Abstract
The three mammalian receptors UNC5H1, UNC5H2, and UNC5H3 (also named UNC5A, UNC5B, and UNC5C in human) that belong to the family of the netrin-1 receptors, UNC5H, were initially proposed as mediators of the chemorepulsive effect of netrin-1 on specific axons. However, they were also recently shown to act as dependence receptors. Such receptors induce apoptosis when unbound to their ligand. We show here that the expression of the human UNC5A, UNC5B, or UNC5C is down-regulated in multiple cancers including colorectal, breast, ovary, uterus, stomach, lung, or kidney cancers. In colorectal tumors, this down-regulation is associated with loss of heterozygosity occurring within UNC5A, UNC5B, and UNC5C genes but may also be partially related to epigenetic processes because histone deacetylase inhibitor increased UNC5C expression in various cancer cell lines. Moreover, sequencing of UNC5C gene in patients with colorectal tumors revealed the presence of missense mutations. The loss/reduction of expression may be a crucial mechanism for tumorigenicity because the expression of UNC5H1, UNC5H2, or UNC5H3 inhibits tumor cell anchorage-independent growth and invasion. Moreover, these hallmarks of malignant transformation can be restored by netrin-1 addition or apoptosis inhibition. Hence, UNC5H1, UNC5H2, and UNC5H3 receptors may represent tumor suppressors that inhibit tumor extension outside the region of netrin-1 availability by inducing apoptosis.
Tessier-Lavigne and his colleagues (1–3) discovered netrin-1 as a chemotropic molecule that mediates axon outgrowth and orientation or neuronal migration during nervous system development. DCC (Deleted in Colorectal Cancer), one of the netrin-1 receptors was, however, not only proposed as a mediator for axon guidance (4, 5) but was originally described as a tumor suppressor whose expression is lost or reduced in a wide variety of cancers (ref. 6; for a review, see ref. 7). Thus, DCC expression appears to represent a negative constraint for tumor expansion. Even though DCC heterozygote knockout mice have not shown an increased number of tumors (5), and although very few mutations of DCC have been detected in cancers (8), it is still a matter of debate whether DCC is a tumor suppressor or not (9, 10). One piece of evidence supporting its tumor suppressor function is the fact that DCC is a dependence receptor (11, 12). Such receptors create cellular states of dependence on their respective ligands by inducing apoptosis in settings in which the ligand is unavailable (13, 14). We and others have then demonstrated that, if in the presence of netrin-1, DCC mediates a “positive signal” of axon outgrowth (3, 4), in the absence of ligand, DCC induces apoptosis through a mechanism involving caspase cleavage of its intracellular domain (11, 12, 15, 16).
The ability of unbound DCC to promote cell death raised the possibility that DCC may act as a tumor suppressor of metastasis and locally invasive growth that would exceed ligand availability (11). Indeed, the pair DCC/ligand (netrin-1 or another known or unknown ligand) may regulate tumor growth outside the region of netrin-1 availability by controlling cell survival/death balance (ref. 11, L.M., C. Bidaud-Bonot, J.-Y.S., and P.M., unpublished work). Interestingly, netrin-1 is a ligand for other receptors that belong to the dependence receptor family (17). Thus, the netrin-1 receptors UNC5H1, UNC5H2, and UNC5H3 that are type 1 transmembrane receptors probably mediate the chemorepulsive activity of netrin-1 (18, 19) but also drive cell death induction in the absence of netrin-1, the latter proapoptotic activity depending on the caspase cleavage of these receptors and the conserved “death domain” located in the C terminus of their intracellular domains (17, 20). Rodent UNC5H1, UNC5H2, and UNC5H3 were first named by sequence homology to the Caenorhabditis elegans UNC5 that was shown to display a clear role in axon guidance through a genetic link with UNC6, the ortholog of netrin-1 (21). Rodent UNC5H1, UNC5H2, and UNC5H3 were then demonstrated to be receptors for netrin-1 (18). Human counterparts were determined by sequence homology and were named UNC5A, UNC5B, and UNC5C. The strong conservation between respectively UNC5H1 and UNC5A, UNC5H2 and UNC5B, and UNC5H3 and UNC5C suggested that the respective rodent UNC5H1, UNC5H2, or UNC5H3 is the ortholog of human UNC5A, UNC5B, or UNC5C (i.e., because of this very complex nomenclature, we will name here UNC5H when UNC5H1, UNC5H2, and UNC5H3, or UNC5A, UNC5B, and UNC5C are both concerned).
