Abstract
The surface density of the triggering receptors responsible for the natural killer (NK)-mediated cytotoxicity is crucial for the ability of NK cells to kill susceptible target cells. In this study, we show that transforming growth factor β1 (TGFβ1) down-regulates the surface expression of NKp30 and in part of NKG2D but not that of other triggering receptors such as NKp46. The TGFβ1-mediated inhibition of NKp30 surface expression reflects gene regulation at the transcriptional level. NKp30 has been shown to represent the major receptor involved in the NK-mediated killing of dendritic cells. Accordingly, the TGFβ1-dependent down-regulation of NKp30 expression profoundly inhibited the NK-mediated killing of dendritic cells. On the contrary, killing of different NK-susceptible tumor cell lines was variably affected, reflecting the differential usage of NKp30 and/or NKG2D in the lysis of such tumors. Our present data suggest a possible mechanism by which TGFβ1-producing dendritic cells may acquire resistance to the NK-mediated attack.
In the past 10 years, two concepts emerged that shed light on how human natural killer (NK) cells function. First, NK cells express a series of inhibitory receptors that on recognition of HLA class I molecules down-regulate their cytolytic activity (1–3). As a consequence, normal cells expressing physiological amounts of HLA class I molecules are protected from NK-mediated killing. Second, NK cells are induced to kill target cells when the interaction between inhibitory receptor and HLA class I does not occur, as in the case of allogeneic cells or in the case of HLA class I-defective targets (such as certain tumor or virally infected cells) (4). Target cell killing depends on the engagement of ligands specifically recognized by activating receptors and coreceptors expressed at the NK cell surface. Among these, the NK-specific NKp46, NKp30, and NKp44, collectively termed natural cytotoxicity receptors (NCR) (5), and NKG2D (6, 7) appear to play a major role in the NK-mediated cytotoxicity. Thus, their simultaneous blocking by specific mAbs results in the virtual abrogation of the NK-mediated cytolytic activity against the majority of target cells. In NK cell populations (both resting and activated) and in NK cell clones derived from healthy individuals, NCR display a coordinated surface expression (8). Moreover, unlike NKG2D, NCR can be expressed at high or low surface density. Although NCRbright and NCRdull NK cells were characterized by a similar cytolytic potential and by a comparable surface expression of NKG2D, they greatly varied in their capability of killing various tumor target cell lines (8).
The relative proportion of NCRbright or NCRdull NK cells is different in different individuals. In healthy donors, NCRdull cells usually represent a minor fraction of the whole NK cell population. On the contrary, in certain pathological conditions, such as acute myeloid leukemia (AML), most patients' NK cells were found to express a homogeneous NCRdull phenotype (9). This, at least in some cases, was also paralleled by an unusually reduced surface expression of NKG2D. It was unclear whether the NCRdull and NKG2Dlow phenotype represented a characteristic of these individuals preexisting the onset of the disease, or was rather consequent to the disease itself. In this case, the decreased surface expression of NCR or NKG2D could result from an effect of the microenvironment, possibly mediated by cytokines. In this context, transforming growth factor β1 (TGFβ1) has been shown to inhibit human cytotoxic T lymphocyte- and, in part, human NK-mediated antitumor cytotoxicity (10–12). On the other hand, no information has been provided so far that cytokines known to exert an immunomodulatory role in immune responses can modulate the surface expression of triggering NK receptors.
In this study we show that in the presence of TGFβ1, a strong down-regulation of the surface expression of NKp30 and, at least in part, of NKG2D occurs in NK cells. The expression of NKp46 and other triggering receptors and coreceptors was not modified. In accordance with the recent finding that NKp30 is the major receptor responsible for the NK-mediated recognition and killing of dendritic cells (DC), the down-regulation of NKp30 resulted in sharp inhibition of DC killing by TGFβ1-treated NK cells.
Methods
mAbs.
The following mAbs, produced in our laboratory, were used in this study: JT3A (IgG2a, anti-CD3), AZ20 and F252 (IgG1 and IgM, respectively, anti-NKp30), ON72 (IgG1, anti-NKG2D), BAB281 and KL247 (IgG1 and IgM, respectively, anti-NKp46), Z231 and KS38 (IgG1 and IgM, respectively, anti-NKp44), MAR206 (IgG1 anti-CD2), PP35 (IgG1, anti-2B4), MA127 (IgG1, anti-NTB-A), MA152 (IgG1, anti-NKp80), c127 (IgG1, anti-CD16), c218 and A6-220 (IgG1 and IgM, respectively, anti-CD56), A6-136 (IgM, anti-HLA class I), and c227 (IgG1 anti-CD69).
Anti-CXCR1 (IgG1, Santa Cruz Biotechnology), anti-CX3CR1 (IgG1, MBL, Nagoya, Japan), anti-CXCR3 and anti-CXCR4 (IgG1 and IgG2b, respectively, R & D Systems), and anti-CCR5 (IgG2a, Becton Dickinson) were obtained from the indicated sources. Anti-CD1a [IgG1, labeled with phycoerythrin (PE)], anti-CD14 (IgG2a), anti-CD83 (IgG2b), and anti-CD86 (IgG2b-PE) were purchased from Immunotech (Luminy, France). D1.12 (IgG2a, anti-HLA-DR) mAb was provided by R. S. Accolla (University of Insubria, Varese, Italy). HP2.6 (IgG2a, anti-CD4) mAb was provided by P. Sanchez-Madrid (Universidad Autonoma de Madrid, Madrid).
