination band (in lane 2, ddG) located opposite the original template C site. Concomitant with the conversion of dC → dU is the presence of bands migrating past the U ← C template site (lane 2), denoted by the bracket to the right of the gel.
Analysis of Deamination Products by Primer Elongation–Dideoxynucleotide Termination (Assay 2).
ssDNA substrates reacted with AID were annealed to a 3-fold excess 18-mer 32P-labeled primer, and the primer was elongated by using T7 sequenase in the presence of three dNTPs and either 2′,3′-dideoxyadenosine (ddA) or 2′,3′-dideoxyguanosine (ddG) triphosphate (see Figs. 1c and 2a). RNA–DNA and dsDNA substrates were heat-denatured, and the separated strands were annealed to 18-mer primer DNA complementary to the DNA strand as described above. The products of reactions were separated on 16% polyacrylamide denaturing gels and visualized and analyzed by phosphorimaging. Deamination efficiencies were calculated from extension reactions with the ddA mix as a ratio of the band intensity opposite the C, U template compared with the integrated band intensities at and past the C template. The efficiencies were also calculated from extension reactions with the ddG mix as a ratio of integrated band intensities past the template C to the integrated band intensities at and past the C template. Backgrounds (≈2–3%), representing a pause band for the ddA mix (or passing bands for the ddG mix) by T7 sequenase at the template C, were subtracted from each calculation. Both methods gave similar values (±0–3%). Deamination on an ssRNA substrate was analyzed similarly by using avian myeloblastosis virus reverse transcriptase instead of T7 sequenase.
Figure 2.
RNase activates AID by digesting AID-associated inhibitor RNA. (a) RNase pretreatment of AID is sufficient to observe AID-catalyzed dC → dU conversion on ssDNA as detected by primer elongation–dideoxynucleotide termination (assay 2). GST-AID bound to glutathione-Sepharose beads was preincubated with RNaseA for 5 min at 37°C and washed extensively to remove the RNaseA. AID-catalyzed dC → dU conversion after RNaseA removal can be observed in lanes 5 and 6 (U ← C template site, indicated at the left of the gel). The fraction of dC → dU conversion is indicated at the bottom of the gel as dC → dU (%). (b) Western blot showing the efficacy of RNaseA removal by washing the GST-AID-bound beads indicated by the absence of a crossreacting band with RNaseA antibody (lane 1). (c) Detection of AID-associated inhibitor RNA. After incubation of AID with proteinase K, a phenol/chloroform/isoamyl extraction was carried out followed by 5′-32P labeling of putative nucleic acids by using T4 polynucleotide kinase and resolution of labeled products by 20% denaturing PAGE. The absence of bands >18 nt in the RNase-treated sample (lane 2) demonstrates the existence of AID-associated RNA. The appearance of bands <18 nt in the RNase-treated sample (lane 2) represents RNase-digested products; RNaseA hydrolyzes ssRNA at C and U sites. Size markers at the left indicate the position to which DNA oligonucleotides have migrated on the gel.
RNA Inhibition Experiment.
AID attached to GST resin was activated by incubation with RNaseA and washed with 15 bed volumes of 1× PBS. The 50 μl of AID attached to GST resin was incubated with 2.5 μg of RNA (mRNA from Ramos cells, total RNA from human heart, tRNA from E. coli, or a 30-nt ssRNA oligo) at 37°C for 3 min and placed on ice for 2 min. Treated beads (25 μl) were used in the deamination reaction with 100 nM ssDNA substrate DDR-2.
CdR and CR Deaminase Activities.
AID was incubated with 3.3 μCi (1 Ci = 37 GBq) of deoxy[5-3H]CR (22.0 Ci/mmol, Amersham Pharmacia) and 250 μM dC in a total volume of 10 μl buffered with 45 mM Tris, pH 7.5. After a 3-h incubation, the reaction was quenched by adding 2 μl of 10 μg/μl each dC and dU. The reaction mixture (4 μl) was applied to a polyethyleneimine-cellulose TLC plate and developed in 7:2 (vol/vol) isopropyl alcohol/10% HCl for 15 h. The dC and dU bands were visualized under UV light, cut out, and quantified by liquid scintillation counting. CR deamination activity was measured similarly by using 3.3 μCi of [5-3H]CR (27.0 Ci/mmol, ICN).