Whereas UNC5H receptors are highly expressed in specific neurons during the development of the nervous system in adult, UNC5H expression is not restricted to the nervous system but can be detected, even though at a lower level, in thyroid, kidney, ovary, uterus, stomach, colon, lung, spleen, bladder, and breast tissues (ref. 22; not shown; and Figs. 1A–3), suggesting a role for these receptors out of the neuronal guidance system. Interestingly, one of the UNC5H genes, UNC5H2/B, was recently shown to be a direct transcriptional target for the tumor suppressor p53 and to mediate p53 proapoptotic activity (20). We then sought to determine whether UNC5H receptors may, similarly to DCC, act as tumor suppressors. We present here evidence that UNC5H expression is strongly reduced in the vast majority of the tested tumors, such reduction being associated with loss of heterozygosity (LOH) occurring within UNC5H loci. Mutations in the UNC5H3/C gene were also observed in patients with colorectal tumors. Moreover, an in vitro study supports the idea that UNC5H receptors are negative regulators of cell transformation.
Figure 1.
UNC5H gene expression is down-regulated in tumors. (A) UNC5H expression analysis on a dot blot membrane containing cDNAs from 241 tumors (T) or their corresponding normal tissues (N). Complete rat UNC5H2 cDNA was used to probe the membrane. This probe hybridizes UNC5A, UNC5B, and UNC5C cDNA. As a control for loading, the same membrane was hybridized with the ubiquitin probe (data shown in Fig. 8, which is published as supporting information on the PNAS web site). (B) Quantitative analysis of A. The dot blot was quantified by using National Institutes of Health image software. The percentage of patients showing a loss of UNC5H expression in tumors >2-, 5-, or 10-fold compared with control tissues is indicated.
Methods
Cells, Transfection Procedures, and Plasmid Constructs.
Transient transfections of 293T or NIH 3T3 were performed as described (2, 11). Similarly, MCF7, LS174T or U373MG cells were transfected by using a Fugene 6 transfection reagent (Roche Molecular Biochemicals). For each cell line, the percentage of transfected cells was determined by measuring after 48 h the number of fluorescent cells in a cell population transfected with a GFP-expressing construct. Rodent UNC5H1–UNC5H2–UNC5H3-expressing constructs have been described (17). Ras-expressing construct was obtained from M. Castellazzi (Institut National de la Santé et de la Recherche Médicale, Lyon, France). pcDNA-p53 and pcDNA-MDM2 were obtained from A. Puisieux (Centre Leon Berard, Lyon, France). UNC5A-B-C probes were obtained by PCR amplification using cDNA from Daudi (for UNC5A and B) or HT29 (for UNC5C) cells as described in the Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org.
Gene Expression in Tumors.
UNC5H gene expression in tumor/normal tissues was monitored by using the Cancer Profiling Array (CLONTECH) following the manufacturer's suggested procedure. UNC5H probes were prepared by using a Megaprime DNA Labeling System (Amersham Pharmacia) and the full-length UNC5H2 cDNA or the UNC5A–UNC5B–UNC5C-specific cDNA (see above) as template. As a control for the samples loading in the array, the ubiquitin probe was also used.
Analysis of UNC5H Loci.
Genomic DNA (50 ng) from matched tumor and corresponding normal tissue was amplified by PCR and analyzed as described (23) by using fluorescently labeled primers for the indicated polymorphic microsatellite markers on chromosomes 5q35, 10q21, or 4q21–23 for, respectively, UNC5A, UNC5B, or UNC5C locus.
Mutation Analysis in UNC5C Gene.
Genomic DNA (50 ng) from 30 colorectal tumors was amplified by PCR using the primers described in the Supporting Text. PCR products were then sequenced on a Megabace 1000 DNA Analysis System (Molecular Dynamics and Amersham Pharmacia) sequencer by using forward and reverse primers.
In Situ Hybridization.
Sense and antisense digoxigenin-labeled RNA probes for UNC5C were prepared by in vitro transcription. In situ hybridization was performed as described (3).
Soft Agar and Matrigel Invasion Assays.
The classical soft agar assay was adapted for transient transfection as described in ref. 24 and the Supporting Text. Biocoat Matrigel invasion chambers (Becton Dickinson) were used to assess the invasiveness of LS174T, U373MG, or Ras-transfected NIH 3T3 cells as described (25).
Results
Human UNC5A, UNC5B, or UNC5C Expression Is Lost/Reduced in Multiple Cancers.