Generation of Polyclonal or Clonal NK Cell Populations.
To obtain peripheral blood lymphocytes, peripheral blood mononuclear cells were isolated on Ficoll–Hypaque gradients and depleted of plastic-adherent cells. Enriched NK cells were isolated by incubating peripheral blood lymphocytes with anti-CD3 (JT3A), anti-CD4 (HP2.6), and anti-HLA-DR (D1.12) mAbs (30 min at 4°C) followed by goat anti-mouse IgG-coated Dynabeads (Dynal, Oslo) (30 min at 4°C) and immunomagnetic depletion (8). CD3− CD4− DR− cells were cultured on irradiated feeder cells in the presence of 100 units/ml recombinant IL-2 (rIL-2; Proleukin, Chiron) and 1.5 ng/ml phytohemagglutinin (GIBCO) to obtain polyclonal NK cell populations or, after limiting dilution, NK cell clones (8).
For TGFβ1 conditioning, freshly isolated or rIL-2-activated polyclonal NK cells were plated at 50,000 cells per well either in the presence of 100 units/ml rIL-2 alone or in the presence of rIL-2 and TGFβ1 or IL-10 or IL-4 (PeproTech, London) at the final concentration of 10 ng/ml.
Generation of DC.
DC were generated as follows: Peripheral blood mononuclear cells were derived from healthy donors, and plastic-adherent cells were cultured in the presence of IL-4 and granulocyte–macrophage colony-stimulating factor (PeproTech) at the final concentration of 20 and 50 ng/ml, respectively. After 6 days of culture, cells were characterized by the CD14−, CD1a+, CD83− phenotype corresponding to immature DC (iDC). To generate CD14−, CD1a+, CD83+, CD86+ mature DC (mDC) iDC were stimulated for 2 days with lipopolysaccharide from Escherichia coli (Sigma) at the final concentration of 1 μg/ml.
Flow Cytofluorimetric Analysis and Cytolytic Activity.
For one- or two-color cytofluorimetric analysis (FACSCalibur, Becton Dickinson), cells were stained with the appropriate mAbs followed by PE- or FITC-conjugated isotype-specific goat anti-mouse second reagent (Southern Biotechnology Associates). NK cells were tested for cytolytic activity in a 4-h 51Cr-release assay (8). The concentrations of the various mAbs added were 10 μg/ml for masking experiments. The effector-to-target cell ratios are indicated in the text. The target cell lines FO-1 (human melanoma) and 721.221 (human B cells transformed by Epstein–Barr virus) (7, 8) were used for cytotoxicity experiments.
RT-PCR Analysis.
Total RNA was extracted from NK cell clones or polyclonal NK cells either untreated or after TGFβ1 treatment, by using RNAClean (TIB-Molbiol, Genoa, Italy). Oligo(dT)-primed cDNA was prepared by the standard technique. The primers used were 5′-ACT CCA TCA TGA AGT GTG ACG (β-actin up) and 5′-CAT ACT CCT GCT TGC TGA TCC (β-actin dw) for β-actin cDNA (249 bp), 5′-CAG ACC CCA GTC CAC CAT G (DAP-10 ORF up) and 5′-GTG CCA CCA CAC ACC ATC (DAP-10 ORF dw) for DAP-10 cDNA (350 bp), 5′-AGG GAA GGA GGT GAG GAA TGG (7A6P-1F) and 5′-GAT TTA TTG GGG TCT TTT GAA G (7A6-138rev) for NKA1/NKp30 cDNA (413 bp), and 5′-GAA GGC TTT TAT CCA CAA (NKG2D up) and 5′-CCC CAG CCC ATC CAC TCT (NKG2D dw) for NKG2D cDNA (1,030 bp). All amplifications were performed for 30 cycles, with a final extension step of 7 min at 72°C. PCR conditions were 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C when β-actin was used. All of the other amplifications have been performed with the same protocol, changing only the annealing temperatures as follows: 55°C for CD16 and DAP-10, 60°C for NKA1/NKp30, and 50°C for NKG2D.
Real-Time RT-PCR.
Relative quantitation of gene expression after TGFβ1 treatment was performed in real-time RT-PCR by using TaqMan probes. To internally standardize the levels of gene expression, we used the GAPDH housekeeping gene. All of the amplifications were performed with the Icycler IQ system (Bio-Rad) in a 25-μl final volume, using primers at 300 nM and probe at 200 nM final concentration for 40 cycles of 30 s at 60°C and 30 s at 95°C. Primers and probes are summarized in Table 1.
Table 1.