Results and Discussion
Because there is no direct biochemical evidence that AID acts on either DNA or RNA, the biologically relevant substrate for AID remains an important open question. To address this problem, we purified GST-AID made in insect cells and used two alternative assays and three nucleic acid substrates (ssDNA, dsDNA, and RNA–DNA hybrids) to investigate AID-catalyzed deamination. In assay 1, UDG converts AID-generated dU to abasic sites that are converted to ss nicks by incubation with APE (as indicated in the sketch accompanying Fig. 1a). In the second assay, the denatured strands were annealed to a DNA primer strand complementary to the separated DNA strand, and a primer-elongation reaction was carried out by using either ddATP or ddGTP chain terminators to detect the presence of either an AID-catalyzed product dU or unreacted dC (Fig. 1c).
Using assay 1, we detected AID-catalyzed dC deamination on the DNA strand of RNA–DNA hybrid molecules, but this turned out to be true only because RNase was added to remove the RNA from the hybrid (data not shown). In addition, assay 2 failed to confirm that deamination had occurred! This initially confusing observation led to the surprising conclusion that preincubation of AID with RNase was necessary to “unlock” deamination activity, suggesting that there might be an RNA inhibitor bound to the enzyme. We therefore investigated the effect of RNase on the purified AID.
AID catalyzes deamination of dC on ssDNA but only when AID is pretreated with RNase by using either assay 1 (Fig. 1 a and b) or assay 2 (Fig. 1c). By using assay 1, when an ssDNA oligomer containing a single dC is incubated with AID and then treated with UDG and APE, a cleaved product band (14 nt) is produced, corresponding to the original location of dC (Fig. 1a, lane 1). No reaction with ssDNA occurs in the absence of either AID or RNase (Fig. 1a, lanes 2 and 3). There is no detectable AID activity on dsDNA (Fig. 1a, lane 5).
AID exhibits ≈3-fold greater activity in two 5′-AGCT SHM hot-spot sequences compared with 5′-ATCT and 5′-GACA non-hot-spot sequences (Fig. 1b). The 3-fold differences are determined by measuring the ratio of integrated gel-band intensities for the upper hot-spot/upper non-hot-spot and lower hot-spot/lower non-hot-spot bands. Note that the intensities of the lower of the two bands in both hot-spot and non-hot-spot lanes are likely to exceed those of the upper bands, because when both Cs are cut, only the lower band is detected in the assay. This explains why the intensity of the lower non-hot-spot band is comparable to that of the upper hot-spot band. This preference for WRCY (W = A or T, R = purine, and Y = pyrimidine) hot-spot motifs reflects the targeting of this motif that has been observed in vivo (11) and suggests that purified AID by itself might play a direct role in this process. Although much more data would be required to substantiate this point, the limited data suggest that AID activity depends on sequence context.
Assay 2, involving primer elongation–dideoxynucleotide termination, provides an independent confirmation of AID-catalyzed deamination of dC on ssDNA (Fig. 1c). The presence of an intense ddA termination band opposite a template C site measures dC → dU conversion (Fig. 1c, lane 1). Conversely, replacement of dC by dU results in a reduced-intensity ddG termination band (Fig. 1c, compare lanes 2 and 4), and concomitant bypass bands reflect synthesis on templates containing dU (Fig. 1c, lane 2 brackets). The low-intensity pause bands arise from dissociation of T7 DNA polymerase when copying relatively short DNA templates. Omission of RNase results in the loss AID-catalyzed dC deamination (Fig. 1c, lanes 3 and 4). The inhibition of RNase extinguishes AID activity (Fig. 1c, lanes 5 and 6). RNase by itself cannot catalyze deamination of dC (Fig. 1c, lanes 7 and 8). AID fails to act on dsDNA (Fig. 1c, lanes 9 and 10), in agreement with assay 1 (Fig. 1a, lanes 5–7). AID is activated by a variety of RNases: pancreatic RNaseA (used in Fig. 1), which digests ssRNA at C and U, RNase T1 (acting at G), and E. coli RNase 1 (acting at all bases) (data not shown). In contrast, AID is inactive in the presence of RNaseH, which digests RNA in RNA–DNA hybrids but not ssRNA (data not shown).