We first surveyed several human tumor types for loss of UNC5H expression. Based on the analysis of 241 tumors and their corresponding normal tissues from a dot blot array by using a full-length cDNA probe that recognizes UNC5A, UNC5B, and UNC5C mRNA, we observed that UNC5H expression was strongly down-regulated in the majority of the tested tumors (Fig. 1A) whereas such down-regulation was not detected with the ubiquitin control probe (see Fig. 8). Whereas no significant change of UNC5H expression was associated with thyroid or prostate tumors, a reduction of expression by a factor of 2 or more was observed in 88% of ovary tumors, 49% of breast tumors, 48% of uterus tumors, 93% of colorectal tumors, 68% of stomach tumors, 74% of lung tumors, and 81% of kidney tumors (Fig. 1B). Along the same line, we failed to detect UNC5H expression in eight of nine cancer cell lines (Fig. 2). These first set of experiments suggested that the global expression of the three UNC5H members was down-regulated. We next analyzed whether the three UNC5H genes were individually shut down with a specific emphasis for colorectal tumors in which the decrease of expression was the more drastic (i.e., >49% of cancer of the colon and 80% of cancer of the rectum showed at least a 5-fold decrease in UNC5H expression). Using dot blot arrays with specific probes for UNC5A, UNC5B, or UNC5C mRNA, we observed that UNC5A, UNC5B, and UNC5C expression was affected in 48%, 27%, and 77% of the tested colorectal tumors, respectively (not shown and Fig. 3A). Studying further UNC5C expression that appeared strongly reduced in colorectal tumors, we performed in situ hybridization in either normal colorectal tissue or well-differentiated adenocarcinoma. In normal tissue, the staining of UNC5C mRNA was restricted to surface and crypt epithelial cells and was observed mainly in a supranuclear position. Interestingly, consistent with the dot blot array analysis, the expression of UNC5H was reduced in neoplastic epithelial cells (Fig. 3B).
Figure 2.
UNC5H gene expression is frequently down-regulated in cancer cell lines. UNC5H, UNC5A, UNC5B, and UNC5C expression analysis on a dot blot membrane containing cDNAs from nine cancer cell lines. The left spot was loaded with cDNA from normal colon mucosa. Complete rat UNC5H2 cDNA (UNC5H) or a specific probe for UNC5A, UNC5B, or UNC5C (see Methods) was used to probe the membrane. The ubiquitin probe was also used as a positive control for loading.
Figure 3.
Down-regulation of UNC5C gene expression in colorectal cancer. (A) UNC5C expression analysis on a dot blot membrane containing cDNAs from 34 colorectal tumors (T) or their corresponding normal tissues (N). Specific UNC5C probe or the control ubiquitin probe was used for hybridization. (B) In situ hybridization was performed on sections from the colorectal mucosa of a patient with a colon adenocarcinoma by using a UNC5C probe in sense or antisense orientation. Villi from the tumor and the adjacent healthy tissue are shown. (Left) Normal mucosa. (Right) Adenocarcinoma. (Upper) With the antisense probe (as). (Lower) With the control sense probe (s). (Scale bar: 100 μm.)
Human UNC5A, UNC5B, and UNC5C Loci Are Subjected to LOH in Colorectal Cancers.
Searching for the mechanisms that may explain such down-regulation, we analyzed whether the three human UNC5H genes may be subjected to LOH. In a panel of 31 colorectal tumors versus corresponding normal tissue, we performed allelic loss analysis by PCR amplification of polymorphic microsatellite markers that neighbor UNC5A, UNC5B, or UNC5C locus. Fig. 4 shows that an important allelic loss was observed within the UNC5A, UNC5B, or UNC5C locus (Fig. 4 A and B). UNC5A and UNC5C genes, whose expression was more frequently reduced, were found to display 52% and 39% of LOH, respectively, whereas 26% of LOH occurred within the UNC5B locus.
Figure 4.
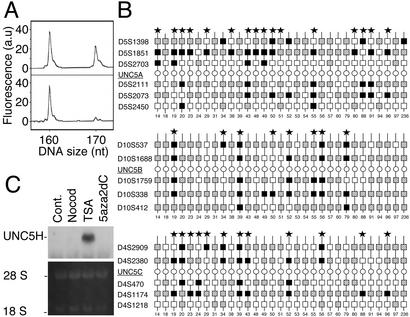
Down-regulation of UNC5H gene expression is associated with allelic loss in UNC5H loci. (A and B) UNC5H allelic loss in colorectal tumors. (A) Fluorescent electropherograms for microsatellite marker D5S1851 on chromosome 5q35.2. The normal (Upper) and tumor (Lower) counterparts of case 14 are shown. a.u., arbitrary units. (B) Summary of LOH analysis for 31 normal/tumor pairs in either UNC5A, UNC5B, or UNC5C locus, showing noninformative (homozygous) markers (white squares), retained markers (shaded squares), and markers with LOH (black squares). Each vertical line indicates a single patient and cases having LOH at one of the UNC5H flanking markers are indicated by *. (C) Effect of trichostatin A and 5aza2dC on UNC5H expression. Northern blot from 5aza2dC-treated (5 μM, 72 h), trichostatin A (TSA)-treated (50 nM, 12 h), or nocodazole (Nocod)-treated (50 ng/ml, 24 h) HT29 cells with an UNC5H2 probe. 28S and 18S RNA are shown as controls for RNA loading.
