Abstract
Oxidative stress is believed to be an important mediator of neurodegeneration. However, the transcriptional pathways induced in neurons by oxidative stress that activate protective gene responses have yet to be fully delineated. We report that the transcription factor Sp1 is acetylated in response to oxidative stress in neurons. Histone deacetylase (HDAC) inhibitors augment Sp1 acetylation, Sp1 DNA binding, and Sp1-dependent gene expression and confer resistance to oxidative stress-induced death in vitro and in vivo. Sp1 activation is necessary for the protective effects of HDAC inhibitors. Together, these results demonstrate that HDAC inhibitors inhibit oxidative death independent of polyglutamine expansions by activating an Sp1-dependent adaptive response.
Oxidative stress has been postulated to be a common mediator of a host of neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, Friedreich's ataxia, Huntington's disease, multiple sclerosis, and stroke (1–9). However, evidence that oxidative stress is an initiator or propagator of any of these diseases is incomplete. Indeed, clinical trials of antioxidants in a host of neurological conditions have not resulted in dramatic clinical improvement (10–12). The limited efficacy of antioxidant treatments results, in part, from our limited understanding of the pathways activated by oxidative stress in neurons that affect cell viability.
Primary cultures of cortical neurons provide a convenient in vitro preparation for examining the mechanism(s) by which neuronal degeneration is induced by oxidative stress (13, 14). Early in their development in culture, embryonic rat cortical neurons exposed continuously to elevated concentrations of extracellular glutamate or the glutamate analog homocysteate (HCA) degenerate over 24 h. Degeneration occurs subsequent to the depletion of intracellular glutathione, an important antioxidant. Under these conditions, glutathione depletion by glutamate occurs as a result of competitive inhibition of cystine (two cysteines joined by a disulfide bond) uptake at its plasma membrane transporter, rather than through the activation of ionotropic glutamate receptors. Although neuronal death associated with decreased glutathione content cannot be blocked by competitive or noncompetitive glutamate receptor antagonists, it can be effectively circumvented by treatment with the antioxidants vitamin E and idebenone (14). Glutathione depletion and hypofunction of glutathione-dependent antioxidant enzymes have been linked directly to the pathogenesis of stroke (15), Huntington's disease (16, 17), and Parkinson's disease (18–21) in studies of rodent models as well as human autopsy tissue.
Glutathione-depletion-induced death in primary neurons has many features of apoptosis (23–25) and is completely suppressed by inhibitors of macromolecular synthesis (24, 26). Although nontranscriptional mechanisms of protection by macromolecular synthesis inhibitors have been proposed (26, 27), recent evidence suggests that these agents may also act by interrupting the de novo expression of “death proteins” (28, 29). The potential requirement for transcription in the proper execution of apoptotic death induced by a host of stimuli, including oxidative stress, has stimulated a search for DNA-binding proteins known as transcription factors that are activated by apoptotic stimuli and that govern expression of putative death proteins. Indeed, several cell transcription factors that are activated by death stimuli and that negatively regulate cell viability have been identified, including p53 (30), c-jun (31–33), and E2F (34, 35). By contrast, several transcription factors that positively regulate neuronal survival have also been identified, including cAMP response element binding protein (25, 36, 37), hypoxia-inducible factor-1 (25, 38), and NF-κB (39–42). Thus, whether a cell survives or undergoes cell death in response to cell stress likely depends on a complex interplay between factors, including the balance between prodeath and prosurvival transcriptional regulators. A more complete understanding of the panoply of transcriptional regulators activated by apoptotic stimuli in neurons will guide attempts to tip the balance of transcriptional activities in favor of survival.
Recent data suggest that transcription factor Sp1 may be added to the list of apoptosis-associated transcription factors. Sp1 is a member of an extended family of DNA-binding proteins that have three zinc finger motifs and bind to GC-rich DNA (43, 44). Although classically thought to regulate the constitutive expression of numerous housekeeping genes, Sp1 transcriptional activities have been found to change in association with differentiation (45–47) and proliferation (48) and to regulate gene expression in association with these as well as other cellular functions (49, 50). Indeed, polyglutamine expansions in the huntingtin protein can induce neuronal toxicity, in part, by sequestering Sp1 and one of its coactivators, TAFII130, suggesting a role for Sp1 in neuronal survival (51, 52). Because mutant huntingtin can induce oxidative stress in vitro (53) and in vivo (3, 54), we chose to examine the role of Sp1 in regulating cell viability in an established in vitro model of oxidative stress. We recently showed that glutathione depletion activates an Sp1-dependent adaptive response in neurons (H.R., J.L., K. Zaman, J. Kubilis, R.J.F., B. D. Ross, R. Neve, and R.R.R., unpublished observations). In this article, we provide evidence that cellular oxidative stress activates Sp1 by enhancing its acetylation. We further demonstrate that histone deacetylase (HDAC) inhibitors prevent oxidative neuronal death, in part, by augmenting this Sp1-dependent adaptive response.
Experimental Methods
Primary Neuronal Culture.
Cell cultures were obtained from the cerebral cortex of Sprague–Dawley rats (day 17 of gestation) as described (14). To evaluate the effects of HDAC inhibitors on HCA-induced cytotoxicity, trichostatin A (TSA, Calbiochem; 10–1,000 ng/ml), suberoyl bis-hydroxamic acid (SAHA; Biomol, Plymouth Meeting, PA; 1–20 μM), and butyrate (1–30 mM) were added at the time cortical neurons were exposed to HCA.
Antisense (AS) Oligonucleotides (ODNs) to Reduce Sp1 Expression.
