Abstract
The structure of a trimeric domain-swapped form of barnase (EC 3.1.27.3) was determined by x-ray crystallography at a resolution of 2.2 Å from crystals of space group R32. Residues 1–36 of one molecule associate with residues 41–110 from another molecule related through threefold symmetry. The resulting cyclic trimer contains three protein folds that are very similar to those in monomeric barnase. Both swapped domains contain a nucleation site for folding. The formation of a domain-swapped trimer is consistent with the description of the folding process of monomeric barnase as the formation and subsequent association of two foldons.
Keywords: protein folding, trimerization
Domain swapping is a possible cause of protein assembly and misassembly into oligomers. It has been defined in a recent review (1) as a process in which one domain of a multidomain protein breaks its noncovalent bonds with other domains, but remains covalently bound through the polypeptide backbone. Its place is taken by the same domain of an identical protein chain. Since the first observation of domain swapping, the term has been used to describe swapping of domains or even single strands or helices. The result is an intertwined dimer or higher oligomer. The interface between domains found both in the monomer and in the domain-swapped oligomer is termed the “closed interface,” and the interface found only in the oligomer is termed the “open interface,” or “trimer interface” in our case. To date, three-dimensional domain swapping has been seen in crystallographic detail in more than 10 dimers (1) and one trimer (2).
We describe a crystal form of barnase in which the barnase molecules form a trimer by three-dimensional domain swapping. Barnase (EC 3.1.27.3) is a 110-residue ribonuclease from Bacillus amyloliquefaciens, whose structure was solved by Mauguen et al. (3) (Fig. 1). The trimer structure presented here was determined at a resolution of 2.2 Å. The observed swapping phenomenon is discussed in the context of the large amount of information available on the folding of barnase (4, 5) and about its reconstitution from fragments (6).
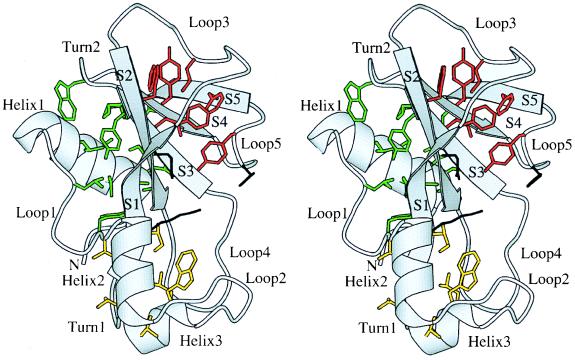
Figure 1.
Overall view of the structure of barnase (3). Secondary structure elements have been labeled. The residues belonging to the hydrophobic cores are shown in green (core 1), yellow (core 2), and red (core 3). The active site residues Lys-27, Glu-73, and His-102 are shown in black. This figure and all subsequent figures have been drawn with the program Bobscript (25).
MATERIALS AND METHODS
Crystallization and Structure Determination.
Crystals were obtained by the hanging drop vapor diffusion method. A protein solution of 10 mg/ml in 50 mM NaOAc, pH 4.5/2 mM exo-guanosine 2′,3′-phosphorothioate (7) was equilibrated against a solution containing 30% (wt/vol) polyethylene glycol 6000 and 0.2 M ammonium sulfate. After several weeks each drop contained two or three small crystals (30 μm in all dimensions) surrounded by a heavy precipitate. Despite their very small size the crystals diffracted to 2.2 Å at the Synchrotron Radiation Source (SRS) in Daresbury, U.K. Data were collected to 3.0 Å by using rotating anode generators equipped either with a MAR image plate (MAR Research, Hamburg) or a FAST detector (Enraf-Nonius Service, Delft, The Netherlands). A high-resolution data set was also collected at the SRS in Daresbury, on station 7.2 (MAR image plate). Details on the data collection are given in Table 1. MAR data were processed with the program mosflm from the CCP4 program suite (8), and FAST data with madness (9). Data were merged with the CCP4 (8) programs scala and agrovata. The unit cells of the different crystals used were very similar, and the final unit cell dimensions used were the average weighted to the number of reflections. The combined data set has a very high multiplicity from 20 to 3 Å, and a high completeness up to 2.2 Å (Table 1). Molecular replacement was performed with the program amore (10), using molecule L of Protein Data Bank entry pdb1brn.ent (11) as a search model. It produced one clear solution, with one molecule per asymmetric unit. The model was refined with xplor (12) using rigid body refinement, simulated annealing procedures, and restrained refinement. Manual model building was performed with o (13). Residues 37–41 did not fit the electron density and were therefore remodeled. A first sulfate ion was modeled in a tetrahedral density in the active site, and a second sulfate ion on the trigonal axis, in the center of the trimer. Water molecules were added in peaks of Fo − Fc density higher than 3σ. The statistics for the final structure are given in Table 1. Molecular surfaces were calculated with the program grasp (14). The structure factors and the coordinates of the final structure have been submitted to the Protein Data Bank.