We used AID attached to glutathione-Sepharose beads to determine whether RNase is able to activate AID directly or whether instead it acts indirectly, perhaps by hydrolyzing an AID-associated RNA inhibitor (Fig. 2). The fraction of dC deamination on ssDNA using bead-linked AID is 37% in the presence of RNase (Fig. 2a, lanes 3 and 4) but remains at background levels (<3%) in the absence of RNase (Fig. 2a, lanes 1 and 2). Preincubation of bead-linked AID with RNase followed by extensive washing to remove the RNase results in a level of deamination (≈42%; Fig. 2a, lanes 5 and 6) similar to when RNase remains present in the reaction (Fig. 2a, lanes 3 and 4). We conclude that the requirement for RNase is indirect, i.e., to hydrolyze an AID-associated inhibitory RNA molecule. The successful removal of RNase during the washing step is shown by the absence of crossreaction with an antibody against RNaseA (Fig. 2b).
The endogenous RNA bound to AID is observed directly by incubating purified AID with proteinase K followed by extraction of the nucleic acid fraction and the addition of a 5′-32P radioactive label (Fig. 2c). The 32P-labeled bands consist of heterogeneously sized, ≈10- to 40-nt RNA, with an average length of ≈25 nt (Fig. 2c, lane 1), which is degradable by RNaseA (Fig. 2c, lane 2). We estimate that the endogenous ssRNA is bound to AID in an ≈1:1 stoichiometry, determined in the following manner. Using phenol-chloroform, we extracted 480 ng of RNA nucleotides from 40 pmol of AID determined by UV absorbance. By assuming that 480 ng of RNA nucleotides corresponds to 40 pmol of RNA molecules (a 1:1 stoichiometry with AID protein), we deduce a predicted average length of the associated RNA as ≈35 ± 10 nt, in close agreement with the radioactive data (Fig. 2c, lane 1).
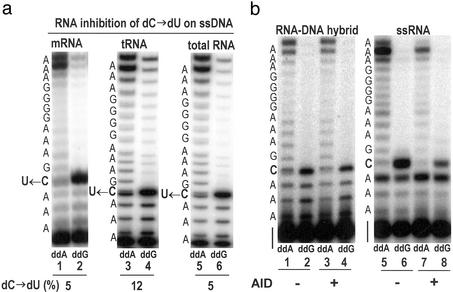
If the inhibitory effect caused by the endogenous RNA is eventually found to be physiologically relevant, then the 1:1 ratio of RNA to AID could become an important observation. However, the addition of much larger amounts (≈10- to 20-fold) of exogenous RNA from a variety of sources also inhibits AID (Fig. 3a). The conversion of dC → dU in the presence of B cell mRNA, E. coli tRNA, or B or heart cell total RNA is 5%, 12%, and 5% (Fig. 3A), respectively, compared with 42% in the absence of RNA (Fig. 2, lanes 5 and 6). RNase-pretreated AID bound to beads (see, e.g., Fig. 2a, sketch at top) did not convert dC → dU in RNA–DNA hybrid molecules (Fig. 3b). The absence of activity on the RNA–DNA hybrid was not caused by inhibition of AID by the presence of a 3-fold excess of RNA over DNA strands used to form the RNA–DNA hybrid, because the deamination reaction on ssDNA was unimpeded in the presence of the same concentration of ssRNA when the annealing heat-renaturation step was not performed (data not shown). Moreover, AID shows no measurable activity on ssRNA (Fig. 3b).
Figure 3.