Primers and probes used in real-time RT-PCR
| Name | Sequence (5′–3′) | Predicted product
|
|
|---|---|---|---|
| Name | Length, bp | ||
| GAPDH f primer | GAA GGT GAA GGT CGG AGT | GAPDH | 155 |
| GAPDH r primer | CAT GGG TGG AAT CAT ATT GGA A | ||
| GAPDH Tq probe | 6FAM-CAA CGG ATT TGG TCG TAT TGG GCG TXTp | ||
| NKA1 f primer | TGA TCA TGG TCC TCC AGG A | NKp30 | 125 |
| NKA1 r primer | AAT GGC CAG TCT CCC TTG G | ||
| NKA1 Tq probe | 6FAM-CCT GTG CTC TCT GGG TGT CCC AGC XTp | ||
| NKG2D f primer | GGC TCC ATT CTC TCA CCC A | NKG2D | 79 |
| NKG2D r primer | TAA AGC TCG AGG CAT AGA GTG C | ||
| NKG2D Tq probe | 6FAM-CCT ACT AAC AAT AAT TGA AAT GCA GAA GGG AGA CTG TXTp | ||
| NKp46 25 f primer | GGC AGA ATC TGA GCG ATG TCT T | NKp46 | 145 |
| NKp46 169 r primer | GCT TTT CCT TTG GAA CCA TGA A | ||
| NKp46 49Tq probe | 6FAM-ACA CTC CCT GCC CTG CTC TGC G TXTp | ||
PCR primers and probes specific for GAPDH, NKA1/NKp30, NKG2D, and NKp46 are listed. Predicted product sizes are indicated on the right. Probes have been labeled with 6-carboxyfluorescein (6FAM) and N,N,N′,N′-tetramethyl-6-carboxyrhodamine (XTp).
Results
TGFβ1 Induces Specific Down-Regulation of NKp30 and NKG2D Surface Expression in Human NK Cells.
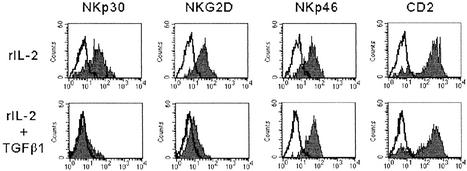
Freshly isolated NK cells, obtained from healthy donors, were cultured in the presence of either rIL-2 alone or rIL-2 in combination with TGFβ1 (hereafter termed TGFβ1-conditioned). Cells were tested at various time intervals for the surface expression of different molecules, including NCR and NKG2D, by using specific mAbs and flow cytofluorimetric analysis. After 7 days of culture in rIL-2, NK cells displayed an increased surface expression of NKp46, NKp30, and NKG2D. In contrast, at the same time interval, TGFβ1-conditioned NK cells showed a sharp down-regulation of NKp30 surface expression (Fig. 1). Although not shown, the surface density of NKp30 in TGFβ1-conditioned NK cells was significantly lower also as compared with freshly isolated NK cells. Also the surface expression of NKG2D was partially inhibited, although the degree of inhibition varied in different experiments and was less marked than for NKp30. In TGFβ1-conditioned NK cells, the surface density of other receptors or coreceptors, including NKp46 and CD2 (Fig. 1) or CD16, 2B4, NTBA, and NKp80 (not shown), was comparable to that detected in cultures supplemented with rIL-2 alone.
Figure 1.
TGFβ1-mediated down-regulation of NKp30 and NKG2D surface molecules in fresh NK cells. Freshly isolated NK cells were cultured either in the presence of rIL-2 alone or in the presence of rIL-2 and TGFβ1. At day 7 cells were analyzed by one-color immunofluorescence and fluorescence-activated cell sorter analysis for the expression of the indicated molecules. PE-conjugated isotype-specific goat anti-mouse IgG was used as second reagent. White profiles represent cells stained with the second reagent alone.
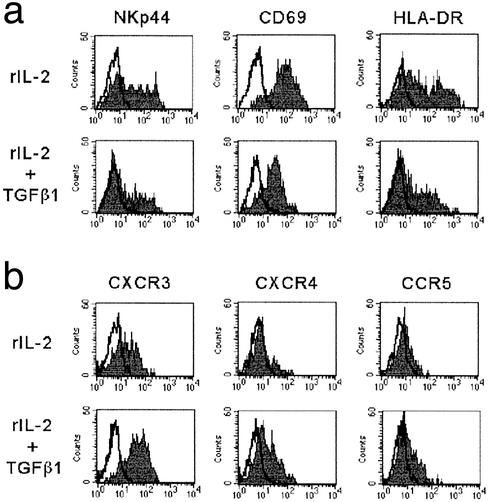
TGFβ1-conditioned fresh NK cells were also analyzed for the surface expression of NKp44, CD69, and HLA-DR, i.e., surface molecules that are normally expressed only after NK cell activation. As shown in Fig. 2a, all these molecules were expressed, although their surface density was lower than in cells cultured in rIL-2 alone. The de novo acquisition of surface molecules that are not expressed in resting NK cells implies that NK cells can undergo activation also in the presence of TGFβ1. These data are in line with previous reports (13) showing that T cells can be activated also in the presence of TGFβ1.
Figure 2.
Expression of activation surface markers and chemokine receptors in TGFβ1-conditioned NK cells. Freshly isolated NK cells were plated either in the presence of rIL-2 alone or in the presence of rIL-2 and TGFβ1. At day 7 cells were analyzed by one-color immunofluorescence and fluorescence-activated cell sorter analysis for the expression of NKp44, CD69, and HLA-DR (a) or of various chemokine receptors (b). PE-conjugated isotype-specific goat anti-mouse IgG was used as second reagent. White profiles represent cells stained with the second reagent alone.
We also analyzed the effect of TGFβ1 on the expression of various chemokine receptors by cultured NK cells. At variance with cytofluorimetric studies but consistent with functional data (i.e., migration assay) (14), we could not detect CXCR3, CXCR4, and CCR5 in freshly isolated NK cells derived from normal individuals, whereas the fractalkine receptor (CX3CR1) as well as the IL-8 receptor (CXCR1) was detected in most CD16+, CD56+ (but not in CD16−, CD56++) fresh NK cells (not shown).