Table 1.
Summary of the structural statistics
| Measurement | Value |
|---|---|
| Space group | R32 |
| Unit cell | |
| a, Å | 70.13 |
| c, Å | 162.30 |
| Resolution range, Å | 21.4–2.17 |
| No. reflections | |
| Observed | 170,163 |
| Unique | 8,045 |
| Rmerge | |
| Total, % | 6.7 |
| Last shell, % | 22.1 |
| Range, Å | 2.20–2.28 |
| Completeness | |
| Total, % | 99.6 |
| Last shell, % | 99.0 |
| I/sig I | |
| Total | 17.8 |
| Last shell | 6.9 |
| Rfree, % | 24.4 |
| Rfinal, % | 18.6 |
| rms deviation on | |
| Bond distance, Å | 0.011 |
| Angles, ° | 1.502 |
| Dihedrals, ° | 24.491 |
| Impropers, ° | 1.229 |
| Ramachandran plot | |
| In core region, % | 89.5 |
| In allowed region, % | 10.5 |
| Average B factors, Å2 | |
| Main-chain atoms | 25.2 |
| Side-chain atoms | 28.3 |
| Residues 37–41 | 23.4 |
| Sulfates | 35.6 |
| Water molecules | 43.3 |
| No. water molecules | 102 |
Gel Filtration Analysis.
The barnase protein used was a kind gift from Plant Genetic Systems (Gent, Belgium). The protein was analyzed on a Superdex 75 HR (10/30) gel-filtration column (Pharmacia) in 50 mM NaOAc, pH 4.5. Samples analyzed were the lyophilized protein dissolved in water, the lyophilized protein dissolved in the crystallization buffer (50 mM NaOAc, pH 4.5), and a sample left to stand for 1 week in 50 mM NaOAc, pH 4.5/0.2 M ammonium sulfate (10 mg/ml protein). Each sample gave a major peak at the position of the monomer (Mr 12,382), and a small peak (3–6%) at a position that corresponds to a molecular weight of 25,000, indicative of the presence of a dimer rather than a trimer. On SDS/PAGE, protein samples from both peaks migrated as a single band of Mr 10,000. The fractions corresponding to the two peaks were collected separately. The protein from the major (monomer) peak produced the same crystals as the starting material. The protein from the minor peak was found to have a molecular weight of 25,700 by ultracentrifugation, showing that the minor peak indeed corresponds to a barnase dimer. As we had only few very small trimer crystals it was not possible to redissolve them and analyze the trimer by gel filtration.
RESULTS
Barnase was crystallized in a crystal form with trigonal symmetry (R32, Table 1), which had not been reported previously. After molecular replacement, refinement converged at high values for R and Rfree (30% and 34%, respectively). Inspection of the electron density maps showed that the chain tracing of residues 37–41 was completely different from that in the model. Starting at residue 37, the electron density extends away from the search model, toward a model molecule related by threefold symmetry (Figs. 2 and 3). Residues 42–110 of that symmetry-related molecule associate with residues 1–36 of a third symmetry-related model molecule, which in its turn is associated with the first molecule. Thus three barnase molecules form a cyclic domain-swapped trimer, with helix 1, loop 1, and helix 2 forming the swapped domain, and loop 2 the hinge loop [the substructure of barnase is defined as in Serrano et al. (15)].
Figure 2.
(A) Closed barnase monomer. Residues 1–36 are shown in orange, the hinge loop 37–41 is shown in white, and residues 42–110 in yellow. (B) Open barnase monomer, as it is found in the trimer (C) Domain-swapped barnase trimer.