UNC5H Expression Is Also Probably Modulated by Epigenetic Mechanisms.
We next checked whether epigenetic processes like DNA methylation may also account for the reduction/loss of UNC5H expression. The possible role of promoter methylation was then assessed in different colorectal cell lines. Fig. 4C shows that a treatment with methylation inhibitor 5-aza-2′-deoxycytidine (5aza2dC) (26) only led to a modest increase of UNC5H expression in HT-29 cells, hence suggesting that promoter methylation may slightly affect UNC5H expression. Putative CpG islets can be found in both UNC5A, UNC5B, or UNC5C promoter but the in vivo methylation of these islets has yet to be determined. Interestingly, if 5aza2dC had only a slight effect on UNC5H expression, treatment with trichostatin A, a specific inhibitor of the histone deacetylases that are known to interact with methylated CpGs and to participate to stable transcriptional repression (27), led to a robust increase of UNC5H expression (Fig. 4C). Taken together, these data suggest that histone deacetylase activity mediates part of the transcriptional repression of UNC5H genes even though ongoing work would have to determine whether this regulation is specific for tumors. Interestingly, UNC5B promoter has been recently presented as a direct target of p53 (20), the prototype tumor suppressor and transcription factor whose expression is regulated by histone deacetylase (28). In that sense, as also described by Tanikawa et al. (20), we observed that p53 overexpression led to an increased level of UNC5B (not shown and see Fig. 6A Inset). Thus, the loss of p53 activity in numerous tumors might also account for part of the loss/reduction of UNC5H expression. Taken together, these results suggest that UNC5H expression may be down-regulated in tumors by at least three mechanisms including allelic loss, promoter silencing, and the absence of p53-dependent transactivation.
Figure 6.
p53-mediated suppression of anchorage-independent growth and invasion in vitro is inhibited by netrin-1. (A) LS174T, U373MG, or MCF7 cells were transfected with p53-, UNC5H2-, netrin-1-, or MDM2-expressing construct as indicated and were assessed for growth in soft agar. (Inset) Using U373MG cells transfected as above, UNC5B expression was then assessed by Western blot using the UNC5B antibody described in ref. 20. (B) Similar experiment as in A but U373MG or LS174T cells were assessed for invasion ability in Matrigel chambers for 2 days. As in Fig. 5, the filters were fixed and stained by using the Diff-Quick procedure (Dade Behring). Standard deviations are indicated (n = 3).
Human UNC5C Gene Is Subjected to Mutations in Patients with Colorectal Cancer.
The loss/reduction of UNC5H expression suggested that UNC5H genes might be putative tumor suppressors. Very often, such tumor suppressors are subjected to mutation in cancers. We then analyzed whether UNC5H genes were targets for mutations. We focused our study on UNC5C because it appears to be strongly affected in colorectal cancer. We then performed in a panel of 30 colorectal tumors PCR amplification and sequencing of the 13 accessible exons covering the complete coding sequence of UNC5C except from amino-acids 200–370 whose genomic region is not available. In 40% of samples analyzed, we failed to amplify at least one of the exons, hence strongly suggesting homozygous deletions in UNC5C gene in these tumors. However, we cannot completely exclude a repeated PCR amplification problem, even though this problem would not be related to (i) the quality of the genomic DNA (because other exons from the same genomic DNA were perfectly amplified) and (ii) the quality of the primers (because genomic DNA from other tumors were easily PCR-amplified with the same primers). More importantly, three different mutations were found in five tumors. All mutations were missense mutations. The first mutation converted a proline (amino acid 57) into a tyrosine. The second mutation converted an alanine (amino acid 628) into a glutamic acid. The third mutation, found in three different tumors, converted an alanine (amino acid 841) into a tyrosine. If the impact of the different mutations in UNC5C activity is to be evaluated, it is interesting to note that the A628E mutation is present in the ZU5 domain of UNC5C and that the frequent (i.e., 10% of the tumors tested) A841Y mutation occurs in the death domain of UNC5C, a domain that was shown to be crucial for UNC5H proapoptotic activity (17). The three mutations detected were not, however, observed only in the tumor but also in the adjacent normal tissue. This finding would argue that the mutations detected are probably not the targeted mechanism for inactivating UNC5H in cancer. However, these mutations may result in a loss/reduction of UNC5H activity that taken together with the decreased UNC5H expression described above can act as a selective advantage for tumor development.
UNC5H Expression Inhibits the Hallmarks of Cell Transformation.
