Abstract
The multiplicity of proteins compared with genes in mammals owes much to alternative splicing. Splicing signals are so subtle and complex that small perturbations may allow the production of new mRNA variants. However, the flexibility of splicing can also be a liability, and several genetic diseases result from single-base changes that cause exons to be skipped during splicing. Conventional oligonucleotide strategies can block reactions but cannot restore splicing. We describe here a method by which the use of a defective exon was restored. Spinal muscular atrophy (SMA) results from mutations of the Survival Motor Neuron (SMN) gene. Mutations of SMN1 cause SMA, whereas SMN2 acts as a modifying gene. The two genes undergo alternative splicing with SMN1, producing an abundance of full-length mRNA transcripts, whereas SMN2 predominantly produces exon 7-deleted transcripts. This discrepancy is because of a single nucleotide difference in SMN2 exon 7, which disrupts an exonic splicing enhancer containing an SF2/ASF binding site. We have designed oligoribonucleotides that are complementary to exon 7 and contain exonic splicing enhancer motifs to provide trans-acting enhancers. These tailed oligoribonucleotides increased SMN2 exon 7 splicing in vitro and rescued the incorporation of SMN2 exon 7 in SMA patient fibroblasts. This treatment also resulted in the partial restoration of gems, intranuclear structures containing SMN protein that are severely reduced in patients with SMA. The use of tailed antisense oligonucleotides to recruit positively acting factors to stimulate a splicing reaction may have therapeutic applications for genetic disorders, such as SMA, in which splicing patterns are altered.
Proximal spinal muscular atrophy (SMA) is an autosomal recessive disorder characterized by the degeneration of the motor neurons in the anterior horn of the spinal cord, resulting in muscular atrophy and weakness. The overall incidence of SMA is 1 in 10,000 live births, with a carrier frequency of 1 in 50. Onset is primarily in childhood, and three different forms are recognized, type I SMA being the most severe form and type III SMA at the milder end of the scale. Children affected by SMA I never sit and usually die within the first year of life, whereas those with type III acquire the ability to walk and have a normal life expectancy. An intermediate category (SMA II) is also recognized, where affected individuals can sit unsupported but cannot walk (1). The gene implicated in SMA is the Survival of Motor Neuron (SMN) gene located on chromosome 5q13 (2). It consists of eight exons, the first seven of which encode a 294-aa protein (3). The human SMN gene exists as a mirror-image duplication with a telomeric (SMN1) and a centromeric (SMN2) copy. Mutations in SMN1 cause SMA (4), whereas the copy number of the residual SMN2 genes is believed to modify the severity of the phenotype, as suggested by the increased copy number in patients with a milder disease course (5). Deletions of both SMN1 and SMN2 have never been observed in humans, and a knockout of the single Smn gene in the mouse results in a nonviable embryo (6–9).
The SMN protein is ubiquitously expressed and localized in the cytoplasm and the nucleus, where it is involved in the process of pre-mRNA splicing. In particular it has a role in the recycling of small nuclear ribonuclear proteins (snRNPs) in the nucleus and also in spliceosomal snRNP assembly in the cytoplasm (3, 10).
The two genes differ by 11 nt, only 2 of which are contained in exons, neither of which alters the coding sequence (11). The reduced efficiency of SMN2 compared with SMN1 in protecting motor neurons from degeneration results from differences in the splicing of exon 7. The SMN1 gene produces primarily full-length SMN transcripts, whereas the predominant transcript derived from SMN2 lacks exon 7. This difference is caused by a single nucleotide, position +6 in exon 7 being a T instead of a C in SMN2 (11, 12). This nucleotide change abolishes the ability of SF2/ASF, a serine/arginine-rich (SR) protein, to bind SMN2 pre-mRNA in this region, thereby reducing the recognition of SMN2 exon 7 by the spliceosome (13).
The retention of intact copies of SMN2 in all patients with SMA has led investigators to devise strategies to enhance the incorporation of exon 7 in pre-mRNA splicing, because this might have therapeutic implications for patients with SMA. Pharmacological agents such as sodium butyrate and aclarubicin are effective, but there are concerns regarding the widespread effect that these substances have on the splicing of other genes; in addition, there is some toxicity associated with their administration (14, 15). More specific splicing modulation was achieved by the use of antisense oligoribonucleotides to block the 3′ splice site of exon 8, producing an increased level of exon 7 incorporation in transfected minigenes (16). However, the pattern of splicing of the endogenous mRNA was not analyzed, and it is quite likely that blocking the 3′ splice site of exon 8 in the endogenous gene would result in the use of cryptic splice sites. In other genes, where the target exon might not be the penultimate exon, blocking the downstream 3′ splice site is likely to lead to skipping of the downstream exons. It would be of much greater value if a method could be devised that would allow the level of inclusion of a specific exon to be increased without compromising the splicing of its neighbors.