NK cells, upon 7 days of culture in rIL-2, expressed both CXCR3 and CCR5. Remarkably, a higher density of these receptors was detected at the same time interval in TGFβ1-conditioned NK cells (Fig. 2b). In addition, also CXCR4 was consistently up-regulated in TGFβ1-conditioned NK cells, whereas it was expressed only by a minor subset of NK cells cultured in rIL-2 (Fig. 2b). Finally, with regard to the expression of CX3CR1 and CXCR1, they were lost both in rIL-2 and TGFβ1-conditioned NK cells (not shown). Thus, the shift in the surface expression of chemokine receptors occurring after NK cell activation is not altered by TGFβ1 conditioning. Rather, it appears to be up-regulated.
Altogether, the above data suggest that TGFβ1-conditioning results in the specific modulation of certain receptors whereas it does not alter that of most surface molecules. A particularly striking effect was the sharp down-regulation of NKp30, a major receptor involved in triggering the NK-mediated cytotoxicity. Moreover, a partial down-regulation of NKG2D was also consistently observed.
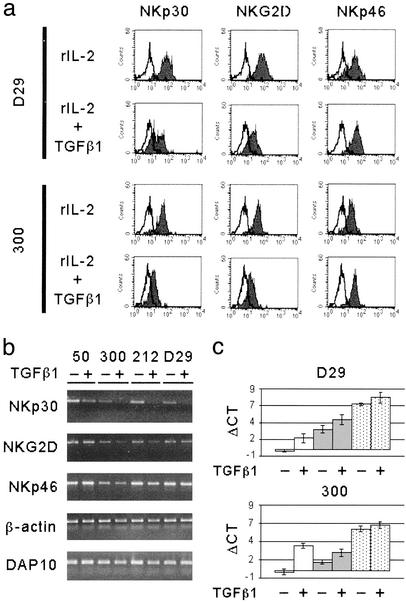
We further analyzed whether TGFβ1-conditioning had any effect on established polyclonal NK cell lines that had been cultured in the presence of rIL-2 for >1 month. These NK cells were further cultured in rIL-2 alone or in the presence of both rIL-2 and TGFβ1 for an additional 3–5 days. Also in this case, TGFβ1 had an inhibitory effect on the surface expression of NKp30 and NKG2D (Fig. 3a). However, this effect was less marked than in TGFβ1-conditioned fresh NK cells. Consistent with data obtained with fresh NK cells, TGFβ1 conditioning did not modify the expression of NKp46, CD16, CD2, 2B4, NTBA, and NKp80 (not shown).
Figure 3.
TGFβ1 effect on NKp30 and NKG2D surface and transcript expression in established NK cell population and clones. (a) Long-term rIL-2 cultured polyclonal or clonal NK cells (see the representative polyclonal NK population D29 and NK cell clone 300) were replated in rIL-2 alone or in the presence of both rIL-2 and TGFβ1. At day 5 cells were analyzed by one-color immunofluorescence and fluorescence-activated cell sorter analysis for the expression of the indicated molecules. PE-conjugated isotype-specific goat anti-mouse IgG was used as second reagent. White profiles represent cells stained with the second reagent alone. (b) End-point RT-PCR analysis was performed on polyclonal or clonal NK cells (see the representative NK cell clones 50, 300, and 212 and the polyclonal NK cell population D29). RNA was extracted from cells either untreated (−) or treated (+) with TGFβ1, as indicated. The transcripts analyzed are indicated on the left. (c) Real-time RT-PCR was performed on polyclonal or clonal NK cells (see the representative polyclonal NK cell population D29 and the NK cell clone 300) either untreated (−) or treated (+) with TGFβ1. Bar histograms indicate the ΔCT calculated as the difference between the PCR threshold cycle number of the analyzed gene and the housekeeping gene GAPDH used as reference. White bars indicate transcription of NKA1/NKp30, gray bars indicate NKG2D transcription, and stippled bars indicate NKp46 expression levels. SD is indicated on each bar.
Finally, the effect of TGFβ1 was analyzed in NK cell clones. Again, upon treatment with TGFβ1, the majority of NK clones analyzed displayed a specific down-regulation of NKp30 and NKG2D (see the representative NK clone 300 in Fig. 3a), whereas some clones were partially resistant to the TGFβ1-mediated effects (not shown).
TGFβ1 Treatment Results in Decrement of NKp30 Transcript Expression.
To test whether TGFβ1 treatment affected not only the surface expression but also the transcription expression levels of NKp30 and NKG2D, classical end-point RT-PCR analysis was performed on various TGFβ1-conditioned NK cell clones and polyclonal NK cell populations. In all instances NK cells displaying, upon TGFβ1 conditioning, a reduced NKp30 surface expression were characterized by a decrease of NKp30 transcript expression (Fig. 3b). In contrast, results obtained on NKG2D transcripts were less homogeneous. Indeed, in some NK clones the reduced levels of NKG2D surface expression appeared to correlate with a reduction of its transcript expression (e.g., see clones 300 and 212 in Fig. 3b). However, in other NK clones as well as in the polyclonal NK cell populations virtually no decrease of NKG2D transcript expression could be detected (Fig. 3b). In these experiments DAP10, NKp46, and β-actin transcripts were used as controls. To analyze more precisely the relative levels of NKp30 and NKG2D transcript expression we used a real-time RT-PCR using TaqMan probes. To this end, different sets of primers and probes were designed (see Table 1) to amplify cDNA fragments of NKp30, NKG2D, and, as control, NKp46. The endogenous control GAPDH was used to standardize the reactions. Quantitation experiments were performed both at clonal and polyclonal cell level. The representative NK cell clone 300 and the D29 polyclonal NK cell population are shown in Fig. 3c.