Sp1 AS ODNs were designed to target single-stranded Sp1 mRNA and specifically reduce its expression. The Sp1 AS sequence was 5′-ATCTTGGTCGCTCATGGTCGC-3′ and the Sp1 mismatch (MM) sequence was 5′-ATCTTGGTCCGTCATGGTCGC-3′. All AS and MM ODNs were modified to have phosphorothiate backbone at the ends (Molecular Research Laboratories, Durham, NC; ref. 55). For cortical neuron cultures, final concentrations in the range of 0.1–1 μM ODNs were incubated for 24–48 h in the presence or absence of HCA ± TSA.
Cell Damage and Death Detection.
Population measurements of neuronal cell viability were measured by using a nonradioactive CellTiter 96 assay kit (Promega). In parallel, lactate dehydrogenase release was performed as described (26).
Immunoblot Analysis.
Cell lysates were obtained by rinsing cortical neurons with cold PBS and adding 100 mM Tris (pH 7.4) buffer containing 1% Triton-X 100, 150 mM NaCl, 1 mM sodium orthovanadate, 5 mM sodium fluoride, 3 mM PMSF, 3 mM DTT, 0.5 μg/ml leupeptin, and 10 μg/ml aprotinin. Proteins were then transferred to nitrocellulose membrane (Bio-Rad). Primary Abs against Sp1 (PEP2, Santa Cruz Biotechnology) were diluted at 1:1,000 in 1% milk TBST and exposed to membranes overnight at 4°C. Immunoreactive proteins were detected according to the enhanced chemiluminescent protocol (Amersham Pharmacia). To monitor the Sp1 acetylation in vivo, neuronal lysates obtained from control and TSA-treated cells were precleared by the addition of 30 μl of protein A-Sepharose (50% vol/vol slurry) for 1 h at 4°C. The precleared lysates were then incubated with 2 μg of Sp1 Ab for 4–6 h. Twenty-five microliters of protein A-Sepharose was added to lysates and left for 1 h at 4°C. The samples were boiled and divided into equal aliquots before separation on SDS/PAGE and proteins immunoreactive to acetyl lysine were detected by using an acetyl lysine-specific Ab (1:1,000 dilution; Upstate Biotechnology, Lake Placid, NY).
Electrophoretic Mobility-Shift Assays (EMSAs) and Supershift Analysis.
We performed EMSAs on nuclear extracts from cortical neurons by using a 32P-labeled ODN containing a WT or mutant Sp-1 binding site (Santa Cruz Biotechnology). The sense strand sequences of the double-stranded WT and mutant ODNs were 5′-ATTCGATCGGGGCGGGGCGAGC-3′ and 5′-ATTCGATCGGTTCGGGGCGAGC-3′, respectively. Parallel EMSAs were performed by using a radiolabeled Oct-1 (5′-TGTCGAATGCAAATGACTAGAA-3′; Santa Cruz Biotechnology) binding site. All subsequent steps were performed as described at 4°C (56). To evaluate the effects of various agents on Sp1 and Sp3 DNA binding, we added TSA (100 ng/ml).
Promoter Activity Assay.
Cortical neurons were plated onto 24-well culture plates. The next day, a transfection mixture was prepared by adding 1.5 μg of the reporter expression vector (pSp1-Luc and pmtSp1-Luc, firefly luciferase plasmid) into 150 μl of DMEM with a combination of pRL-cytomegalovirus or thymidine kinase vector (1 μg, containing Renilla luciferase gene) (57). Twenty minutes after the addition of DMRIE-C reagent (6 μl; Life Technologies, Grand Island, NY), the transfection mixture was combined with 1.0 ml of fresh medium and placed onto the cells in 24-well plates. The cells were incubated for 24 h, and then cells were treated with the designated concentrations of HDAC inhibitors. The next day, cells were washed with PBS and then lysed with luciferase assay buffer (Promega).
In Vitro Protein Acetylation Assay.
GST, GST-Sp1, GST-Sp3, GST-p53, and GST-Zta proteins were synthesized in Escherichia coli and purified with glutathione beads. Fusion proteins were eluted by using 20 mM glutathione in 1×HAT buffer [50 mM Tris · HCl, pH 8.0/10% glycerol/1 mM PMSF/0.1 mM EDTA/1 mM DTT/0.05 M NaCl/0.01 M sodium butyrate (Calbiochem)]. The acetylation assay was performed as described (63, 64).
3-Nitroproprionic Acid (3-NP) Toxicity in Mice in Vivo.
Male mice (n = 14) from the B6CBA background strain were housed under standard conditions with free access to water and food. Equal numbers of mice (n = 7) were administered sodium butyrate (1.2 g/kg) via i.p. injection each day for 7 days starting at 40 days of age. After 1 week, 3-NP (Sigma) was administered to both sodium butyrate- and PBS-treated mice (50 mg/kg i.p. twice per day for 10 doses). During 3-NP treatment, both sodium butyrate and PBS administration continued. The brain specimens were cut on a cryostat at 50 microns and stained for Nissl substance (cresyl violet) and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL). Striatal lesion volumes were computed in serial sections through the rostro-caudal extent of each brain by videomicroscopic capture of brain sections and subsequent volume analysis by using neurolucida (Microbrightfield, Williston, VT) image analysis software. The Mann–Whitney U test was used to analyze differences between groups.
Results
Oxidative Stress Induces Sp1 Acetylation.