Here we describe a strategy to modify the splicing of SMN2 that would, in principle, be widely applicable whenever mutations that affect exonic splicing enhancers (ESEs) have produced suboptimal levels of incorporation during splicing. We designed oligonucleotides that, although complementary to the target exon, do not block reactions at their binding sites like conventional antisense RNA. Instead, the oligonucleotides incorporate a noncomplementary “tail” consisting of sequences that mimic ESEs (17). We show here that these tailed oligonucleotides induce the inclusion of SMN2 exon 7 with high efficiency in a cell-free splicing assay. We also show that this approach was successful in vivo: the proportion of exon 7 inclusion in mRNA from the endogenous SMN2 gene was increased in fibroblasts from patients with SMA to match the levels seen in control fibroblasts, and the formation of gems, intranuclear structures containing SMN, was partially rescued.
Methods
β-Globin/SMN Constructs for Use in Cell-Free Splicing Assays.
Rabbit β-globin exon 2, intron 2, and the beginning of exon 3 were amplified with the primers BGEX2F and BGEX3R (see end of Methods for sequences) and cloned into the pCR-Blunt II-TOPO cloning vector (Invitrogen). SMN1 and SMN2 exons 7 and flanking regions were amplified from previously sequenced clones with the primers SALRIIF and SALSMNR designed to create SalI sites. SalI-digested PCR products were then cloned into the similarly digested TOPO/β-globin vector created previously. The SalI site is situated within the intronic region between the two β-globin exons such that SMN1 or SMN2 exon 7 and intronic regions will be situated between the two β-globin exons. The stop codon at the end of SMN exon 7 was then altered to allow read-through of the β-globin/SMN constructs.
In Vitro Transcription Mix.
Novel primers with the forward primer incorporating the T7 promoter sequence, T7BGEX2F, and BGEX3R, situated in β-globin exon 3, were used to amplify SMN exon 7 and flanking β-globin exons from the β-globin/SMN1 and β-globin/SMN2 constructs, resulting in an 800-bp product. One hundred nanograms of the PCR products were then combined in an in vitro transcription mix and the transcripts were labeled with [α-32P]GTP at 37°C for 3–4 h. F-dyes (10 μl) were then added and the mixture was run on a 5% polyacrylamide gel at 30 W.
Visualization of Transcription Products.
The gel plates were separated and the gel was exposed to Biomax x-ray film (Kodak) for 1–5 min before developing. The transcript bands were cut out of the gel, placed in SDS lysis buffer, and left at 4°C overnight.
In Vitro Splicing.
The transcripts were ethanol-precipitated and resuspended in 20 μl of TE buffer (10 mM Tris/1 mM EDTA, pH 8) containing 0.1% RNase inhibitor (RNasin, Promega). A stock splicing mix (18) was made containing 0.5 μl of 100 mM ATP, 4 μl of 0.5 M creatine phosphate, 4 μl of 80 mM MgCl2, 2 μl of Hepes buffer, 0.3 μl of RNasin, and 17 μl of 13% polyvinyl alcohol. Finally, 40 μl of HeLa nuclear extract and 20 μl of DGlu buffer (buffer D with 80 mM potassium glutamate replacing 0.1 M KCl) (19) were added. Splicing mix (4.5 μl) was added to 0.5 μl of each labeled transcript. A timed assay was carried out with reactions incubated at 30°C for 0 min, 30 min, and 1, 2, and 3 h before being frozen. Proteinase K stop mix (50 μl) was added to the thawed reactions and placed at 37°C for 10 min. The samples were then ethanol-precipitated and resuspended in 10 μl of F-dyes, and 3 μl was run on a 5% denaturing polyacrylamide gel. The fixed and dried gel was then exposed to a phosphor screen and imagequant software was used for quantification.
ESE-Tailed Antisense Oligonucleotides.
A series of tailed antisense oligonucleotides were designed. They all contained both 2′-O-methyl and phosphorothioate modifications and were obtained from Eurogentec (Brussels). These oligonucleotides were complementary to the 5′ end of SMN2 exon 7 and, in addition, contained tails designed to mimic ESE sequences. The oligonucleotides were incorporated at concentrations of 0, 50, 100, 200, and 250 nM and preincubated for 10 min with the SMN2 transcript at 30°C before the addition of the splicing mix. The reactions were allowed to proceed for 3 h at 30°C. All experiments were repeated in triplicate, and the relative abundance of the spliced products was normalized against SMN1 readings (included as an internal control) and the means plotted on a graph.
Oligonucleotide Sequences.
In these sequences, the tail region is given in lowercase and the complementary (antisense) RNA sequence is in uppercase. The letters “o” and “s” refer to 2′-O-methyl and phosphorothioate chemical groups, respectively. 5′GGA, 5′-asgsgsasgsgacggaggacggaggacaGoAoUosUosUosUosGosUoCoUoAoAosAosAosCo-3′; 5′polypyrimidine tract binding protein (PTB), 5′-asuscsususuccucuuuccucuuuccaGoAoUosUosUosUosGosUoCoUoAoAosAosAosCo-3′; 5′A1, 5′-asusasgsgsgcaggcuagggcaggccaGoAoUosUosUosUosGosUoCoUoAoAosAosAosCo-3′; NT (no tail), 5′-GosAosUosUosUosUoGoUoCoUosAosAosAosAosCo-3′; Scram, 5′-oAosoCosoCosoCosoUosoGoUoCoUoUosoAosoGosoGosoUoso-3′; and RevGGA, 5′-asgsgsasgsgacggaggacggaggacacgosAosAosAosUoCoUoGoUoUoUoUosAosGosUosUosU-3′.