The very frequent loss/reduction of UNC5H expression in the majority of the tested tumors suggest that the presence of UNC5H is a negative constraint for tumor development. We then analyzed whether UNC5H might actually inhibit in vitro the classical hallmarks of malignant transformation that are the anchorage-independent growth and the ability to invade (25). Colorectal LS174T, Ras-transformed NIH 3T3, or large T-transformed 293 cells were then transiently transfected with UNC5H-expressing constructs and allowed to grow in soft agar or invade through a reconstituted 3D basement membrane gel (Matrigel). Expression of one of the three UNC5H receptors in both cell lines (Fig. 5A and data not shown) was sufficient to drastically inhibit growth in soft agar and invasion in Matrigel (Fig. 5 and not shown). Interestingly, when netrin-1 was added, both growth in soft agar and invasion were restored (Fig. 5 and not shown). To further investigate the role of UNC5H/netrin-1 in the regulation of these malignant properties, we sought to determine whether endogenous UNC5H, as opposed to ectopically expressed UNC5H, might also regulate anchorage-independent growth and invasive ability. We took advantage of the recent observation that p53 transactivates the UNC5B gene (20). Thus, colorectal LS174T or glioblastoma U373MG cells were transfected with p53 and analyzed for growth in soft agar and invasion in Matrigel. As described, p53 expression led to enhanced UNC5B expression in these cell lines (ref. 20 and Fig. 6A Inset). In these conditions, we detected a reduction of growth in soft agar (Fig. 6A) and a reduction of invasion in Matrigel (Fig. 6B). Remarkably, netrin-1 addition was sufficient to block this reduction (Fig. 6), hence demonstrating that, in these experimental conditions, p53-induced inhibition of these hallmarks of cell transformation is mediated via UNC5B (Fig. 6). Moreover, these data support the fact that the endogenous expression of UNC5B, provoked by p53, is sufficient to inhibit in vitro cell transformation. Along the same line, when the p53 transactivity was inhibited in MCF7 cells by MDM2 overexpression, an increase of growth in soft agar was observed, such an increase being inhibited by UNC5H expression (Fig. 6). Taken together, these data support the role of the pair netrin-1/UNC5H as regulators of cell transformation.
Figure 5.
UNC5H expression suppresses anchorage-independent growth and invasion in vitro. (A) UNC5H3 expression in transfected 293T cells. 293T cells were transiently transfected with either a mock plasmid or UNC5H3-expressing construct in the presence or absence of netrin-1. UNC5H3 expression was monitored by Western blot using an anti-Flag M2 antibody. (B and C) UNC5H expression inhibits growth in soft agar assays. 293T cells were transiently transfected with UNC5H1, UNC5H2, UNC5H3, or netrin-1 expressing constructs (17). Cells were allowed to grow for 10.5 days in soft agar. (B) A set of representative plates is shown. (C) The colonies number was determined as the number of isolated clones with a diameter >100 μm. (D) UNC5H expression inhibits Ras-dependent cell invasion in vitro. NIH 3T3 cells transfected with Ras, UNC5H1, UNC5H2, UNC5H3, and/or netrin-1 were cultured in Matrigel chambers for 2 days. The filters were fixed and stained by using the Diff-Quick procedure (Dade Behring, Düdingen, Switzerland). The percentage of invasion was obtained from the ratio between the number of cells in the coated inserts to the number of cells in the uncoated inserts. Standard deviations are indicated (n = 3).
UNC5H Expression Inhibits the Hallmarks of Cell Transformation by Regulating Apoptosis.
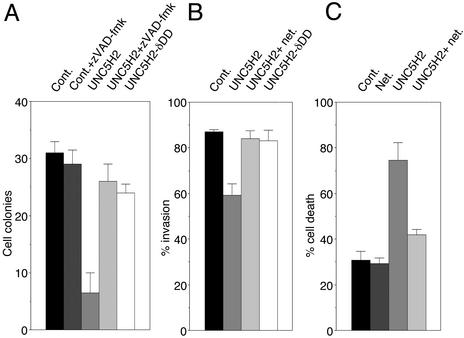
We finally analyzed whether the regulation of apoptosis by the pair netrin-1/UNC5H may be involved in the inhibition of anchorage-independent growth and invasive ability. We observed that a treatment with the general and potent caspase inhibitor zVAD-fmk completely suppress both UNC5H proapoptotic activity (17) and the UNC5H-mediated inhibition of anchorage-independent growth or invasion (Fig. 7 and not shown). Moreover, we described that UNC5H-mediated apoptosis requires the presence of the UNC5H death domain (17). Interestingly a mutant form of UNC5H2 deleted of its death domain was not able to suppress the hallmarks of malignant transformation (Fig. 7). Moreover, expression of UNC5H2 induced a marked increase of trypan blue staining in nascent soft agar cell colonies (Fig. 7C), strongly suggesting that the transformation-suppressing properties of UNC5H are related to the proapoptotic activity of UNC5H.