Activity of RNase-activated AID attached to GST-Sepharose beads. (a) Inhibition of AID-catalyzed dC → dU deamination on ssDNA substrate by exogenous RNA. AID attached to GST resin was activated by RNaseA as described for Fig. 2. Activated AID beads (25 μl) were preincubated with 1.25 μg of mRNA (from Ramos cells), tRNA (E. coli), or total RNA (heart cells) for 3 min at 37°C and used in the deamination reaction with ssDNA substrate. The fraction of dC → dU conversion is indicated at the bottom of the gel as dC → dU (%). (b) AID does not deaminate dC in RNA–DNA hybrid or C in ssRNA. RNA–DNA hybrid or ssRNA was incubated in the reaction buffer at 37°C for 5 min in the presence or absence of activated AID-GST beads. The deamination product for ssRNA was analyzed by assay 2, in which avian myeloblastosis virus reverse transcriptase was used instead of T7 sequenase.
Previous to this study, the sole activity observed for AID was with a free nucleoside (CR) or deoxynucleoside CdR substrate (12). This activity did not require pretreatment with RNase. To confirm and extend the previous data, we find that ≈100% dC → dU conversion is achieved at “high” and “low” RNase concentrations on ssDNA (Fig. 4a). Preincubation of RNaseA with RNaseA inhibitor strongly inhibits AID activity on ssDNA (Fig. 1c, lanes 5 and 6). In contrast, the reaction proceeds to completion with unaltered kinetics when RNase inhibitor is added 5 min after the reaction has begun (Fig. 4a, see arrow). The 5-min reaction is thus sufficient to allow for degradation of the RNA bound to AID, which agrees with the RNase-pretreated AID-bound bead data (Fig. 2a, lanes 5 and 6). AID exhibits ≈10-fold higher specific activity on ssDNA for the deamination of dC → dU compared with 5-methylcytosine → T (Fig. 4b). AID is most active when deaminating the free deoxynucleoside, CdR → deoxyuridine (Fig. 4b Inset). However, in marked contrast to its activity on ssDNA, AID does not require the presence of RNase when acting on free deoxy or ribonucleoside substrates (Fig. 4b Inset). We speculate that these much smaller substrate molecules are able to diffuse into the active site of the AID, whereas larger nucleic acid substrates are blocked by the presence of RNA bound to AID. AID contains bound zinc, and its activity on both ssDNA and free deoxy and ribonucleosides is inhibited by the Zn inhibitor 1,10-o-phenanthroline (data not shown) as documented for inhibition of CR (12).
Figure 4.
AID-deamination activities. (a) Time course for AID (200 nM)-catalyzed dC → dU conversion on ssDNA (100 nM) in the presence of 1 μg of RNaseA (filled circles) or 0.1 μg of RNaseA, for which either RNase inhibitor was absent (open circles) or absent at t = 0 and added to the reaction at t = 5 min (see arrow) (filled triangles). (b) AID-catalyzed deamination rates as a function of AID concentration for various cytosine-containing substrates. Filled circles, dC → dU on ssDNA; filled triangles, 5-MeC → T on ssDNA (in 5′-CpG sequence context); open circles, 5-MeC → T on ssDNA (in non-5′-CpG sequence context). (Inset) AID-catalyzed deamination rates as a function of AID concentration for the free deoxynucleoside, CdR (open circles), and free nucleoside, CR (filled circles), in the absence of RNase.
Because SHM occurs only in the presence of RNA transcription (13–15), we modeled variable-length transcription-like bubbles and measured the activity of AID in comparison to ssDNA (Table 1). AID activity on dsDNA substrates composed of 3- and 4-nt bubbles is less than that for ssDNA by ≈4- and 2.5-fold, respectively (Table 1). However, its activity on 5- and 9-nt bubbles is significantly greater than on ssDNA by ≈2- and 3-fold, respectively, with the 9-nt bubble corresponding to a typical transcription bubble size. AID has no measurable deamination activity on a dC:dA single base mismatch (Table 1).
Table 1.