SF2/ASF Binding Assays.
Biotinylated SMN2 RNA was produced by transcription of a PCR product that comprised exon 7 with an additional 12-nt 3′ extension that provided a strong U1 small nuclear ribonuclear protein binding site. RNA (10 pmol) was incubated with 10 pmol of the 5′GGA oligonucleotide at 30°C for 10 min in Dglu buffer. The RNA was added to 10 μl of streptavidin agarose beads (Sigma) prewashed in 20 mM Hepes (pH 8), 150 mM NaCl, and 0.05% Triton X-100. After 2 h at 4°C, the beads were washed three times by centrifugation in the same buffer for 2 minutes in a microfuge at 3,000 rpm (850 × g). A standard splicing reaction mixture (74 μl) containing HeLa cell nuclear extract was added. After incubation at 30°C for 10 min, the beads were washed three times as above but without centrifugation, and the proteins were eluted and separated by 12% SDS/PAGE. The separated proteins were transferred to nitrocellulose membrane and detected with anti-SF2 Ab and protein A/G peroxidase (Pierce). Chemiluminescence was detected on film, and the intensity was measured by using a Kodak EDAS 290 camera system. Quantification of the image used optiquant (Packard). For the rabbit β-globin control, the biotinylated RNA contained exon 2, a truncated version of intron 2, and 50 nucleotides of exon 3, amounting to ≈380 nucleotides.
Cell Culture and Transfections.
SV-40-transformed human SMA type I fibroblast cell lines derived from two different patients were grown in DMEM containing 10% (vol/vol) FCS and 2 mM glutamine. Cells were plated at 3 × 104 cells per well in 24-well plates 18–24 h before transfection. Each well was treated with 50, 100, 250, or 500 nM oligonucleotide complexed with jetPEI (Qbiogene, Nottingham, U.K.) transfection reagent. After a 5-h incubation, 10% FCS media was added. Transfections using the 5′GGA oligonucleotides were carried out three to five times, whereas single transfections were performed with control oligonucleotides (5′A1 and NT).
Semiquantitative RT-PCR of SMN Transcripts.
Total RNA was extracted 24 h after transfection by means of the Qiagen RNeasy kit. First-strand cDNA synthesis was carried out with Superscript II reverse transcriptase (Invitrogen). The endogenous SMN transcripts were amplified by using the primers 541C618 and 541C1120 situated in exons 6 and 8, respectively, of the SMN gene (20). The PCR consisted of 20 cycles and was carried out in the presence of [α-32P]dATP. The resulting PCR products were boiled and run on a 6% denaturing polyacrylamide gel, and imagequant software was used for quantification.
To exclude the possibility of unequal efficiency of cDNA amplification arising during RT-PCR because of differing concentrations of input cDNA, an 8-fold range of concentrations of starting cDNA was tested by using cDNA from the patient-derived fibroblast cell line. The ratios of the PCR products derived from mRNA produced by inclusion and skipping of exon 7 were constant across the range. Without transfection, the percentage of spliced mRNA that had incorporated the exon was 52.6 (SD = 0.7). After transfection with 250 nM GGA oligonucleotide, the percentage was 83.3 (SD = 5.1). There was no detectable effect of starting cDNA concentrations.
The validity of the PCR amplification was also checked by cycle curves. The amplification efficiency of the shorter isoform (excluding exon 7) was calculated to be higher by a factor of ≈1.02 (18). Thus, the maximum possible distortion of the ratio over 20 cycles is ≈1.5-fold, whereas the ratio of included/excluded isoforms detected by RT-PCR changes after transfection from 1.1 to 4.9, a difference of >4-fold. We conclude that RT-PCR is an insignificant source of errors.
Immunohistochemistry.
The SMA fibroblast cell lines were plated on collagen-coated (Nutacon, Leimuiden, The Netherlands) coverslips in 24-well plates, and the oligonucleotide transfections were performed the following day. Twenty-four hours after transfection, immunofluorescent staining was carried out as described (21). The anti-SMN mAb MANSMA2 (22) was diluted 1:100, and a fluorophore-labeled donkey anti-mouse IgG diluted 1:2,000 was used to visualize the anti-SMN Ab staining. The coverslips were mounted with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories), and the cells were visualized on a Leica confocal microscope by using the ×100 objective.
Primer Sequences.
BGEX2F, GGGCTGCTGGT TGTCTACCCA; BGEX3R, AACTTACCTGC CAAAAATGATGAGACA; SALRIIF, ATTCATGTTATATGGTCGACAGACT ATCAACTTAATTTCTG; SALSMNR, GTTAGCAGAGTCGACGTGATATAA AATGGCATATC; and T7BGEX2F, AAATTAATACGACTCACTATAGGGCTGCTGGTTGTCTACCCA.
Results
Replication of SMN Alternative Splicing Within an in Vitro System.