Figure 7.
UNC5H expression suppresses anchorage-independent growth and invasion in vitro by inducing apoptosis. (A) 293T cells were transfected with a mock plasmid or expressing constructs for UNC5H2 full-length or UNC5H2 deleted for its death domain (UNC5H2-δDD) and were assessed for growth in soft agar as in Fig. 5. To assay the role of apoptosis in the blockade of anchorage-independent growth by full-length UNC5H2, 20 μM zVAD-fmk was added repeatedly. (B) Similar experiment as in A but LS174T cells were assessed for invasion ability in Matrigel chambers for 2 days. As in Fig. 5 the filters were fixed and stained by using the Diff-Quick procedure (Dade Behring). Standard deviations are indicated (n = 3). (C) UNC5H2 induces cell death. Similar experiment as in A was performed except after 48 h of growth in soft agar, cells were stained with trypan blue and numerated as described (11). The percentage of cell death was obtained as ratio between the number of trypan blue-positive cells and the total number of cells. Standard deviations are indicated (n = 3).
Discussion
Here we present evidence that UNC5H receptors, initially described as receptors involved in axon guidance, are expressed in other tissues than the nervous system in which they may act as negative regulators of tumor growth probably by inducing cell apoptosis. Interestingly, the majority of the tumors examined and >90% of colorectal tumors show a loss/reduction of the expression of UNC5H. This reduction is associated with allelic loss in the UNC5A, UNC5B, or UNC5C gene in colorectal cancer. Interestingly, deletions of the chromosome regions where the UNC5A, UNC5B, or UNC5C locus have been mapped (5q35, 10q21–22, and 4q21–23, respectively) have been associated with various cancers (29–31). Further study will have to analyze whether the reduction of UNC5H expression in other types of cancer is also associated with LOH. In colorectal cancer, LOH frequency in each UNC5H gene correlates with the frequency of reduction/loss of expression, even though it is not known whether the loss of one allele is sufficient to explain the loss/reduction of expression. Along this line, the fact that >80% of rectal tumors and 49% of colon tumors show a very important reduction of UNC5H expression (i.e., >5-fold) cannot be explained only with the LOH frequency (52%, 26%, and 39% for the UNC5A, UNC5B, or UNC5C locus, respectively). This finding would argue for an alternative mechanism for the observed reduction of UNC5H expression. We propose that at least two other mechanisms can regulate UNC5H expression. The first mechanism may be related to epigenetic processes because we observed that in colorectal cancer cell lines, UNC5H expression is regulated by histone deacetylase activity. However we do not know yet whether this activity on UNC5H genes occurs specifically in tumors and not in normal tissues and whether this activity affects directly UNC5H promoters or affects other genes that in turn regulate UNC5H. Interestingly, p53 activity was shown to be modulated by histone deacetylase activity. The second mechanism may then be promoter regulation by the tumor suppressor p53 because Arakawa and colleagues (20) have demonstrated that UNC5B is a direct transcriptional target of p53 (20). The loss of p53 activity in multiple cancers may then account for the loss/reduction of UNC5H expression.
Interestingly, we detected mutations in the UNC5C gene. Further work will have to determine whether UNC5A and UNC5B are also subjected to mutation and whether the identified mutations affect UNC5C activity. However, it is interesting to note that three patients harbored a mutation in the death domain of UNC5C, a domain involved in the proapoptotic activity of UNC5H proteins. Such activity was shown here to be crucial for the UNC5H-mediated suppression of the hallmarks of cell transformation. Hence, it is tempting to speculate that inhibition of UNC5H expression and/or inhibition of UNC5H proapoptotic activity would lead to a selective advantage for tumor development. In any case, in view of the frequency of tumors with a reduced UNC5H expression or with mutations in UNC5C compared with the known frequency of mutation/loss of Apc, p53, and Ras in colorectal cancer, UNC5H genes may turn out to be very important tumor suppressors.