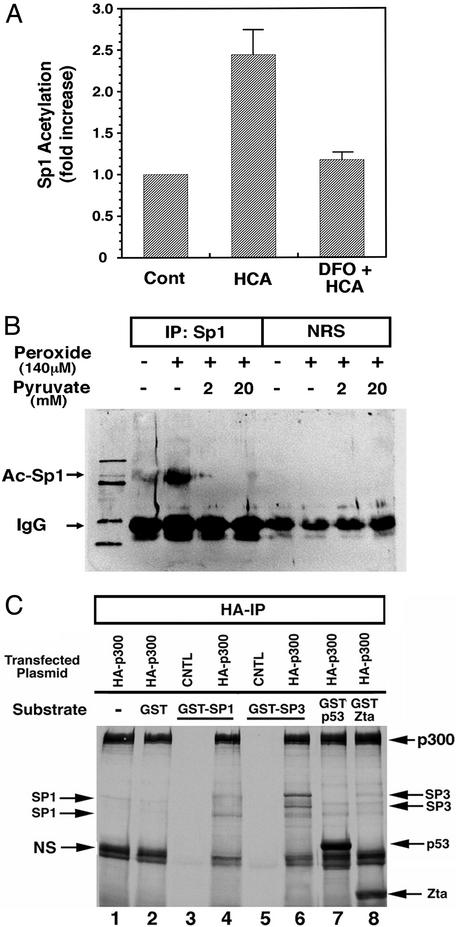
Recent studies have suggested that Sp1-dependent regulation of gene expression can be repressed by HDACs, specifically HDAC1 (58). These studies have suggested, but not demonstrated directly, that HDAC1 or other HDAC family members can repress gene expression by deacetylation of the N-acetyl lysine resides of tissue factors such as Sp1. Indeed, acetylation of lysine residues in transcription factors has been described for a host of factors, including p53 (59), EKLF (60), and NF-κB (61). We therefore examined whether glutathione depletion in cortical neurons could lead to an increase in Sp1 acetylation. Immunoprecipitation with an Sp1 Ab followed by Western blot analysis with an acetyl lysine Ab revealed a statistically significant increase in Sp1 acetylation in response to oxidative stress in cortical neurons (Fig. 1A). This increase in acetylation could be suppressed by antioxidants such as deferoxamine mesylate (Fig. 1A), which abrogate glutathione depletion-induced neuronal death (25). Acetylation of Sp1 appears to represent a general response of cells to oxidative stress as addition of peroxide to U373 glioblastoma cells also leads to increases in Sp1 acetylation (Fig. 1B). Moreover, peroxide-induced Sp1 acetylation in glioblastoma cells can be completely abrogated by the peroxide scavenger pyruvate (Fig. 1B; ref. 62).
Figure 1.
Sp1 acetylation is increased by oxidative stress. (A) Immunoprecipitation of Sp1 followed by immunoblotting with an Ab to acetyl lysine residues reveals an increase in Sp1 acetylation in response to glutathione-depletion-induced oxidative stress. Coadministration of the antioxidant deferoxamine mesylate prevented Sp1 acetylation. Studies have established that deferoxamine (DFO) inhibits oxidative neuronal death distal to glutathione depletion (25). Results are mean fold increase of densitometric units ± SE for three separate experiments (P < 0.03). (B) U373 glioblastoma cell cultures were subjected to hydrogen peroxide with or without sodium pyruvate, harvested after 4 h, immunoprecipitated with anti-Sp1 Ab or normal rat serum (NRS), and analyzed by Western blots with anti-acetyl lysine Ab as described in Experimental Methods. (C) In vitro acetylation of recombinant Sp1 and Sp3 fusion proteins. Autoradiogram of SDS/PAGE gel of fractionated proteins subjected to the in vitro acetylation reaction. Arrows point to respective transcription factors containing radioactive acetyl groups in presence of the acetyl transferase hemagglutinin (HA)-p300. Differences in levels of acetylation by p300 among GST-Sp1, GST-Sp3, GST-p53, and GST-Zta principally reflect differing amounts of full-length protein loaded on gel as determined by Coomassie staining (data not shown).
To determine whether cellular acetyl transferases can directly acetylate Sp1 and its related family member Sp3, we examined whether p300, a transcriptional coactivator with acetyl transferase activity, could acetylate GST-Sp1 or GST-Sp3 purified from bacteria. Consistent with two recent reports (63, 64), we found that purified, recombinant Sp1 and Sp3 could be acetylated by immunoprecipitated p300 (Fig. 1C). We also confirmed observations that recombinant p53 and Zta could be acetylated in vitro by a p300-containing complex (65). These results suggest that Sp1 and Sp3 can be directly acetylated by known transcriptional coactivators with acetyl transferase activity.