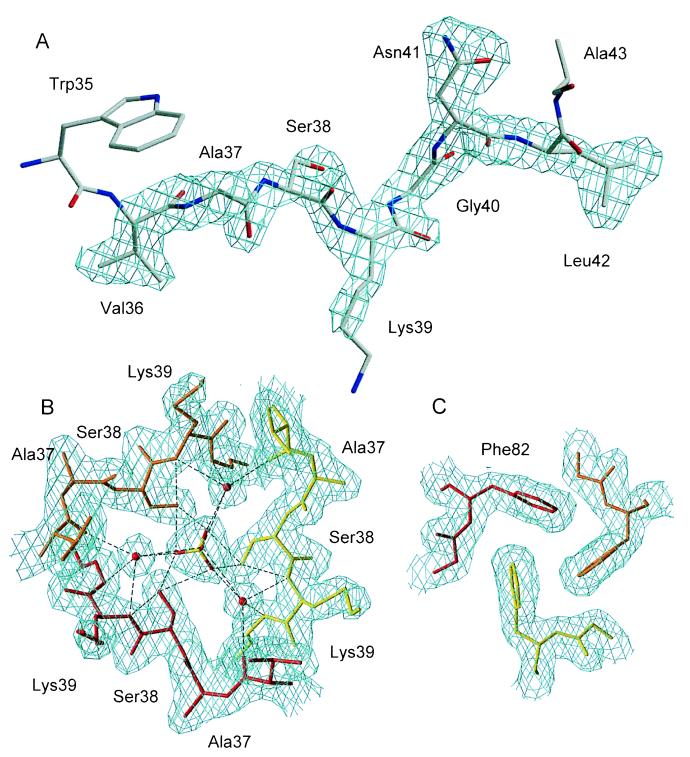
Figure 3.
(A) Fo − Fc simulated annealing omit map for residues 36–42 in the trimeric, swapped form of barnase. The map is contoured at 3σ. (B) 2Fo − Fc map of the trimer interface contoured at 1σ. The network of hydrogen bonds linking Ser-38, the sulfate, and water molecules is shown. (C) 2Fo − Fc map of the trimer interface contoured at 1σ: Stacking interactions between symmetry-equivalent Phe-82 side chains.
The interface between the two domains, or closed interface (1), is identical to that found in the native monomer. It runs through the major hydrophobic core formed by the packing of the first α-helix against the β-sheet (Fig. 1) and through core 2 formed by helix 2, the beginning of loop 2, helix 3, and the first β-strand. It has a molecular surface of 800 Å2 per monomer, which is within the range found for dimer contacts (16, 17). The open interface, corresponding to the new interface created by the trimerization, is limited to Phe-82 and the hinge loop. The surface of the open interface (not taking the hinge loop into account) is quasizero, which implies that a nonswapped trimer would not be stable, as the contact surface between monomers would be small or zero.
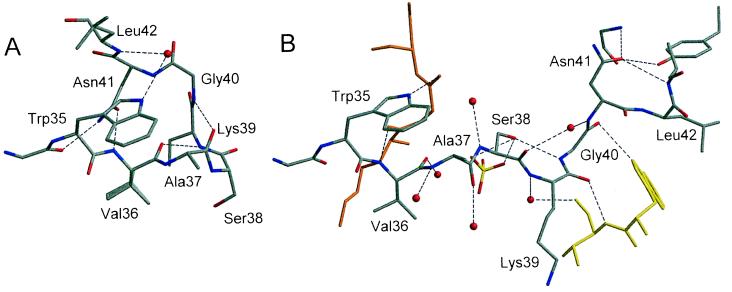
The structures of the two individual domains are conserved; domain 1 (residues 1–36) and domain 2 (residues 42–110) superimpose on the equivalent domains of the normal monomer with rms distances between main chain atoms of 0.25 and 0.30 Å, respectively. The loop 37–41 changes from a helical conformation in the native monomer to a more extended conformation in the swapped molecule (Fig. 4). The most notable difference occurs in Gly-40, where the φ angle changes from 51° in the normal monomer to −85° in the domain-swapped structure. In the native monomer the side chain of Asn-41 stabilizes the “closed” loop conformation by forming hydrogen bonds with Gly-34 O and Val-36 NH. In the trimer the side chain of Asn-41 points in a different direction and replaces a water molecule that interacts with Leu-43 NH, Tyr-78 OH, and Gly-81 NH in the native monomer. This water molecule can be considered as an intrinsic structural component of the monomeric barnase. It is conserved in seven of eight barnase structures with different packing environments, as well as in all structures of the homologous binase (18). This is a clear example of the mimicry of a conserved water molecule by a side-chain oxygen atom. The “open” conformation of the 37–41 loop found in the trimer is further stabilized by hydrogen bonds between Ser-38 Oγ and Gly-40 N, between Lys-39 O and Val-36 N of a symmetry-equivalent molecule, and between Gly-40 O and Trp-35 Nɛ1 of the same symmetry-related molecule. The hinge loop is in contact with the solvent and is hydrogen bonded to a sulfate and several clearly defined water molecules.