The observation that UNC5H expression is lost/reduced in multiple cancers suggests that a selective process to permit the development of tumors is the inhibition of these receptors expression. It is then interesting to correlate these results with the fact that DCC, another receptor inducing apoptosis in the absence of netrin-1 (11, 12, 16), is lost in numerous tumors, including >50% of colorectal tumors (6–8). DCC was initially presented as a putative tumor suppressor because the DCC gene is present in the minimal region of allelic loss in chromosome 18q in two-thirds of colorectal tumors (6–9). However, despite a series of interesting follow-up reports that have supported its tumor suppressor activity, the rarity of point mutations identified in DCC coding sequences, the lack of a tumor predisposition phenotype in DCC hemizygous mice, and the presence in 18q of other candidate tumor suppressor genes have raised questions about DCC's role as a tumor suppressor (ref. 8; P.M. and E. R. Fearon, unpublished work). One of the arguments that supported the tumor suppressor activity of DCC was the observation that DCC induces apoptosis in settings in which netrin-1 is absent (11). The regulation of cell death by the pair DCC/netrin-1 may then represent a key mechanism for controlling tumor development. An hypothesis would be that tumor cells that express DCC would be committed to die as soon as netrin-1 reaches a limited concentration (i.e., after tumor cell proliferation) or as soon as netrin-1 is no more available as would be the case for metastatic cancers.
Netrin-1 may then appear not only as a guidance molecule but also as a survival factor that controls cell fate. Interestingly, in the small intestine, netrin-1 mRNA is detected at the base of the crypt but is no more observed at the villus tip (L.M., C. Bidaud-Bonot, J.-Y.S., and P.M., unpublished work). This restricted expression may then support the role of netrin-1 as a survival molecule whose presence gradually decreases whereas epithelial cells age and move toward the villus tip where they are known to be shed in the intestinal lumen and die (32, 33). Such loss of the netrin-1 receptors like DCC or, as we show here, UNC5H may then affect the naturally occurring cell death induction and may then become a selective advantage for tumorigenesis. This restricted expression of netrin-1 also supports the overall idea of the proapoptotic activity of these receptors being switched off in normal UNC5H/DCC-expressing cells located in an adequate environment whereas it would be switched on when abnormal UNC5H/DCC-expressing cells grow in inadequate settings in which netrin-1 is unavailable (e.g., during metastasis). Further work will have to determine whether modulation of netrin-1 level in vivo affects tumor development.
Interestingly, one of the UNC5H receptors, UNC5B, is a direct transcriptional target for the tumor suppressor p53 (20). Moreover, Tanikawa et al. (20) have elegantly suggested that, in various cell lines, p53-induced apoptosis is mediated via expression of UNC5B and can be antagonized by netrin-1 addition. Here, we propose that the p53-mediated suppression of the classical hallmarks of malignant transformation such as anchorage-independent growth or invasive ability can also be inhibited by the presence of netrin-1. Taken together, these data suggest that the balance between netrin-1 and UNC5H expression may be a crucial parameter for controlling tumor development. Whether netrin-1 represents an interesting target for cancer therapy is now an intriguing question.
Supplementary Material
Acknowledgments
We thank M. Castellazzi, A. Puisieux, and H. Arakawa for reagents, N. Forey and C. Guix for excellent technical assistance, and R. Corvi for preliminary observations on loss of UNC5H expression. This work was supported by the Ligue Contre le Cancer, The Schlumberger Fondation, and National Institutes of Health Grant NS0045093-01).
Abbreviations
- DCC
Deleted in Colorectal Cancer
- LOH
loss of heterozygosity
- 5aza2dC
5-aza-2′-deoxycytidine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Serafini T, Kennedy T E, Galko M J, Mirzayan C, Jessell T M, Tessier-Lavigne M. Cell. 1994;78:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 2.Serafini T, Colamarino S A, Leonardo E D, Wang H, Beddington R, Skarnes W C, Tessier-Lavigne M. Cell. 1996;87:1001–1014. doi: 10.1016/s0092-8674(00)81795-x. [DOI] [PubMed] [Google Scholar]
- 3.Forcet C, Stein E, Pays L, Corset V, Llambi F, Tessier-Lavigne M, Mehlen P. Nature. 2002;417:443–447. doi: 10.1038/nature748. [DOI] [PubMed] [Google Scholar]