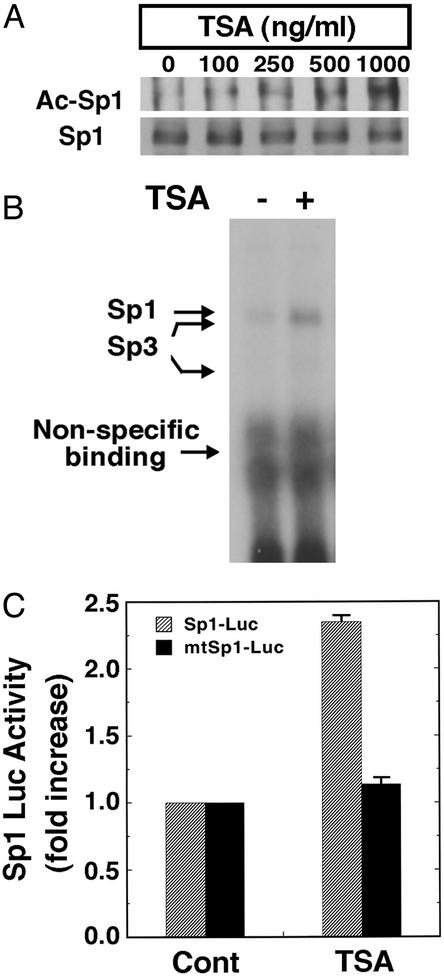
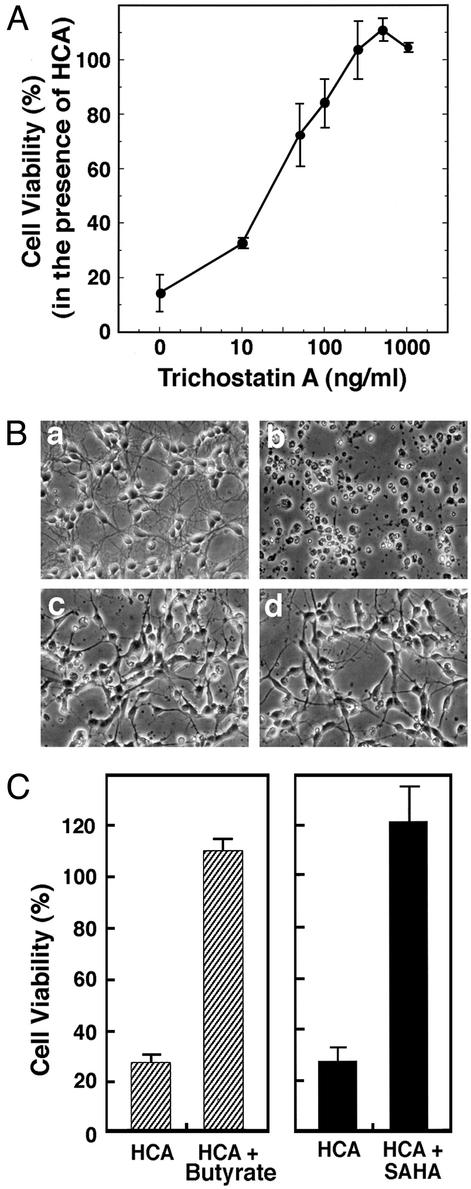
The net amount of acetylation of a protein is a balance between acetyl transferase activity and histone-deacetylating activity (66). To determine whether small molecule, HDAC inhibitors can increase Sp1 acetylation and Sp1 transcriptional activity in neurons, we treated cortical neurons with the prototypic HDAC inhibitor TSA (67, 68). TSA is an organic hydroxamic acid that can potently inhibit the zinc hydrolase activity of HDACs by chelating zinc (69). TSA was added exogenously to cortical neurons, resulting in a concentration-dependent increase of Sp1 acetylation (Fig. 2A). TSA-induced Sp1 acetylation correlated with an increase in Sp1 DNA binding (Fig. 2B) and reporter gene expression driven by two Sp1 consensus response elements (Fig. 2C). Because results from our laboratory (H.R., J.L., K. Zaman, J. Kubilis, R.J.F., B. D. Ross, R. Neve, and R.R.R., unpublished observations) and others (51, 52) predicted that agents that enhanced Sp1 transcriptional activity would abrogate oxidative stress-induced death, we examined the effects of TSA on oxidative cell death induced by the glutamate analog HCA. As expected, we found that TSA prevented oxidative stress-induced death in a concentration-dependent manner (Fig. 3 A and B). Protection by HDAC inhibition was associated with larger neuronal cell bodies and more extensive neurites (Fig. 3B) and was sustained for at least 7–10 days after removal of HCA and HDAC inhibitors (data not shown). We attributed this protective effect to the factor or histone acetylating function of TSA; the structurally distinct HDAC inhibitors butyrate (5 mM; ref. 70) and suberoanilide hydroxamic acid (20 μM; ref. 71) also completely abrogate oxidative death (Fig. 3C).
Figure 2.
HDAC inhibitors augment Sp1 acetylation, Sp1 DNA binding activity, and Sp1-dependent reporter gene expression in cortical neuronal cultures. (A) Sp1 acetylation levels in cortical neurons treated with the prototypic HDAC inhibitor TSA as determined by immunoprecipitation with an Sp1 Ab followed by immunoblotting with acetyl lysine Ab (Ac-Sp1) or Sp1 Ab alone (Sp1). Note that levels of Sp1 do not change with increasing concentrations of TSA. (B) TSA enhances binding of Sp1 and Sp3 to a canonical Sp1 DNA binding site. The electrophoretic mobility-shift assay was performed by using nuclear extracts from cortical neurons treated with and without TSA (100 ng/ml) for 60 min. The presence of Sp1 and Sp3 in each of the induced complexes was verified by supershift analysis. During this short period of TSA exposure, Sp1 or Sp3 protein levels did not change. (C) Sp1-dependent luciferase activity in control and TSA (100 ng/ml)-treated cortical neurons (gray bars). Note that luciferase activity does not change in the presence of TSA when the Sp1 response element has been mutated (black bars).
Figure 3.
HDAC inhibitors inhibit neuronal death induced by glutathione-depletion-induced oxidative stress. (A) TSA inhibits HCA-induced apoptosis in a concentration-dependent manner. Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction. Lactate dehydrogenase (LDH) release assay and TUNEL were performed in parallel to verify that MTT changes reflect changes in viability. Each point is the mean ± SD of three to five independent experiments. (B) Phase-contrast microscopy of cortical neurons: (a) control; (b) 1 mM HCA; (c) 100 ng/ml TSA (note the change in the morphology of the cell bodies); and (d) 100 ng/ml TSA plus 1 mM HCA (note how TSA-treated neurons maintain their cell body and neurite morphology in the presence of 1 mM HCA). (Magnifications: ×200.) (C) Structurally distinct HDAC inhibitors, butyrate (5 mM) and SAHA (5 μM), also inhibit HCA-induced death. Cell viability was measured by using the MTT reduction, LDH release, or TUNEL as described in Experimental Methods. All methods gave quantitatively similar results.