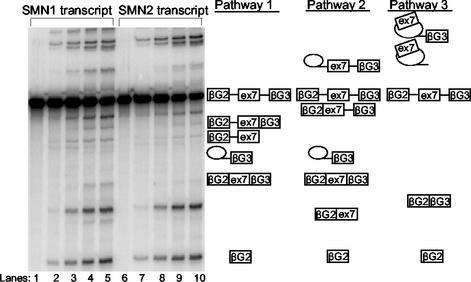
To perform our initial experiments, a well characterized β-globin splicing construct was chosen into which SMN1 and SMN2 exons 7 and flanking intronic regions were inserted. The results of the SMN1 and SMN2 in vitro splicing assays can be seen in Fig. 1, where the SMN sequences were set between exons 2 and 3 of rabbit β-globin. Splicing reactions with three exons follow three possible pathways for splicing: skipping, inclusion via splicing of intron 1 before intron 2, and inclusion via splicing of intron 2 before intron 1. Most of the bands could be assigned by direct side-by-side comparisons of the two reactions, with the exception of the faint bands containing a single intron in a lariat. Based on these assignments, we were able to identify the mRNA derived by skipping and inclusion of exon 7. The faint band below the skipped mRNA, i.e., β-globin exons 2 and 3 spliced together, is an intermediate in the inclusion pathway that proceeds via splicing of intron 1 before intron 2. In subsequent experiments, a shorter transcript, containing a 3′ exon that was 85 nt smaller, was used because of its higher efficiency of splicing.
Figure 1.
In vitro splicing assay incorporating SMN1 and SMN2 transcripts shows alternative splicing. Denaturing polyacrylamide gel (5%) showing an in vitro timed splicing assay using α-32P-labeled SMN1 and SMN2 transcripts. Symbols identifying the various splicing intermediates and products are shown. Lanes 1–5 and 6–10 represent different time points. Lanes 1 and 6, 0 min; lanes 2 and 7, 30 min; lanes 3 and 8, 1 h; lanes 4 and 9, 2 h; lanes 5 and 10, 3 h. Also shown are the three different splicing pathways that occur; pathways 1 and 2 promote exon 7 inclusion, whereas the third pathway skips exon 7.
The splicing efficiencies of both the longer and shorter transcripts recapitulated the splicing pattern of endogenous human SMN genes. When comparing the intensities of the bands representing exon 7 inclusion with the total amount of spliced product, for the shorter transcript, we found that SMN1 exon 7 inclusion was on average 3.5-fold higher (24.6 ± 6.3% mean inclusion) than that of SMN2 (7 ± 0.74% mean inclusion). The readings varied between 19% and 32.3% for the SMN1 and between 6.5% and 8.1% for the SMN2 transcripts. Exon inclusion seemed to result from both possible pathways, but the majority of transcripts seemed to follow the route in which intron 1 was removed first.
“Tailed” Antisense Oligonucleotides Increase Exon 7 Inclusion.
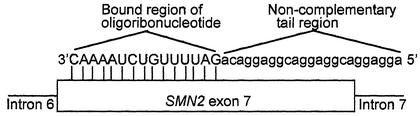
Antisense oligoribonucleotides were designed that were complementary to SMN2 exon 7 and contained additional noncomplementary sequences (tails) that were predicted to mimic ESE sequences (Fig. 2). One oligonucleotide (named 5′GGA) contained a tail of GGA repeats, the sequence GGAGGA being known to be an active enhancer sequence (23). Control oligonucleotides contained either no tail or consisted of a scrambled sequence.
Figure 2.
Diagrammatical representation of the tailed oligoribonucleotides bound to SMN2 exon 7. The complementary RNA sequence of the oligoribonucleotide is uppercase, whereas the tail region containing sequences that mimic ESEs are lowercase. The oligonucleotide binds via a complementary region to the first part of SMN2 exon 7 (diagram not to scale) indicated by the vertical lines, from position +1 on exon 7 to position +16, thereby leaving 38 nucleotides of the exon unannealed by the oligonucleotide. The noncomplementary tail region remains unbound and is available to bind to splicing proteins in the in vitro splicing reaction mix.
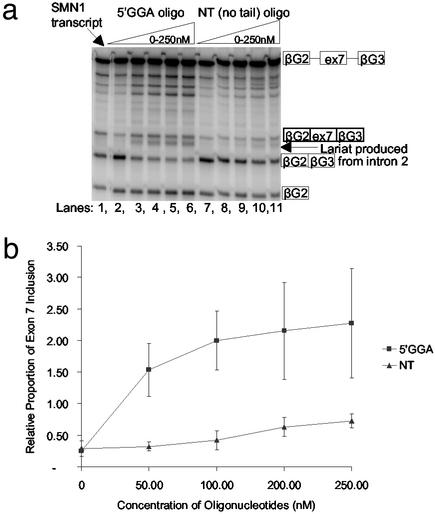
The oligonucleotides were incubated with the pre-mRNA substrate and then mixed with a splicing reaction mixture. The 5′GGA oligonucleotides reduced the level of the SMN2 exon 7 skipped product even at the lowest concentration (50 nM) and increased the level of the mRNA product including exon 7 (Fig. 3). In contrast, the NT oligonucleotide, lacking the ESE sequence, had very little effect.
Figure 3.