Histograms of ΔCT for TGFβ1-untreated vs. TGFβ1-treated cells revealed that NK cells displaying, upon TGFβ1 conditioning, a reduced NKp30 surface expression were always characterized by a statistically significant decrement of NKp30 transcript expression (≈12-fold for the NK cell clone 300 and 3-fold for the D29 polyclonal NK cell population; Fig. 3c). On the other hand, no statistically significant decrements of NKp30 transcript could be detected in NK cell clones that did not display TGFβ1-mediated reduction of NKp30 surface expression (not shown). The fact that the polyclonal NK cell population displayed a less marked decrease of transcript expression as compared with the NK clones may indeed reflect the heterogeneity of NK cells in their susceptibility to TGFβ1 treatment. Regarding the levels of NKG2D transcript expression, the decrement was statistically significant in the case of some NK cell clones (Fig. 3c, clone 300) but not in other NK clones (data not shown) or in polyclonal NK cell populations (Fig. 3c). In line with the cytofluorimetric analysis, the levels of NKp46 transcription, used as control, were essentially unchanged after TGFβ1 treatment. Thus, both classical end-point and real-time RT-PCR suggest that TGFβ1 down-regulates NKp30 surface expression by acting mainly on its transcription. On the other hand, the TGFβ1-mediated inhibitory effect on the NKG2D surface expression may depend only in part on the regulation of transcription levels.
TGFβ1 Inhibits the NK-Mediated Killing of DC.
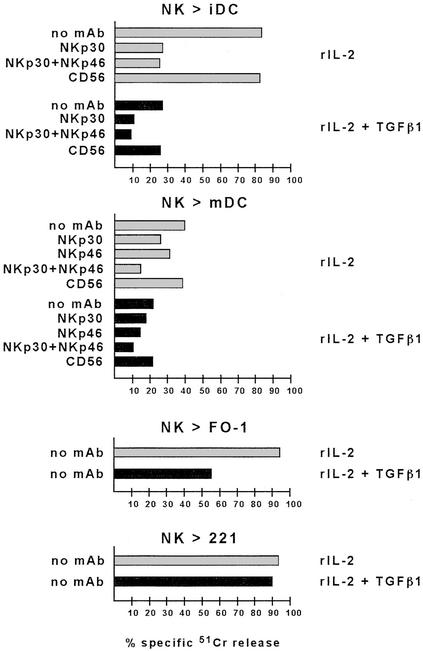
It has recently been shown that NK cells can interact with DC, leading to bidirectional cross-talk that may result in NK cell priming and proliferation as well as in DC maturation (15–17). In addition, activated NK cells efficiently kill iDC, whereas mDC are virtually NK resistant. Remarkably, the NK-mediated killing of DC has the unique property to be mostly NKp30 dependent, whereas other surface molecules (e.g., NKp46) play a marginal role (15). Our present data that TGFβ1 selectively inhibited the surface expression of NKp30 suggested a possible effect on the NK-mediated killing of DC. iDC were obtained from plastic-adherent peripheral blood mononuclear cells cultured in the presence of granulocyte–macrophage colony-stimulating factor and IL-4 as described (15–17). These cells, characterized by the typical CD14−, CD1a+, CD83− surface phenotype, were analyzed for their susceptibility to lysis by polyclonal NK cell populations cultured in rIL-2 either alone or in the presence of TGFβ1.
As shown in Fig. 4, iDC were efficiently lysed by rIL-2-cultured NK cells and, in agreement with previous data (15), the cytolytic activity was strongly inhibited by the addition of blocking anti-NKp30 mAb. On the other hand, mAb-mediated blocking of NKp46 had only a little inhibitory effect, thus confirming that NKp46 has only a marginal role in the process of iDC killing (data not shown). Moreover, in agreement with the lack of MHC class I chain-related (MIC)A/B and UL16 binding protein (ULBP) (i.e., the known ligands for NKG2D) on iDC (ref. 15 and unpublished data), mAb-mediated blocking of NKG2D had no effect on cytotoxicity (data not shown). Importantly, TGFβ1-conditioned NK cells displayed a dramatic reduction of cytotoxicity against iDC. The residual cytotoxicity was mostly dependent on NKp30 because a further reduction was detected upon mAb-mediated masking of NKp30 (Fig. 4). Moreover the simultaneous mAb-mediated masking of NKp46, the expression of which was not modified by TGFβ1 treatment, had virtually no additional effect. These data further reinforce the concept that NKp46 is poorly efficient in mediating killing of iDC, even when expressed in much higher surface density than NKp30.
Figure 4.
Effect of TGFβ1 on NK-mediated cytotoxicity against different target cells. Polyclonal NK cells were plated in rIL-2 alone or in the presence of both rIL-2 and TGFβ1. At day 5 cells were analyzed for cytolytic activity in a 4-h 51Cr-release assay against iDC or mDC, or against the FO-1 or LCL 721.221 target cell lines either in the absence of mAb or in the presence of the mAbs to the indicated molecules. The effector-to-target cell ratio was 6:1. Masking experiments were performed by using mAbs of IgM isotype. The result are representative of three independent experiments; the SD of the mean of the triplicates was <5%.