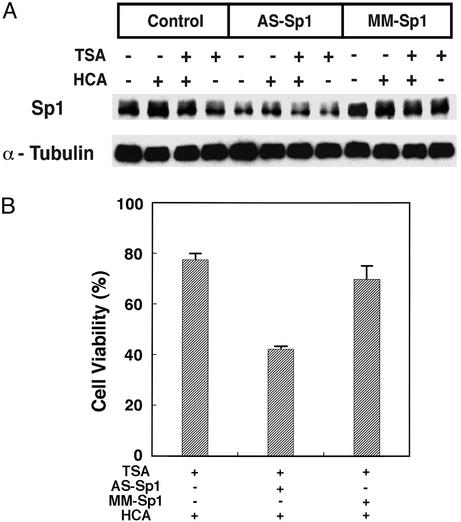
To determine whether Sp1 expression is necessary for the protective effects of HDAC inhibitors, we diminished neuronal Sp1 expression in the presence of TSA by using an AS ODN specific for Sp1 as well as its corresponding MM ODN control. We first confirmed that Sp1 was reduced by 1 μM Sp1 AS ODN but not 1 μM MM ODN in the presence of TSA (50 ng/ml) and HCA (Fig. 4A). Densitometric analysis of three independent experiments revealed that Sp1 AS ODN reduced Sp1 expression by 40%, whereas the MM ODN control reduced Sp1 expression by only 8%. We also found that levels of α-tubulin (Fig. 4A) and the closely related Sp1 family member, Sp3, were unaffected by both ODNs (data not shown). In parallel, we measured viability and as expected, we found that protection by submaximal concentrations of TSA (50 ng/ml) is reduced significantly by the Sp1 ODN (P < 0.05) but not the MM ODN (Fig. 4B). Together, these results are consistent with the hypothesis that HDAC inhibitors prevent neuronal death in this and possible other neurodegenerative paradigms (72), in part, by activating Sp1-dependent transcriptional activity.
Figure 4.
Sp1 is necessary for the protective effects of HDAC inhibitors. (A) Sp1 AS ODNs, but not MM ODNs, deplete Sp1 protein levels in cortical neurons. Sp1 ODNs do not alter the levels of α-tubulin. (B) Sp1 AS ODNs reverse TSA-induced prevention of HCA-induced death; MM ODNs do not. Results are mean ± SE for three separate experiments.
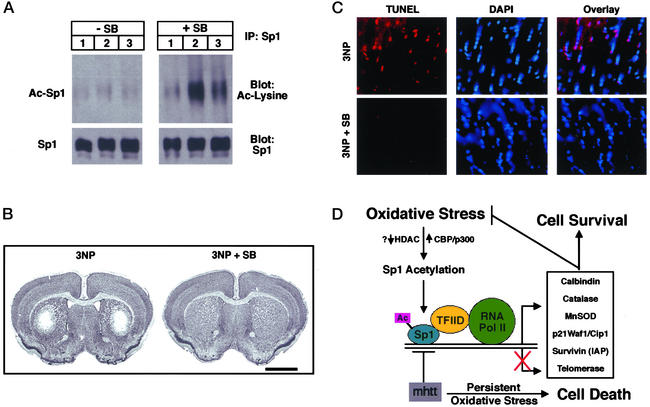
To evaluate whether HDAC inhibitors can induce Sp1 acetylation and prevent neuronal injury caused by oxidative stress in vivo independent of expanded polyglutamine repeat proteins, we examined the effects of sodium butyrate on 3-NP toxicity in rodents. 3-NP is a mitochondrial toxin that produces striatal lesions that closely mimic Huntington's disease and are mediated by excitotoxic-induced oxidative stress mechanisms (73, 74). Animals were pretreated for 1 week with 1.2 g/kg per day sodium butyrate and then exposed to 3-NP as described (73). Animals treated with sodium butyrate and 3-NP had increased levels of acetylated Sp1 as compared with 3-NP-treated animals alone (Fig. 5A). The increased acetylation of Sp1 induced by sodium butyrate treatment was associated with nearly complete protection from striatal 3-NP toxicity as determined by Nissl staining (Fig. 5B) and TUNEL (Fig. 5C). The histopathological evaluation of 3-NP-induced striatal lesion volumes showed significantly less tissue damage in sodium butyrate-treated mice than in PBS-treated mice. (Sodium butyrate-treated 3-NP mice: 1.59 ± 0.38 mm3; PBS-treated 3-NP mice: 9.62 ± 2.24 mm3; P < 0.001.) There was an 83.5% reduction in lesion volume in the sodium butyrate-treated mice (Fig. 5B).
Figure 5.
The HDAC inhibitor sodium butyrate enhances Sp1 acetylation and inhibits 3-NP-induced oxidative neuronal death in vivo. (A) Sp1 acetylation levels in brains of mice (n = 3 for each group) treated without (Left) and with (Right) the HDAC inhibitor sodium butyrate (SB;1.2 g/kg per day) along with 3-NP (50 mg i.p. twice a day for 5 days) as determined by immunoprecipitation with an Sp1 Ab followed by immunoblotting with acetyl lysine Ab (Ac-Sp1) or Sp1 Ab alone (Sp1). Note that levels of Sp1 do not change with SB treatment. (B) Nissl staining of a representative tissue section from mice treated as described in A. Note the loss of Nissl staining induced by 3-NP administration is completely inhibited by SB treatment. (C) TUNEL of a representative tissue section from mice treated as described in A. TUNEL labels double-stranded breaks in DNA and reflects DNA damage. DAPI (4′6-diamidino-2-phenylindole) intercalates into DNA and stains nuclei. Note overlay between TUNEL and DAPI in mice treated with 3-NP alone but not those treated with 3-NP plus SB. (Magnifications: ×400.) (D) Model for redox activation of Sp1 and adaptive responses to oxidative stress. Oxidative stress leads to enhanced acetylation of Sp1. Increased Sp1 acetylation could accrue from decreases in HDAC activity or increases in histone acetyl transferase activity. Hyperacetylated Sp1 binds DNA more avidly, leading to recruitment of coactivators such as TAFII130 in the TFIID complex. This putative Sp1-associated complex can then recruit RNA polymerase II to the promoter of genes such as catalase (22), MnSOD, and p21 waf1/cip1 and increase expression of their mRNA and protein. Up-regulation of the expression of these genes permits the cell to counter oxidative stress and oxidative damage and promotes cell survival. Mutant huntingtin protein can sequester Sp1 and TAFII130 and prevent appropriate adaptation to oxidative stress. The absence of these adaptive responses can lead to persistent oxidative stress and cell dysfunction and ultimately death.