Figure 4.
(A) Residues 36–42 in the monomeric form of barnase. (B) Residues 34–43 in the trimeric form of barnase. Symmetry-equivalent molecules are shown in orange and yellow. Hydrogen bonds are shown as dashed lines.
The trimer interface is centered on the crystallographic threefold axis. It has a remarkably layered structure. The first layer consists of seven well-ordered water molecules. Next is a hydrophilic layer consisting of three Ser-38 residues from the three equivalent monomers, a sulfate that lies on the threefold axis, and three water molecules (Fig. 3B). The Ser-38 residues, the sulfate, and the three water molecules are connected by an intricate hydrogen bonding network. The next layer is hydrophobic and consists of three symmetry-equivalent phenylalanine side chains (Phe-82) that participate in stacking interactions with each other (Fig. 3B). The last layer again consists of water molecules.
In the trimer the active site is formed by residues from two distinct polypeptide chains. The general acid Glu-73 and general base His-102 are part of one chain, and the electrostatic catalyst Lys-27 is from another. Despite this, the structure of the active site remains very similar to that of the monomeric barnase. The exo-guanosine 2′,3′-phosphorothioate added in the crystallization setups was, unlike the situation with RNase T1 (19), not bound to the active site of barnase. Instead a sulfate anion makes contacts similar to those of the active site phosphate in the barnase⋅d(CGAC) complex (11). The guanine recognition site and first subsite (11) are solvent accessible, so trimer formation is not expected to sterically inactivate the enzyme.
DISCUSSION
The domain-swapped barnase trimer was formed during the crystallization process, at relatively low pH (pH 4.5) and high protein concentrations. Gel-filtration studies showed that the protein used in the study was initially monomeric, and it remained essentially monomeric in the crystallization buffer solution. Apparently the very high protein concentrations in the actual crystallization experiments are necessary for the formation of the trimer. As only very few, very small crystals grew from each drop, and a heavy precipitate was present in the drops, it appears that only a fraction of the protein crystallized out. It may well be that the cyclic trimeric form selectively crystallized from a mixture of oligomers. Such a mixture of multimers has been observed previously for barnase by Sanz and colleagues (20), who studied the “A-state” of barnase. They found that the thermal unfolding in acidic conditions (pH 2.7) of certain mutants (I4A/I51V and I4A/Y78F) involves slow, multimolecular equilibria. These mutants had been designed to accumulate the folding intermediate of barnase. From fluorimetric and CD studies they concluded that a species with little tertiary structure, but with accessible hydrophobic patches and significant secondary structure, is involved in these processes. They proposed that these mutants form multimers via a pathway that branches off from the folding intermediate of monomeric barnase. They did not observe any multimerization with wild-type barnase, but the concentrations they used were significantly lower than those used in our crystallization setups. Domain swapping can explain many of the observations of Sanz and colleagues. Indeed, the formation of a trimer from monomeric barnase at some stage involves the formation of an “open” form of barnase, in which the two domains are dissociated and the tertiary structure is lost to a major extent (Fig. 2). The closed interface, which is mainly hydrophobic in character, would then become solvent accessible. This “open” form of barnase can form dimers, trimers, and higher multimers, which may explain the heavy precipitate we observe in our crystallization setups.
In retrospect, it is not surprising that this well-studied protein swaps domains in certain conditions. Protein engineering studies have shown that barnase contains two nucleation sites for folding, situated in the first helix and in the β-pleated sheet (21, 22). Both domains involved in the swapping process contain one of these nucleation sites. Studies on mutants of barnase show that loop 2, linking the two domains, is formed very late in the folding process, and that the hydrophobic core 1 is formed throughout the folding reaction (23). Fersht (24) describes the folding of barnase as a process in which two foldons form independently, and form a stable intermediate. The rate-determining step is then the docking and rearrangement of the foldons. Sancho and Fersht (6) have shown that the two folding domains of barnase do not need to be covalently linked to form a functional molecule. All these results directly suggest that the domain-swapped form of barnase derives from the same folding intermediate. At high protein concentrations and acidic conditions the association of foldons from different monomers becomes more favorable, resulting in stable oligomers.
With the barnase trimer presented here we add another member to the growing group of proteins for which domain swapping has been observed. It is also the structure of a genuine domain-swapped trimer for which both the monomeric and the domain-swapped forms are known. Domain swapping, the misassembly of proteins, and alternate folded conformations have recently received much interest, as they are thought to be linked to diseases such as Alzheimer’s and prion-transmissible encephalopathies. The swapped form of barnase could serve as a model system for the study of these phenomena. Fortunately, the monomeric form of the protein, including its folding process, is extremely well characterized.