We also analyzed the effect of TGFβ1-conditioning on the ability of NK cells to kill mDC characterized by the CD14−, CD1a+, CD83+, CD86+ phenotype. It is of note that, in line with previous reports (15), autologous mDC, unlike iDC, are resistant to NK-mediated killing caused by the interactions between HLA class I molecules (on mDC) and HLA class I-specific inhibitory receptors (on NK cells) (data not shown). Thus, to prevent these inhibitory interactions, cytolytic assays were performed by using allogeneic mDC. In this experimental setting, mDC were susceptible to NK-mediated killing (Fig. 4). It is of note, however, that also in this case, lysis of mDC was higher in the presence of anti-HLA class I mAb (data not shown). This reflected the existence, in the effector NK cell population, of a fraction of cells expressing inhibitory receptors still recognizing HLA class I molecules on allogeneic mDC. Unlike NK-mediated lysis of iDC, the cytotoxicity against mDC was only partially inhibited by anti-NKp30 mAb, thus suggesting that additional receptors expressed at the NK cell surface may be involved in the NK cell triggering upon interaction with mDC. Indeed, different from iDC, lysis of mDC could be inhibited, at least in part, also by mAb-mediated masking of NKp46 (Fig. 4). According to this observation, TGFβ1-conditioning only partially affected the NK-mediated lysis of mDC. Moreover mAb-mediated masking of both residual NKp30 and NKp46 molecules was necessary to obtain a significant reduction of mDC lysis (Fig. 4).
Taken together these data provide evidence that the NK-mediated killing of iDC, which is mostly dependent on the engagement of NKp30, is strongly affected by TGFβ1 treatment of NK cells, whereas killing of mDC is less affected because of the involvement of additional triggering receptors.
Finally, we evaluated the effect of TGFβ1 conditioning on the ability of NK cells to kill tumor target cells. In this case, multiple triggering NK receptors are involved in tumor cell lysis. Accordingly, the NK-mediated lysis of these cells was abrogated only by the simultaneous blocking of different triggering receptors (4). For example, rIL-2-activated NK cells efficiently lysed the (HLA class I-negative) FO-1 melanoma cell line (Fig. 4). Although mAb-mediated masking of individual activating receptors (i.e., NKp30) did not significantly inhibit the NK-mediated lysis (7), the simultaneous mAb-mediated blocking of NKp30, NKp46, and NKG2D resulted in the abrogation of cytotoxicity (data not shown). In line with these data, killing of FO-1 by TGFβ1-conditioned NK cells (characterized by low NKp30 and NKG2D surface densities) was only partially reduced. The residual cytolytic activity was mainly due to the engagement of NKp46 (data not shown), a receptor that is unaffected by TGFβ1 treatment. Along this line, lysis of LCL 721.221 target cells, which is mostly NKp46 dependent (8), was not significantly reduced in TGFβ1-conditioned NK cells. These results indicate that, at variance with DC, the NK-mediated killing of tumors may or may not be affected by TGFβ1, depending on the type of triggering receptor–ligand interactions required for killing of a given target cell.
These data also imply that the overall cytolytic potential of TGFβ1-conditioned NK cells is not significantly reduced as compared with controls.
Discussion
In this study, we provide experimental evidence that the surface expression and function of triggering receptors responsible for NK-mediated recognition and killing of various target cells can be regulated by cytokines. In particular, TGFβ1 selectively down-regulates the surface expression of NKp30 and NKG2D. As a consequence of the down-regulation of NKp30, TGFβ1-conditioned NK cells fail to lyse iDC. On the other hand, the cytolytic activity against different HLA class I-negative tumor target cells is variably affected, depending on which triggering receptors are primarily used by NK cells against a given target. It is of note that, although in TGFβ1-treated NK cells the down-regulation of NKp30 surface expression is clearly consequent to the decrease of the NKp30 transcript expression, that of NKG2D may not be solely dependent on mechanisms of transcript regulation. TGFβ1 is released by tumors of different histotypes, including melanomas, neuroblastomas, carcinomas, and leukemias (18–22). This release may be viewed as a mechanism of tumor escape from immune surveillance as TGFβ1 affects, at various levels, the ability of immune cells to detect and clear cancer cells. For example, TGFβ1 has been shown to induce the surface expression of the inhibitory receptor CD94/NKG2A in cytolytic T cells (23). This inhibition may affect the T cell-mediated killing of HLA-E+ target cells. No data are available so far on the effect of TGFβ1 on the expression and function of receptors involved in triggering or inhibition of human NK cells. However, recent data suggested that tumor-derived cytokines could inhibit the NK cell function by acting on the expression of their receptors. Thus, Costello et al. (9) showed that NK cells isolated from patients affected by AML frequently expressed an unusually low surface density of triggering receptors including NKp30, NKp44, NKp46, and, in some patients, NKG2D. No evidence, however, was provided for specific factor(s) capable of down-regulating the NK receptor expression. The finding that such NCRdull phenotype characterizes virtually all NK cells of AML patients, whereas in normal individuals it is frequently absent or confined to a small NK subset, suggested that the leukemia itself could be responsible for modulating the expression of the NK receptors. Our present data suggest a possible role for TGFβ1 in leukemic patients, as AML cells frequently produce TGFβ1 (20). Because B chronic lymphocytic leukemia and HTLV+ T cell leukemia can release TGFβ1 (21, 22), it will be interesting to evaluate the NCR phenotype also in patients affected by these leukemias. It is noteworthy that TGFβ1 does not affect the expression of NKp46 and NKp44 while these receptors are down-regulated in AML together with NKp30. Our present findings may imply that other yet-undefined factors are responsible for the down-regulation of NKp46 and NKp44. The TGFβ1-induced modulation of NKp30 and NKG2D was not sufficient to prevent killing of the various tumor cell lines analyzed. This insufficiency reflects the fact that multiple triggering NK receptors participate in tumor recognition and killing (4). Thus, one could speculate that tumor cells have evolved a more complex mechanism of escape based on the release of multiple immunomodulating cytokines acting each on different, functionally relevant, molecules of immunocompetent effector cells. In this context, we also analyzed the effect on NK cells of IL-4 or IL-10, i.e., two cytokines that, similar to TGFβ1, exert an immunomodulatory activity (24–26). In no instance, however, could we detect variations of the surface expression of triggering NK receptors (unpublished data).