Discussion
Oxidative Stress Induces Sp1 Acetylation.
Recent studies have focused attention on the role of decreased activity of acetyl transferases such as cAMP response element-binding protein or TAFII130 in mediating the toxic, transcriptional, repressive effects of mutated proteins, including proteins such as huntingtin, androgen receptor, and atrophin, with expanded polyglutamine repeats, in neurons (72, 75–77). These findings, along with the established ability of Sp1-dependent transcription to be activated by acetyl transferases and repressed by HDACs (58), raised the possibility that cellular stresses such as oxidative stress might stimulate adaptive Sp1-dependent transcription by altering the balance of activity of acetylation and deacetylation in favor of acetylation. Indeed, we found that glutathione-depletion-induced or peroxide-induced oxidative stress significantly increased Sp1 acetylation in neurons (Fig. 1A) and glioblastoma cells (Fig. 1B), respectively. The results here also represent direct evidence that Sp1 can be acetylated in an intact cell. We confirmed the ability of Sp1 to be acetylated by p300, a known Sp1 coactivator that contains acetyl transferase enzyme activity by in vitro acetylation assays (Fig. 1C). However, whether oxidative stress, like nerve growth factor, induces Sp1-dependent gene expression by recruitment of the coactivator p300 to Sp1 or Sp3 bound to DNA or alternatively, by dissociation of HDAC1 from an Sp1/Sp3 complex, is unclear (Fig. 5D).
The increase in steady-state levels of acetylated Sp1 by oxidative stress is analogous to hyperacetylation of p53 in response to a host of stresses. DNA damage or oxidative stress induce p53 acetylation. The increased p53 acetylation seems to be necessary for p53-dependent transcriptional responses. The NAD-dependent histone and nonhistone deacetylase, Sir2α, can deacetylate p53 at its lysine (382) residue and inhibit p53-dependent p21 waf1/cip expression in mouse embryo fibroblasts (65). Defining the precise residues in the Sp1 protein that are acetylated in response to oxidative stress will be an important step toward understanding how changes in the redox state of a neuron are transduced into increases in Sp1 acetylation.
HDAC Inhibitors Abrogate Oxidative Stress-Induced Death Induced by Glutathione Depletion in Vitro and 3-NP in Vivo: Implications for Therapy of Neurological Diseases.
Enhanced acetylation of Sp1 and enhanced neuronal survival in response to glutathione depletion by three structurally distinct HDAC inhibitors (TSA, butyrate, and SAHA) suggest that glutathione depletion-induced Sp1 acetylation in the absence of HDAC inhibitors represents a frustrated attempt of the cells to protect themselves. HDAC inhibitors have been found to up-regulate a number of genes with putative neuroprotective actions including calbindin D28 (78), metallothionine (79), telomerase (80), and p21 waf1/cip1 (71). Of note, induction of the telomerase (80), MnSOD (81), and p21 waf1/cip1 (82) promoters by butyrate, TSA, or SAHA requires Sp1 and/or Sp3 binding sites. Together, these studies are congruent with a model in which Sp1 is necessary for the protective effects of HDAC inhibitors in cortical neurons subjected to glutathione depletion. The finding here that AS suppression of Sp1 inhibits the ability of HDAC inhibitors to completely inhibit oxidative death (Fig. 4) is also consistent with such a model (Fig. 5D). Several groups have now identified protective roles for HDAC inhibitors in combating the toxicity of mutant huntingtin or mutant androgen receptors, proteins with expanded polyglutamine repeats in vitro (83, 84) and in vivo (85). Moreover, HDAC inhibitors can also abrogate cellular toxicity caused by X-linked adrenoleukodystrophy; the protective effects of these agents in this paradigm have been correlated with increased mitochondrial activity and/or mass (86). Future studies will clarify whether the prevention of neuronal injury and/or death in response to proteins with expanded polyglutamine repeats or peroxisomal disorders by HDAC inhibitors requires, in part, Sp1 binding to GC boxes in DNA. Independent of the precise mechanism of action of HDAC inhibitors, our findings in vitro (Fig. 3) and in vivo (Fig. 5) suggest that these agents are able to abrogate the deleterious effects of oxidative stress in neurons independent of expanded polyglutamine repeats and thus may be propitious therapeutic agents for a host of neurological diseases, including Huntington's disease, amyotrophic lateral sclerosis, Parkinson's disease, and stroke, which have been associated in some cases with decreased histone acetyl transferase activity and in all cases with increased levels of oxidative damage.
Acknowledgments
We thank Rini Ratan, Don DeFranco, and David Ginty for careful review of the manuscript. This work was supported by National Institutes of Health Grants R01 NS39170 (to R.R.R.); RO1 CA71019 (to J.A.-C.); and R01 NS37102, R01 NS35255, and R01 NS045242 (to R.J.F.). R.J.F. was also supported by the Department of Veterans Affairs.