Acknowledgments
We thank Remy Loris and Freddy Poortmans for help with the data collection, Maria Vanderveken for excellent technical assistance, Joris Messens for help with the gel filtration analysis, Jan Steyaert and Tom Transue for their careful reading of the manuscript, and Yves Geunes for help with the figures. We thank the Fonds voor Wetenschappelijk Onderzoek (FWO) for grants to I.Z. and J.D.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1yvs).
References
- 1.Schlunegger M, Bennett M, Eisenberg D. Adv Protein Chem. 1997;50:61–122. doi: 10.1016/s0065-3233(08)60319-8. [DOI] [PubMed] [Google Scholar]
- 2.Pei X Y, Holliger P, Murzin A G, Williams R L. Proc Natl Acad Sci USA. 1997;94:9637–9642. doi: 10.1073/pnas.94.18.9637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauguen Y, Hartley R W, Dodson G, Bricogne G, Chothia C, Jack A. Nature (London) 1982;297:162–164. doi: 10.1038/297162a0. [DOI] [PubMed] [Google Scholar]
- 4.Fersht A R. FEBS Lett. 1993;325:5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- 5.Fersht A R, Oliveberg M. J Mol Biol. 1998;276:625–646. doi: 10.1006/jmbi.1997.1546. [DOI] [PubMed] [Google Scholar]
- 6.Sancho J, Fersht A R. J Mol Biol. 1992;224:741–747. doi: 10.1016/0022-2836(92)90558-2. [DOI] [PubMed] [Google Scholar]
- 7.Zegers I, Haikal A F, Palmer R, Wyns L. J Biol Chem. 1994;269:127–133. [PubMed] [Google Scholar]
- 8.Collaborative Computing Project Number 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 9.Messerschmidt A, Pflugrath J W. J Appl Crystallogr. 1987;20:306–311. [Google Scholar]
- 10.Navaza J. Acta Crystallogr D. 1994;50:157–163. [Google Scholar]
- 11.Buckle A M, Fersht A R. Biochemistry. 1994;33:1644–1653. doi: 10.1021/bi00173a005. [DOI] [PubMed] [Google Scholar]
- 12.Brünger A T. xplor; Version 3.1, A System for Crystallography and NMR. New Haven, CT: Yale Univ.; 1992. [Google Scholar]
- 13.Jones T, Zou J, Cowan S, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 14.Nicholls A, Kim S A, Honig B. Proteins Struct Funct Genet. 1998;11:282–290. [Google Scholar]
- 15.Serrano L, Kellis J T, Cann P, Matouschek A, Fersht A R. J Mol Biol. 1992;224:783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- 16.Janin J, Miller S, Chothia C. J Mol Biol. 1988;204:155–164. doi: 10.1016/0022-2836(88)90606-7. [DOI] [PubMed] [Google Scholar]
- 17.Jones S, Thornton J. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loris, R., Langhorst, U., Devos, S., Bouckaert, J., Decanniere, K., Maes, D., Transue, T., Steyaert, J. (1999) Protein Sci., in press. [DOI] [PubMed]
- 19.Zegers I, Loris R, Dehollander G, Haikal A F, Poortmans F, Steyaert J, Wyns L. Nature Struct Biol. 1998;5:280–283. doi: 10.1038/nsb0498-280. [DOI] [PubMed] [Google Scholar]
- 20.Sanz J M, Johnson C M, Fersht A R. Biochemistry. 1994;33:11189–11199. doi: 10.1021/bi00203a015. [DOI] [PubMed] [Google Scholar]
- 21.Arcus V L, Vuilleumier S, Freund S M, Bycroft M, Fersht A R. J Mol Biol. 1995;254:305–321. doi: 10.1006/jmbi.1995.0618. [DOI] [PubMed] [Google Scholar]
- 22.Freund S M, Wong K B, Fersht A R. Proc Natl Acad Sci USA. 1996;93:10600–10603. doi: 10.1073/pnas.93.20.10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matouschek A, Serrano L, Fersht A R. J Mol Biol. 1992;224:819–835. doi: 10.1016/0022-2836(92)90564-z. [DOI] [PubMed] [Google Scholar]
- 24.Fersht A R. Curr Opin Struct Biol. 1997;7:3–9. doi: 10.1016/s0959-440x(97)80002-4. [DOI] [PubMed] [Google Scholar]
- 25.Esnouf R. J Mol Graphics. 1997;15:133–138. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]