Tailed 5′GGA oligonucleotides promote exon 7 inclusion. (a) Cell-free in vitro splicing assay using α-32P-labeled SMN2 transcripts combined with the 5′GGA and NT oligonucleotides at concentrations between 0 and 250 nM. The SMN1 transcript is included in the first lane. (b) Graph showing exon 7 inclusion relative to the SMN1 level of splicing with increasing concentrations of oligonucleotides. The SMN1 transcript was included in all gels as an internal control, enabling successive gels to be directly correlated. The “relative proportion” label on the y axis indicates that the readings have been normalized against the readings obtained for the SMN1 transcripts in each gel. The results of three experiments were combined to produce these data with SDs varying from 0.03 to 0.86. The readings were not corrected for the differing amounts of labeled radionucleotides.
A comparison of the bands produced by splicing in the presence of the 5′GGA oligonucleotide with the pattern of SMN1 splicing shows that the oligonucleotide produces a relatively high level of bands corresponding to pathway 1 linear intermediates containing the first intron (compare Fig. 1). These bands suggest that pathway 1 is promoted, i.e., that removal of intron 2 is accelerated and that the pathway intermediates accumulate because splicing of intron 1 is limiting. Another relatively abundant band can be seen underneath the inclusion mRNA (Fig. 3a), which may represent intron 2, but this has not been formally verified.
The Increase in SMN2 Exon 7 Inclusion at Low Concentrations of Oligonucleotide Requires an Enhancer-Like Sequence and Base Pairing to Exon 7.
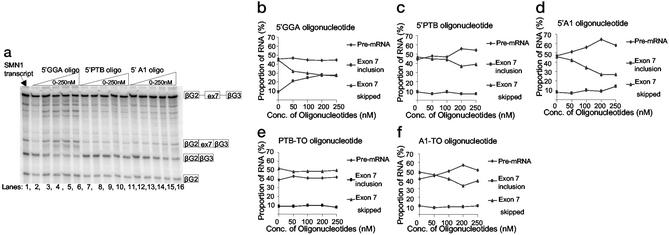
To determine whether the increase in exon 7 inclusion was a specific consequence of attaching the GGA repeat sequence to SMN2 exon 7, we tested other tailed oligonucleotides that lacked ESE sequences but would anneal to the same target site. In these oligonucleotides (called 5′PTB and 5′A1), the ESE sequences were replaced with sequences that bind to inhibitors of splicing such as PTB or hnRNP A1, respectively (24–27). In contrast to the 5′GGA oligonucleotide, both these oligonucleotides and the NT oligonucleotide demonstrated that annealing at the 5′ end of SMN2 exon 7 was insufficient to stimulate incorporation (Fig. 4a). The 5′PTB oligonucleotide had no effect at any concentration (Fig. 4c), whereas the 5′A1 oligonucleotide inhibited skipping of SMN2 exon 7 at the higher concentrations (Fig. 4d).
Figure 4.
Application of the 5′GGA, 5′PTB, 5′A1, PTB-TO, and A1-TO oligonucleotides to SMN2 transcripts. (a) Denaturing polyacrylamide gel (5%) with 5′GGA, 5′PTB, and 5′A1 oligonucleotides incorporated at increasing concentrations with the SMN2 transcripts. The SMN1 transcript is included in the first lane. (b–f) Graphs showing the percentage of RNA in the initial pre-mRNA transcript, the exon 7-included product, and the skipped product at increasing concentrations of each separate oligonucleotide. The products have been corrected for the numbers of labeled radionucleotides in each form of RNA. These graphs were plotted from a single experiment, but the results were reproducible in at least three different experiments; however, the error bars have been excluded to avoid confusion of the results shown.
We also tested oligonucleotides consisting only of the tail regions of 5′PTB and 5′A1, called PTB-tail only (TO) and A1-TO. In the absence of the sequence complementary to SMN2 exon 7, the A1-TO oligonucleotide again inhibited splicing, suggesting that this oligonucleotide was sequestering protein in the nuclear extract without annealing to SMN2 exon 7 (Fig. 4f).
To determine whether base pairing of the oligonucleotide to the substrate was required, we tested an oligonucleotide containing the GGA repeat ESE sequence in which the sequence complementary to SMN2 exon 7 was reversed (called RevGGA). This oligonucleotide produced only a small and nonsaturable change up to concentrations of 1 μM, irrespective of the presence of the ESE sequence (data not shown).
5′GGA Oligonucleotide Mediates Binding of SF2/ASF.
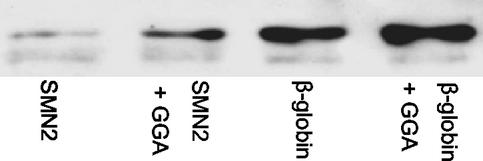
The increased use of the exon produced by annealing to 5′GGA should be associated with recruitment of splicing proteins known to bind ESE sequences. One such protein is SF2/ASF, which is both limiting for SMN2 exon 7 (13) and able to bind GGA repeat sequences (23). Thus, we tested whether there was increased recruitment of SF2/ASF to SMN2 exon 7 in the presence of the 5′GGA oligonucleotide. Biotinylated SMN2 exon 7 was incubated with the oligonucleotide, retained on beads, and then incubated in nuclear extract. Bound SF2/ASF was detected after washing by Western blotting. In three separate experiments, the binding rose by 150–280% when the 5′GGA oligonucleotide was present (Fig. 5), whereas, in a parallel reaction with β-globin, binding rose by just 50%. We conclude that the 5′GGA oligonucleotide does mediate an increase in the binding of at least one protein that might be expected to bind to an ESE.