Abbreviations
- HDAC
histone deacetylase
- TSA
trichostatin A
- HCA
homocysteate
- ODN
oligonucleotide
- AS
antisense
- MM
mismatch
- SAHA
suberoyl bis-hydroxamic acid
- 3-NP
3-nitroproprionic acid
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Sagara Y, Dargusch R, Chambers D, Davis J, Schubert D, Maher P. Free Radical Biol Med. 1998;24:1375–1389. doi: 10.1016/s0891-5849(97)00457-7. [DOI] [PubMed] [Google Scholar]
- 2.Atwood C S, Huang X, Moir R D, Tanzi R E, Bush A I. Met Ions Biol Syst. 1999;36:309–364. [PubMed] [Google Scholar]
- 3.Browne S E, Ferrante R J, Beal M F. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castagne V, Gautschi M, Lefevre K, Posada A, Clarke P G. Prog Neurobiol. 1999;59:397–423. doi: 10.1016/s0301-0082(99)00012-x. [DOI] [PubMed] [Google Scholar]
- 5.Estevez A G, Crow J P, Sampson J B, Reiter C, Zhuang Y, Richardson G J, Tarpey M M, Barveito L, Beckman J S. Science. 1999;286:2498–2500. doi: 10.1126/science.286.5449.2498. [DOI] [PubMed] [Google Scholar]
- 6.Floyd R A. Proc Soc Exp Biol Med. 1999;222:236–245. doi: 10.1046/j.1525-1373.1999.d01-140.x. [DOI] [PubMed] [Google Scholar]
- 7.Mattson M P, Pedersen W A, Duan W, Culmsee C, Camandolsa S. Ann NY Acad Sci. 1999;893:154–175. doi: 10.1111/j.1749-6632.1999.tb07824.x. [DOI] [PubMed] [Google Scholar]
- 8.Ratan R R. In: Cell Death and Disease of the Nervous System. Koliatsos V E, Ratan R R, editors. Totowa, NJ: Humana; 1999. pp. 649–666. [Google Scholar]
- 9.Albers D S, Beal M F. J Neural Transm. 2000;59,Suppl.:133–154. doi: 10.1007/978-3-7091-6781-6_16. [DOI] [PubMed] [Google Scholar]
- 10.Shoulson I. Ann Neurol. 1998;44:S160–S166. [PubMed] [Google Scholar]
- 11.Lee J M, Zipfel G J, Choi D W. Nature. 1999;399:A7–A14. doi: 10.1038/399a007. [DOI] [PubMed] [Google Scholar]
- 12.Hickenbottom S L, Grotta J. Semin Neurol. 1998;18:485–492. doi: 10.1055/s-2008-1040901. [DOI] [PubMed] [Google Scholar]
- 13.Murphy T H, Miyamoto M, Sastre A, Schnaar R L, Coyle J T. Neuron. 1989;2:1547–1558. doi: 10.1016/0896-6273(89)90043-3. [DOI] [PubMed] [Google Scholar]
- 14.Murphy T H, Schnaar R L, Coyle J T. FASEB J. 1990;4:1624–1633. [PubMed] [Google Scholar]
- 15.Crack P J, Taylor J M, Flentjar N J, de Haan J, Hertzog P, Ianello R C, Kola I. J Neurochem. 2001;78:1389–1399. doi: 10.1046/j.1471-4159.2001.00535.x. [DOI] [PubMed] [Google Scholar]
- 16.Tkac I, Keene C D, Pfeuuffer J, Low W C, Gruetter R. J Neurosci Res. 2001;66:891–898. doi: 10.1002/jnr.10112. [DOI] [PubMed] [Google Scholar]
- 17.Klivenyi P, Andreassen O A, Ferrante R J, Dedeoglu A, Mueller G, Lancelot E, Bodgonav M, Andersen J K, Jiang D, Beal M F. J Neurosci. 2000;20:1–7. doi: 10.1523/JNEUROSCI.20-01-00001.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry T L, Godin D V, Hansen S. Neurosci Lett. 1982;33:305–310. doi: 10.1016/0304-3940(82)90390-1. [DOI] [PubMed] [Google Scholar]
- 19.Dexter D T, Sian J, Rose S, Hindmarsh J G, Mann V M, Copper F R, Wells S E, Daniel A J, Lee A H, Schapira A H. Anal Neurol. 1994;35:38–44. doi: 10.1002/ana.410350107. [DOI] [PubMed] [Google Scholar]
- 20.Jenner P, Olanow J P. Neurology. 1996;47:S161–S170. doi: 10.1212/wnl.47.6_suppl_3.161s. [DOI] [PubMed] [Google Scholar]
- 21.Jha N, Kumar M J, Boonplueang R, Andersen J K. J Neurochem. 2002;80:555–561. doi: 10.1046/j.0022-3042.2001.00009.x. [DOI] [PubMed] [Google Scholar]
- 22.Nenoi M, Ichimura S, Mita K, Yukawa O, Cartwright I L. Cancer Res. 2001;61:5885–5894. [PubMed] [Google Scholar]
- 23.Ratan R R, Murphy T H, Baraban J M. J Neurochem. 1994;62:376–379. doi: 10.1046/j.1471-4159.1994.62010376.x. [DOI] [PubMed] [Google Scholar]
- 24.Tan S, Sagara Y, Liu Y, Maher P, Schubert D. J Cell Biol. 1998;141:1423–1432. doi: 10.1083/jcb.141.6.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaman K, Ryu H, Hall D, O'Donovan K, Lin K, Miller M P, Marquis J C, Baraban J M, Semenza G L, Ratan R R. J Neurosci. 1999;19:9821–9830. doi: 10.1523/JNEUROSCI.19-22-09821.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ratan R R, Murphy T H, Baraban J M. J Neurosci. 1994b;14:4385–4392. doi: 10.1523/JNEUROSCI.14-07-04385.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furukawa K, Estus S, Fu W, Mark R J, Mattson M P. J Cell Biol. 1997;136:1137–1149. doi: 10.1083/jcb.136.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Maher P, Schubert D. J Cell Biol. 1997a;139:1317–1324. doi: 10.1083/jcb.139.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Maher P, Schubert D. Neuron. 1997b;19:453–463. doi: 10.1016/s0896-6273(00)80953-8. [DOI] [PubMed] [Google Scholar]