- 4.Keino-Masu K, Masu M, Hinck L, Leonardo E D, Chan S S, Culotti J G, Tessier-Lavigne M. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- 5.Fazeli A, Dickinson S L, Hermiston M L, Tighe R V, Steen R G, Small C G, Stoeckli E T, Keino-Masu K, Masu M, Rayburn H, et al. Nature. 1997;386:796–804. doi: 10.1038/386796a0. [DOI] [PubMed] [Google Scholar]
- 6.Fearon E R, Cho K R, Nigro J M, Kern S E, Simons J W, Ruppert J M, Hamilton S R, Preisinger A C, Thomas G, Kinzler K W, et al. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- 7.Cho K R, Fearon E R. Eur J Cancer. 1995;31:1055–1060. doi: 10.1016/0959-8049(95)00128-6. [DOI] [PubMed] [Google Scholar]
- 8.Fearon E R. Biochem Biophys Acta. 1996;1288:M17–M23. doi: 10.1016/0304-419x(96)00023-6. [DOI] [PubMed] [Google Scholar]
- 9.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 10.Hilgers W, Song J J, Haye M, Hruban R R, Kern S E, Fearon E R. Genes Chromosomes Cancer. 2000;27:353–357. [PubMed] [Google Scholar]
- 11.Mehlen P, Rabizadeh S, Snipas S J, Assa-Munt N, Salvesen G S, Bredesen D E. Nature. 1998;395:801–804. doi: 10.1038/27441. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Yao F, Wu R, Morgan M, Thorburn A, Finley R L, Jr, Chen Y Q. J Biol Chem. 2002;277:26281–26285. doi: 10.1074/jbc.M204679200. [DOI] [PubMed] [Google Scholar]
- 13.Rabizadeh S, Oh J, Zhong L T, Yang J, Bitler C M, Butcher L L, Bredesen D E. Science. 1993;261:345–348. doi: 10.1126/science.8332899. [DOI] [PubMed] [Google Scholar]
- 14.Bredesen D E, Ye X, Tasinato A, Sperandio S, Wang J J, Assa-Munt N, Rabizadeh S. Cell Death Differ. 1998;5:365–371. doi: 10.1038/sj.cdd.4400378. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y Q, Hsieh J T, Yao F, Fang B, Pong R C, Cipriano S C, Krepulat F. Oncogene. 1999;18:2747–2754. doi: 10.1038/sj.onc.1202629. [DOI] [PubMed] [Google Scholar]
- 16.Forcet C, Ye X, Granger L, Corset V, Shin H, Bredesen D E, Mehlen P. Proc Natl Acad Sci USA. 2001;98:3416–3421. doi: 10.1073/pnas.051378298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llambi F, Causeret F, Bloch-Gallego E, Mehlen P. EMBO J. 2001;20:2715–2722. doi: 10.1093/emboj/20.11.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonardo E D, Hinck L, Masu M, Keino-Masu K, Ackerman S L, Tessier-Lavigne M. Nature. 1997;386:833–838. doi: 10.1038/386833a0. [DOI] [PubMed] [Google Scholar]
- 19.Hong K, Hinck L, Nishiyama M, Poo M M, Tessier-Lavigne M, Stein E. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- 20. Tanikawa, C., Matsuda, K., Nakanashi, H., Fukuda, S., Nakamura, Y. & Arakawa, H. (2003) Nat. Cell. Biol., in press. [DOI] [PubMed]
- 21.Tessier-Lavigne M, Goodman C S. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 22.Ackerman S L, Kozak L P, Przyborski S A, Rund L A, Boyer B B, Knowles B B. Nature. 1997;386:838–842. doi: 10.1038/386838a0. [DOI] [PubMed] [Google Scholar]
- 23.Canzian F, Salovaara R, Hemminki A, Kristo P, Chadwick R B, Aaltonen L A, de la Chapelle A. Cancer Res. 1996;56:3331–3337. [PubMed] [Google Scholar]
- 24.Slack A, Cervoni N, Pinard M, Szyf M. J Biol Chem. 1999;274:10105–10112. doi: 10.1074/jbc.274.15.10105. [DOI] [PubMed] [Google Scholar]
- 25.Fenrick R, Wang L, Nip J, Amann J M, Rooney R J, Walker-Daniels J, Crawford H C, Hulboy D L, Kinch M S, Matrisian L M, Hiebert S W. Mol Cell Biol. 2000;20:5828–5839. doi: 10.1128/mcb.20.16.5828-5839.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soengas M S, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, McCombie R, Herman J G, Gerald W L, Lazebnik Y A, et al. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- 27.Bird A P, Wolffe A P. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 28.Cameron E E, Bachman K E, Myohanen S, Herman J G, Baylin S B. Nat Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 29.Petersen S, Aninat-Meyer M, Schluns K, Gellert K, Dietel M, Petersen I. Br J Cancer. 2000;82:65–73. doi: 10.1054/bjoc.1999.0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adra C N, Donato J L, Badovinac R, Syed F, Kheraj R, Cai H, Moran C, Kolker M T, Turner H, Weremowicz S, et al. Genomics. 2000;69:162–173. doi: 10.1006/geno.2000.6281. [DOI] [PubMed] [Google Scholar]
- 31.Mendes-da-Silva P, Moreira A, Duro-da-Costa J, Matias D, Monteiro C. Mol Pathol. 2000;53:184–187. doi: 10.1136/mp.53.4.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall P A, Coates P J, Ansari B, Hopwood D. J Cell Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 33.Potten C S. Cancer Metastasis Rev. 1992;11:179–195. doi: 10.1007/BF00048063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.