Comparing AID-catalyzed dC → dU conversion on ssDNA, dsDNA, and dsDNA bubble substrates
| Substrate | ssDNA | dsDNA | Bubble substrate size, nt
|
||||
|---|---|---|---|---|---|---|---|
| 1 | 3 | 4 | 5 | 9 | |||
| dC → dU, % | 20 ± 7 | 0† | 0 | 5 ± 1 | 8 ± 2 | 35 ± 4 | 56 ± 10 |
Reactions were carried out by using 100 nM substrate DNA and 75 ng of AID for 5 min at 37°C, and the fraction of dC → dU conversion for each substrate was calculated by using assay 2 as described in Materials and Methods. dsDNA bubble substrates refer to partially complementary dsDNA (27-mer) containing centrally located noncomplementary regions forming ss bubbles of 1, 3, 4, 5, and 9 nt.
0, no dC → dU was detected above background in the absence of AID.
Our data showing that AID catalyzes deamination of dC on ssDNA, a prerequisite for generation of abasic moieties required for both class-switch recombination and SHM, provides a biochemical basis for these enigmatic processes (16). The hypermutations per se are presumably made by error-prone DNA polymerases (17), pol η (18, 19), pol ι (20), and pol ζ (21), possibly while filling in base excision repair-generated gaps (22) or when error-prone or replicative polymerases copy unrepaired lesions (23). The action of AID on ssDNA might target mutations to V-gene hot spots by providing sites for nicks (24) or ds breaks (25, 26) proximal to 3′-YCRW SHM hot-spot motifs. There are several noteworthy aspects of this study. First, a nucleic acid substrate (ssDNA) has been identified for AID. Second, AID activity on ssDNA is inhibited by small ssRNA molecules (≈20–40 nt) bound to the enzyme with 1:1 stoichiometry such that treatment with RNase is required to activate AID. This observation explains why it has been so difficult to do any biochemistry with AID. It also raises potentially important questions about whether the relationship between AID and its bona fide RNA-editing homologues has allowed AID to be regulated by inhibitory RNA in vivo and if so how the activation of AID occurs. These studies also again raise the question of whether an AID-compatible ssDNA substrate might be generated during RNA transcription (Table 1), a well documented but poorly understood requirement for SHM (13–15), or whether an ssDNA substrate is generated as RNA polymerase encounters hot-spot pause sites (27). Our data showing that AID catalyzes dC → dU deamination activity most avidly on dsDNA substrates containing a 9-nt “transcription-like” ssDNA bubble offers a hint of a targeting mechanism for AID in initiating SHM.
Acknowledgments
We thank Dr. Alberto Martin and Dr. John Petruska for reading the manuscript and Dr. Kathy Hanley for generous technical assistance. This work was supported by National Institutes of Health (NIH) grants to M.F.G. (GM21422 and GM42554) and M.D.S. (CA72649 and AI53362). P.P. and R.B. were supported by NIH, National Institute on Aging, Grant T32 AG00093.