A different scenario may be envisaged when NK cells interact with DC in response to pathogens (bacteria, viruses, or parasites) or tumors (27, 28). In this case, NK cells, which are recruited together with DC precursors from the blood stream to the inflammatory sites, may either kill iDC, thus limiting their further maturation and function, or favor their maturation program into efficient antigen-presenting cells. DC, in turn, may provide signals (e.g., IL-15 and IL-12) for priming NK cells and inducing increases of both cytolytic activity and IFN-γ production. The preferential occurrence of one or another effect in the course of the NK–DC interaction is likely to depend on the type or the strength of signals delivered to DC (27). This may be related to the type of pathogen encountered and/or may be related to the different stages of DC maturation and function. For example, the NK-dependent killing of iDC, (15–17, 29–31) (which may play a role in the selection of DC undergoing maturation) is primarily allowed by the reduced surface density of HLA-class I molecules and induced by the expression of the still undefined NKp30 ligand(s) at the iDC cell surface. Indeed, it has been shown that expression of HLA class I is very low in iDC whereas it is strongly up-regulated in mDC. This difference explains why, in an autologous setting, iDC but not mDC are susceptible to NK-mediated lysis. Our present findings suggest that the TGFβ1-mediated down-regulation of NKp30 expression represents a possible mechanism to prevent the NK-mediated killing of iDC, whereas it would have only marginal effects on the killing of mDC. Along this line, TGFβ1 production by iDC has been detected in response to the uptake of apoptotic bodies that may occur during physiological tissue regeneration (13). Thus, in the absence of suitable proinflammatory signals, the TGFβ1 released by iDC would not only prevent DC maturation by an autocrine mechanism (27) but also down-regulate the NKp30 expression, thus preventing their NK-mediated killing. In line with this hypothesis, we observed that iDC expressing the CD86low CD83− surface phenotype were characterized by a constitutive release of TGFβ1 in the culture supernatant (data not shown). Consistent with these data, murine TGFβ1-producing DC were enriched in the CD86low subset corresponding to iDC (32). On the other hand, some TGFβ1 production could be detected also in human mDC expressing the CD83+ phenotype (33). In the presence of tumors characterized by the production of high levels of TGFβ1, the sustained down-modulation of NKp30 may impair the ability of NK cells to select iDC that, upon antigen uptake in injured tissues, undergo maturation and migration. Thus, the altered regulation of the NK/DC cross-talk could also affect the downstream events (in secondary lymphoid compartments) that are necessary for the generation of optimal cytotoxic T lymphocyte responses (27).
Acknowledgments
We thank Ms. Tiziana Baffi for secretarial assistance. This work was supported by grants awarded by Associazione Italiana per la Ricerca sul Cancro, Istituto Superiore di Sanità, Ministero della Sanità, Ministero dell'Università e della Ricerca Scientifica e Tecnologica, and Compagnia San Paolo (Turin, Italy). R. Castriconi and E. Marcenaro are recipients of a fellowship awarded by Fondazione Italiana per la Ricerca sul Cancro.