Figure 5.
Recruitment of SF2/ASF to SMN2 exon 7 by the 5′GGA oligonucleotide. Biotinylated RNA (SMN2 exon 7 or β-globin, as indicated) was bound to streptavidin beads and incubated in nuclear extract. The proteins associated with the RNA were separated by SDS/PAGE, and SF2/ASF was detected by Western blotting. Lanes “+GGA” indicate that the RNA was incubated with the GGA oligonucleotide before addition to the beads.
Increase in Exon 7 Inclusion Within the Endogenous SMN2 Gene.
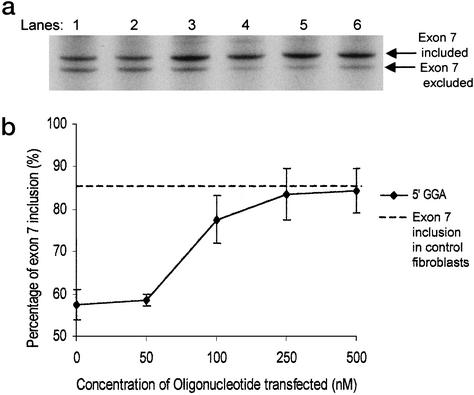
To verify that our in vitro data were reproducible in an in vivo system, the tailed antisense oligonucleotides were transfected into SMA type I patient fibroblasts at various concentrations between 50 and 500 nM. By using a semiquantitative RT-PCR to analyze the splicing pattern of the 5′GGA oligonucleotide-transfected cells, a clear dose-dependent increase in exon 7 inclusion could be seen 24 h after transfection (Fig. 6a). This increase in exon 7 inclusion changed from 57% exon 7 inclusion in untransfected cells to 84% in cells transfected with 500 nM 5′GGA oligonucleotide. This increase matched the level of exon 7 inclusion seen in control fibroblasts (Fig. 6b). The same effect was seen in both SMA patient cell lines. These results strengthen our hypothesis that the tailed oligonucleotides are capable of acting as ESE sequences in trans to create a positive effect on splicing of the SMN2 gene. The transfection efficiency of the oligonucleotides was investigated by means of a fluorescently tagged oligonucleotide of similar length to the oligonucleotides used here. An effective transfection efficiency of 90% was calculated.
Figure 6.
Transfection of type I SMA patient fibroblasts. (a) Denaturing polyacrylamide gel (6%) showing the results of a semiquantitative RT-PCR, using primers situated in exons 6 and 8 of the SMN gene, was carried out on cDNA from cells transfected with increasing concentrations of the 5′GGA oligonucleotide. Lane 1, untransfected cells; lane 2, 50 nM oligonucleotide-transfected; lane 3, 100 nM oligonucleotide-transfected; lane 4, 250 nM oligonucleotide-transfected; lane 5, 500 nM oligonucleotide-transfected; lane 6, normal control. (b) Graph showing the percentage of exon 7 inclusion in the transcripts derived from the 5′GGA transfected cells. The results of three different transfection experiments were combined to produce the graph. The readings were corrected for the amounts of labeled radionucleotides, and the percentage of exon 7 inclusion was calculated by exon 7 inclusion mRNA/total mRNA. The horizontal dashed line represents the percentage of exon 7 inclusion obtained in control fibroblasts.
To investigate the specificity of the above results, the 5′A1 and NT oligonucleotides were also transfected into the SMA type I fibroblasts and the splicing patterns were examined. Both the 5′A1 and NT oligonucleotide produced little or no effect on exon 7 inclusion (data not shown), indicating a clear necessity for an appropriate ESE sequence.
Increased SMN Protein Expression in SMA Type I Fibroblasts.
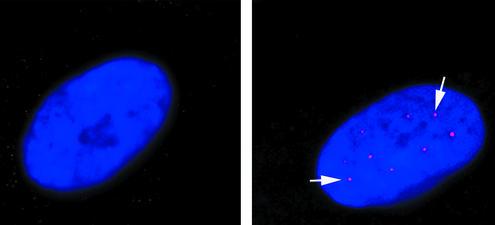
The physiological significance of the increased exon 7 inclusion in vivo was examined by the analysis of gems. These are intranuclear structures in which SMN protein accumulates. Gem numbers in fibroblasts have been shown to correlate with phenotypic severity and are thus an indication of the amount of SMN protein present (21). Our control fibroblasts showed gem staining in 52% of the nuclei, with three to six gems per nucleus. In the SMA type I fibroblasts used, gems were observed in only 2–3% of nuclei, and each positive nucleus contained only a single gem. After transfection of the SMA fibroblasts, the number of positive nuclei rose to 13% and there was a very striking increase in the number of gems per nucleus, which rose to WT levels of three to six (Fig. 7). The number of gem-positive nuclei we observed was intermediate between that found in type I SMA and in carrier parent fibroblast cell lines (25% gem-positive nuclei) (21).