Abbreviations
- CR
cytidine
- AID
activation-induced CR deaminase
- SHM
somatic hypermutation
- CdR
deoxycytidine
- UDG
uracil DNA glycosylase
- APE
apurinic endonuclease
- ss
single-stranded
- ds
double-stranded
- ddA
2′,3′-dideoxyadenosine
- ddG
2′,3′-dideoxyguanosine
References
- 1.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, et al. Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 3.Martin A, Bardwell P D, Woo C J, Fan M, Shulman M J, Scharff M D. Nature. 2002;415:802–806. doi: 10.1038/nature714. [DOI] [PubMed] [Google Scholar]
- 4.Martin A, Scharff M D. Proc Natl Acad Sci USA. 2002;99:12304–12308. doi: 10.1073/pnas.192442899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshikawa K, Okazaki I M, Eto T, Kinoshita K, Muramatsu M, Nagaoka H, Honjo T. Science. 2002;296:2033–2036. doi: 10.1126/science.1071556. [DOI] [PubMed] [Google Scholar]
- 6.Petersen-Mahrt S K, Harris R S, Neuberger M S. Nature. 2002;418:99–103. [PubMed] [Google Scholar]
- 7.Harris R S, Petersen-Mahrt S K, Neuberger M S. Mol Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 8.Martin A, Scharff M D. Nat Rev Immunol. 2002;2:605–614. doi: 10.1038/nri858. [DOI] [PubMed] [Google Scholar]
- 9.Gerber A P, Keller W. Trends Biochem Sci. 2001;26:376–384. doi: 10.1016/s0968-0004(01)01827-8. [DOI] [PubMed] [Google Scholar]
- 10.Poltoratsky V, Goodman M F, Scharff M D. J Exp Med. 2000;192:F27–F30. doi: 10.1084/jem.192.10.f27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogozin I B, Kolchanov N A. Biochim Biophys Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu M, Sankaranand V S, Anant S, Sugai M, Kinoshita K, Davidson N O, Honjo T. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 13.Maizels N. Cell. 1995;83:9–12. doi: 10.1016/0092-8674(95)90227-9. [DOI] [PubMed] [Google Scholar]
- 14.Storb U. Immunol Rev. 1998;162:5–11. doi: 10.1111/j.1600-065x.1998.tb01424.x. [DOI] [PubMed] [Google Scholar]
- 15.Fukita Y, Jacobs H, Rajewsky K. Immunity. 1998;9:105–114. doi: 10.1016/s1074-7613(00)80592-0. [DOI] [PubMed] [Google Scholar]
- 16.Storb U, Stavnezer J. Curr Biol. 2002;12:R725–R727. doi: 10.1016/s0960-9822(02)01247-2. [DOI] [PubMed] [Google Scholar]
- 17.Goodman M F. Annu Rev Biochem. 2002;71:17–50. doi: 10.1146/annurev.biochem.71.083101.124707. [DOI] [PubMed] [Google Scholar]
- 18.Zeng X, Winter D B, Kasmer C, Kraemer K H, Lehmann A R, Gearhart P J. Nat Immunol. 2001;2:537–541. doi: 10.1038/88740. [DOI] [PubMed] [Google Scholar]
- 19.Rogozin I B, Pavlov Y I, Bebenek K, Matsuda T, Kunkel T A. Nat Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 20.Faili A, Aoufouchi S, Flatter E, Gueranger Q, Reynaud C A, Weill J C. Nature. 2002;419:944–947. doi: 10.1038/nature01117. [DOI] [PubMed] [Google Scholar]
- 21.Diaz M, Verkoczy L K, Flajnik M F, Klinman N R. J Immunol. 2001;167:327–335. doi: 10.4049/jimmunol.167.1.327. [DOI] [PubMed] [Google Scholar]
- 22.Rada C, Williams G T, Nilsen H, Barnes D E, Lindahl T, Neuberger M S. Curr Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 23.Faili A, Aoufouchi S, Gueranger Q, Zober C, Leon A, Bertocci B, Weill J C, Reynaud C A. Nat Immunol. 2002;3:815–821. doi: 10.1038/ni826. [DOI] [PubMed] [Google Scholar]
- 24.Kong Q, Maizels N. Genetics. 2001;158:369–378. doi: 10.1093/genetics/158.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papavasiliou F N, Schatz D G. Nature. 2000;408:216–221. doi: 10.1038/35041599. [DOI] [PubMed] [Google Scholar]
- 26.Bross L, Fukita Y, McBlane F, Demolliere C, Rajewsky K, Jacobs H. Immunity. 2000;13:589–597. doi: 10.1016/s1074-7613(00)00059-5. [DOI] [PubMed] [Google Scholar]
- 27.Storb U, Peters A, Kim N, Shen H M, Bozek G, Michael N, Hackett J, Jr, Klotz E, Reynolds J D, Loeb L A, Martin T E. Cold Spring Harbor Symp Quant Biol. 1999;64:227–234. doi: 10.1101/sqb.1999.64.227. [DOI] [PubMed] [Google Scholar]