Abbreviations
- NK
natural killer
- NCR
natural cytotoxicity receptors
- TGFβ1
transforming growth factor β1
- DC
dendritic cell
- iDC
immature DC
- mDC
mature DC
- AML
acute myeloid leukemia
- rIL-2
recombinant IL-2
- PE
phycoerythrin
References
- 1.Moretta A, Bottino C, Vitale M, Pende D, Biassoni R, Mingari M C, Moretta L. Annu Rev Immunol. 1996;14:619–648. doi: 10.1146/annurev.immunol.14.1.619. [DOI] [PubMed] [Google Scholar]
- 2.Lanier L L. Annu Rev Immunol. 1998;16:359–393. doi: 10.1146/annurev.immunol.16.1.359. [DOI] [PubMed] [Google Scholar]
- 3.Long E O. Annu Rev Immunol. 1999;17:875–904. doi: 10.1146/annurev.immunol.17.1.875. [DOI] [PubMed] [Google Scholar]
- 4.Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari M C, Biassoni R, Moretta L. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- 5.Moretta A, Biassoni R, Bottino C, Mingari M C, Moretta L. Immunol Today. 2000;21:228–234. doi: 10.1016/s0167-5699(00)01596-6. [DOI] [PubMed] [Google Scholar]
- 6.Bauer S, Groh V, Wu J, Steinle A, Phillips J H, Lanier L L, Spies T. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 7.Pende D, Cantoni C, Rivera P, Vitale M, Castriconi R, Marcenaro S, Nanni M, Biassoni R, Bottino C, Moretta A, Moretta L. Eur J Immunol. 2001;31:1076–1086. doi: 10.1002/1521-4141(200104)31:4<1076::aid-immu1076>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 8.Sivori S, Pende D, Bottino C, Marcenaro E, Pessino A, Biassoni R, Moretta L, Moretta A. Eur J Immunol. 1999;29:1656–1666. doi: 10.1002/(SICI)1521-4141(199905)29:05<1656::AID-IMMU1656>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 9.Costello R J, Sivori S, Marcenaro E, Lafage-Pochiutaloff M, Mozziconacci M J, Reviron D, Gastaud J A, Pende D, Olive D, Moretta A. Blood. 2002;99:3661–3667. doi: 10.1182/blood.v99.10.3661. [DOI] [PubMed] [Google Scholar]
- 10.Rook A H, Kehrl J H, Wakefield L M, Roberts A B, Sporn M B, Burlington D B, Lane H C, Fauci A S. J Immunol. 1986;136:3916–3920. [PubMed] [Google Scholar]
- 11.Kehrl J H, Wakefield L M, Roberts A B, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn M B, Fauci A S. J Exp Med. 1986;163:1037–1050. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellone G, Aste-Amezaga M, Trinchieri G, Rodeck U. J Immunol. 1995;155:1066–1073. [PubMed] [Google Scholar]
- 13.Gorelik L, Flavell R A. Nat Rev Immunol. 2002;2:46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 14.Campbell J J, Qin S, Unutmaz D, Soler D, Murph K E, Hodge M R, Wu L, Butcher E C. J Immunol. 2001;166:6477–6482. doi: 10.4049/jimmunol.166.11.6477. [DOI] [PubMed] [Google Scholar]
- 15.Ferlazzo G, Tsang M L, Moretta L, Melioli G, Steinman R M, Münz C. J Exp Med. 2002;195:343–351. doi: 10.1084/jem.20011149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piccioli D, Sbrana S, Melandri E, Valiante N M. J Exp Med. 2002;195:335–341. doi: 10.1084/jem.20010934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerosa F, Baldani-Guerra B, Nisii C, Marchesini V, Carra G, Trinchieri G. J Exp Med. 2002;195:327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berking C, Takemoto R, Schaider H, Showe L, Satyamoorthy K, Robbins P, Herlyn M. Cancer Res. 2001;61:8306–8316. [PubMed] [Google Scholar]
- 19.Pasche B J. J Cell Physiol. 2001;186:153–168. doi: 10.1002/1097-4652(200002)186:2<153::AID-JCP1016>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 20.Bergmann L, Schui D K, Brieger J, Weidmann E, Mitrou P S, Hoelzer D. Exp Hematol. 1995;23:1574–1580. [PubMed] [Google Scholar]
- 21.Schena M, Gaidano G, Gottardi D, Malavasi F, Larsson L G, Nilsson K, Caligaris-Cappio F. Leukemia. 1992;6:120–125. [PubMed] [Google Scholar]
- 22.Kim S J, Kehrl J H, Burton J, Tendler C L, Jeang K T, Danielpour D, Thevenin C, Kim K Y, Sporn M B, Roberts A B. J Exp Med. 1990;172:121–129. doi: 10.1084/jem.172.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertone S, Schiavetti F, Bellomo R, Vitale C, Ponte M, Moretta L, Mingari M C. Eur J Immunol. 1999;29:23–29. doi: 10.1002/(SICI)1521-4141(199901)29:01<23::AID-IMMU23>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 24.Feldmann M, Brennan F M, Maini R N. Annu Rev Immunol. 1996;14:397–440. doi: 10.1146/annurev.immunol.14.1.397. [DOI] [PubMed] [Google Scholar]
- 25.Moore K W, de Waal-Malefyt R, Coffman R L, O'Garra A. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 26.Naohiro S, Yoshiki T, Masahiro T, Kohji E. J Immunol. 1999;163:242–249. [Google Scholar]
- 27.Moretta A. Nat Rev Immunol. 2003;2:957–965. doi: 10.1038/nri956. [DOI] [PubMed] [Google Scholar]
- 28.Zitvogel L. J Exp Med. 2002;195:9–14. doi: 10.1084/jem.20012040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson J L, Heffler L C, Charo J, Scheynius A, Bejarano M T, Ljunggren H G. J Immunol. 1999;163:6365–6370. [PubMed] [Google Scholar]
- 30.Spaggiari G M, Carosio R, Pende D, Marcenaro S, Rivera P, Zocchi M R, Moretta L, Poggi A. Eur J Immunol. 2000;31:1656–1665. doi: 10.1002/1521-4141(200106)31:6<1656::aid-immu1656>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 31.Carbone E, Terrazzano G, Ruggiero G, Zanzi D, Ottaiano A, Manzo C, Karre K, Zappacosta S. Eur J Immunol. 1999;29:4022–4029. doi: 10.1002/(SICI)1521-4141(199912)29:12<4022::AID-IMMU4022>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 32.Morelli A E, Zahorchak A F, Larregina A T, Colvin B L, Logar A J, Takayama T, Falo L D, Thomson A W. Blood. 2001;98:1512–1523. doi: 10.1182/blood.v98.5.1512. [DOI] [PubMed] [Google Scholar]
- 33.Zhou L J, Tedder T F. Blood. 1995;86:3295–3301. [PubMed] [Google Scholar]