Figure 7.
Restoration of gems in SMA patient fibroblasts. Images show untransfected (a) and transfected (b) SMA type I fibroblasts. 4′,6-Diamidino-2-phenylindole staining highlights the nuclei in blue, and the white arrows indicate the gems (red dots in nucleus). Untransfected cells show 2–3% of their nuclei containing gems, whereas transfected cells show 13% gem-positive nuclei.
Discussion
The two SMN genes are ≈99% similar (2). A single nucleotide difference in exon 7 results in the different splicing characteristics of the SMN1 and SMN2 genes. The SMN2 gene generates a smaller proportion of full-length RNA transcripts and a low level of SMN protein that only partially compensates for the lack of SMN1-derived protein. Correcting the deficient splicing of the SMN2 gene should increase SMN protein production and provide a therapeutic benefit to patients with SMA. Alteration of SMN2 splicing has been attempted by various investigators, some of whom have used inhibitory antisense oligonucleotides (16), but the magnitude and specificity of effects have so far been very low, making their possible therapeutic use unlikely.
ESE sequences are predominantly found in exons flanked by weak splice sites (28). In one model for their effects, they are bound by SR or other proteins that promote spliceosome formation, aiding the recognition of nearby splice sites and activating splicing (29–33). We have devised a strategy that takes advantage of an antisense oligonucleotide approach but, in contrast to the conventional use of such oligonucleotides as physical obstructions of a reaction at a target site or as mediators of RNase H degradation, we have used these oligonucleotides to attach potent enhancer sequences to SMN2 exon 7, which is then incorporated in the mature transcript.
Our results suggest that the use of tailed oligonucleotides containing binding motifs for splicing activators might be a useful approach for controlling splicing efficiency. We have shown that the 5′GGA tailed antisense oligonucleotide results in very significant changes in the relative proportions of skipping and inclusion of exon 7 within SMN2 both in vitro and in vivo. The level of exon 7 incorporation obtained with the 5′GGA oligonucleotide in our in vitro experiments reached approximately that seen with the SMN1 transcripts, whereas the proportion of full-length mRNA produced from SMN2 genes in patient fibroblasts reached that of control fibroblasts at oligonucleotide concentrations as low as 250 nM. Furthermore, a significant increase in the number of gems was observed, indicating a partial restoration of full-length fully functional SMN. However, because gems were not completely restored to the levels found in control individuals, it is possible that the overall level of expression from the SMN2 gene is still limiting. Further investigations will be required to resolve this issue and improve the efficiency of protein restoration. Nevertheless, these results are encouraging, because the level of gem restoration in a patient with type I SMA was similar to those observed in patients with a milder form of SMA, indicating that these levels are likely to be functional.
The use of these antisense oligonucleotides to enhance expression of latent exons may ultimately be of therapeutic use. Various phosphorothioate oligonucleotides have reached phases I and II in clinical trials for the treatment of viral infections and cancer (34). In particular, a drug which is composed of a phosphorothioate oligonucleotide, designed to inhibit human cytomegalovirus replication, has been licensed recently (34). However, with regard to our method of specific up-regulation using tailed antisense oligonucleotides, there are three critical issues that need to be addressed. First, we are investigating strategies for optimizing the response to the oligonucleotide, initially by looking at finding the optimal position for the trans-acting enhancers and also by optimizing the enhancer sequences. Second, the type of chemical modification might be important. Recent studies have suggested that there are a number of alternatives to phosphorothioate oligonucleotides that possesses lower toxicity. Finally, the systemic administration of these oligonucleotides might not be successful because of their inability to cross the blood–brain barrier. However, progress is being made in this field with synthetic chemistries that are able to cross the blood–brain barrier (35, 36), a factor that will be of great importance in terms of therapeutic possibilities for patients with SMA.
Approximately 15% of point mutations identified produce splicing abnormalities resulting in increased exon inclusion or exclusion ultimately culminating in genetic disease (31). Our approach of using “tailed” oligonucleotides to alter splicing thus represents a promising new therapeutic approach not only for SMA but for a variety of genetic disorders. In therapeutic practice, systemic or generalized administration of SR or SR-related proteins might have a detrimental effect because of their action on multiple genes, as suggested by the toxicity observed in the experiments to produce stable transfectants expressing SR proteins (15). Instead, the method we describe here should allow specific exons to be activated at very low concentrations of oligonucleotide, especially when the issue of transport across the blood–brain barrier is resolved. The method may also have more immediate practical benefits for research in that the ability to induce incorporation of latent exons in vivo might be useful in studies of splicing mechanisms or the functions of protein isoforms.
Acknowledgments
We thank A. R. Krainer (Cold Spring Harbor) for the mAb against SF2/ASF and for discussions and exchange of results before publication. We also thank K. Davies and N. Owen (University of Oxford) for the patient cell lines and G. Morris (MRIC, Wrexham, U.K.) for the mAb against SMN. We also thank S. Brown and S. Torelli for their help in the protein studies performed. The Motor Neuron Disease Association (U.K.), the Wellcome Trust, and the Families of SMA (U.S.) supported this work. The principle of the method described here is the subject of a patent application by the University of Leicester and Imperial College London.
Abbreviations
- SMA
spinal muscular atrophy
- ESE
exonic splicing enhancer
- SR
serine/arginine-rich
- NT
no tail
- TO
tail only
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dubowitz V. Muscle Disorders in Childhood. 2nd Ed. London: Saunders; 1995. [Google Scholar]
- 2.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 3.Liu Q, Dreyfuss G. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 4.McAndrew P E, Parsons D W, Simard L R, Rochette C, Ray P N, Mendell J R, Prior T W, Burghes A H M. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taylor J E, Thomas N H, Lewis C M, Abbs S J, Rodrigues N R, Davies K E, Mathew C G. Eur J Hum Genet. 1998;6:467–474. doi: 10.1038/sj.ejhg.5200210. [DOI] [PubMed] [Google Scholar]
- 6.Battaglia G, Princivalle A, Forti F, Lizier C, Zeviani M. Hum Mol Genet. 1997;6:1961–19711. doi: 10.1093/hmg/6.11.1961. [DOI] [PubMed] [Google Scholar]
- 7.Schrank B, Gotz R, Gunnersen J, Ure J, Toyka K, Smith A, Sendtner M. Proc Natl Acad Sci USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsieh-Li H M, Chang J, Jong Y, Wu M, Wang N M, Tsai C H, Li H. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 9.Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- 10.Mattaj I W. Curr Biol. 1998;8:93–95. doi: 10.1016/s0960-9822(98)70055-7. [DOI] [PubMed] [Google Scholar]
- 11.Monani U R, Lorson C L, Parsons D W, Prior T W, Androphy E J, Burghes A H M, McPherson J D. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 12.Lorson C L, Hahnen E, Androphy E J, Wirth B. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartegni L, Krainer A R. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 14.Chang J G, Hsieh-Li H M, Jong Y J, Wang N M, Tsai C H, Li H. Proc Natl Acad Sci USA. 2001;98:9808–9813. doi: 10.1073/pnas.171105098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreassi C, Jarecki J, Zhou J, Coovert D D, Monani U R, Chen X, Whitney M, Pollok B, Zhang M, Androphy E, et al. Hum Mol Genet. 2001;10:2841–2849. doi: 10.1093/hmg/10.24.2841. [DOI] [PubMed] [Google Scholar]
- 16.Lim S R, Hertel K J. J Biol Chem. 2001;276:45476–45483. doi: 10.1074/jbc.M107632200. [DOI] [PubMed] [Google Scholar]
- 17.Cartegni L, Chew S L, Krainer A R. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 18.Eperon I C, Krainer A R. In: RNA Processing: A Practical Approach. Higgins S J, Hames B D, editors. Vol. 1. Oxford: IRL; 1994. pp. 57–101. [Google Scholar]
- 19.Black D L. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 20.Parsons D W, McAndrew P E, Innaccone S T, Mendell J R, Burghes A H M, Prior T W. Am J Hum Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coovert D D, Le T T, McAndrew P E, Strasswimmer J, Crawford T O, Mendell J R, Coulson S E, Androphy E J, Prior T W, Burghes A H M. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 22.Young P J, Le T T, Man N, Burghes A H M, Morris G E. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 23.Liu H X, Zhang M, Krainer A R. Genes Dev. 1998;12:1998–2012. doi: 10.1101/gad.12.13.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chabot B. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 25.Cartegni L, Maconi M, Morandi E, Cobianchi F, Riva S, Biamonti G. J Mol Biol. 1996;259:337–348. doi: 10.1006/jmbi.1996.0324. [DOI] [PubMed] [Google Scholar]
- 26.Del Gatto-Konczak F, Olive M, Gesnel M C, Breathnach R. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu J, Mayeda A, Krainer A R. Mol Cell. 2001;8:1351–1361. doi: 10.1016/s1097-2765(01)00409-9. [DOI] [PubMed] [Google Scholar]
- 28.Fairbrother W G, Yeh R F, Sharp P A, Burge C B. Science. 2002;9:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 29.Zhu J, Krainer A R. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graveley B R. RNA. 2000;6:1197–1211. doi: 10.1017/s1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blencowe B J. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 32.Graveley B R, Hertel K J, Maniatis T. RNA. 2001;7:806–818. doi: 10.1017/s1355838201010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guth S, Tange T O, Kellenberger E, Valcarcel J. Mol Cell Biol. 2001;21:7673–7681. doi: 10.1128/MCB.21.22.7673-7681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galderisi U, Cascino A, Giordano A. J Cell Physiol. 1999;181:251–257. doi: 10.1002/(SICI)1097-4652(199911)181:2<251::AID-JCP7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 35.Mercatante D R, Sazani P, Kole R. Curr Cancer Drug Targets. 2001;3:211–230. doi: 10.2174/1568009013334124. [DOI] [PubMed] [Google Scholar]
- 36.Estibeiro P, Godfray J. Trends Neurosci. 2001;24:S56–S62. doi: 10.1016/s0166-2236(00)01968-8. [DOI] [PubMed] [Google Scholar]