- 30.Lowe S W, Ruley H E, Jacks T, Housman D E. Cell. 1993;74:957–967. doi: 10.1016/0092-8674(93)90719-7. [DOI] [PubMed] [Google Scholar]
- 31.Estus S, Zaks W J, Freeman R S, Gruda M, Bravo R, Johnson EM., Jr J Cell Biol. 1994;127:1717–1727. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ham J, Babij C, Whitfield J, Pfarr C M, Lallemand D, Yaniv M, Rubin L L. Neuron. 1995;14:927–939. doi: 10.1016/0896-6273(95)90331-3. [DOI] [PubMed] [Google Scholar]
- 33.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 34.O'Hare M J, Hou S T, Morris E J, Cregan S P, Xu Q, Slack R S, Park D S. J Biol Chem. 2000;275:25358–25364. doi: 10.1074/jbc.M001725200. [DOI] [PubMed] [Google Scholar]
- 35.Liu D X, Greene L A. Neuron. 2001;32:425–438. doi: 10.1016/s0896-6273(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 36.Walton M, Woodgate A M, Muravlev A, Xu R, During M J, Dragunow M. J Neurochem. 1999;73:1836–1842. [PubMed] [Google Scholar]
- 37.Riccio A, Ahn S, Davenport C M, Blendy J A, Ginty D D. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 38.Carmeliet P, Dor Y, Herbert J M, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 39.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 40.Lin K I, DiDonato J, Hoffman A, Hardwick J M, Ratan R R. J Cell Biol. 1998;141:1479–1487. doi: 10.1083/jcb.141.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mattson M P, Culmsee C, Yu Z F. Cell Tissue Res. 2000;301:173–187. doi: 10.1007/s004419900154. [DOI] [PubMed] [Google Scholar]
- 42.Digicaylioglu M, Lipton S A. Nature. 2001;412:641–647. doi: 10.1038/35088074. [DOI] [PubMed] [Google Scholar]
- 43.Briggs M R, Kadonaga J T, Bell S P, Tjian R. Science. 1986;234:47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- 44.Suske G. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 45.Leggett R W, Armstrong S A, Barry D, Mueller C R. J Biol Chem. 1995;270:25879–25884. doi: 10.1074/jbc.270.43.25879. [DOI] [PubMed] [Google Scholar]
- 46.Yan G Z, Ziff E B. J Neurosci. 1997;17:6122–6132. doi: 10.1523/JNEUROSCI.17-16-06122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krainc D, Bai G, Okamoto S, Carles M, Kusiak J W, Brent R N, Lipton S A. J Biol Chem. 1998;273:26218–26224. doi: 10.1074/jbc.273.40.26218. [DOI] [PubMed] [Google Scholar]
- 48.Black A R, Jensen D, Lin S Y, Azizkhan J C. J Biol Chem. 1999;274:1207–1215. doi: 10.1074/jbc.274.3.1207. [DOI] [PubMed] [Google Scholar]
- 49.Lania L, Majello B, De Luca P. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 50.Yang Y, Hwang C K, Junn E, Lee G, Mouradian M M. J Biol Chem. 2000;275:38863–38869. doi: 10.1074/jbc.M007906200. [DOI] [PubMed] [Google Scholar]
- 51. Dunah, A. W., Jeong, H., Griffin, A., Kim, Y. M., Standaert, D. G., Hersch, S. M., Mouradian, M. M., Young, A. B., Tanese, N. & Krainc, D. (2002) 296, 2238–2243. [DOI] [PubMed]
- 52.Li S H, Cheng A L, Zhou H, Lam S, Rao M, Li H, Li X J. Mol Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo A P, Rubinsztein D C. Hum Mol Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- 54.Bogdanov M B, Andreassen O A, Dedeoglu A, Ferrante R J, Beal M F. J Neurochem. 2001;79:1246–1249. doi: 10.1046/j.1471-4159.2001.00689.x. [DOI] [PubMed] [Google Scholar]
- 55.Patzel V, Steidl U, Kronenwett R, Haas R, Sczakiel G. Nucleic Acids Res. 1999;27:4328–4334. doi: 10.1093/nar/27.22.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatterjee S, Zaman K, Ryu H, Conforto A, Ratan R R. Anal Neurol. 2001;49:345–354. [PubMed] [Google Scholar]
- 57.Hsiao M, Tse V, Carmel J, Tsai Y, Felgner P L, Haas M, Silverberg G D. Biochem Biophys Res Commun. 1997;233:359–364. doi: 10.1006/bbrc.1997.6459. [DOI] [PubMed] [Google Scholar]
- 58.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Mol Cell Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 60.Zhang W, Bieker J J. Proc Natl Acad Sci USA. 1998;95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen C J, Deng Z, Kim A Y, Blobel G A, Lieberman P M. Mol Cell Biol. 2001;21:476–487. doi: 10.1128/MCB.21.2.476-487.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin K I, Pasinelli P, Brown R H, Hardwick J M, Ratan R R. J Biol Chem. 1999;274:13650–13655. doi: 10.1074/jbc.274.19.13650. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki T, Kimura A, Nagai R, Horikoshi M. Genes Cells. 2000;5:29–41. doi: 10.1046/j.1365-2443.2000.00302.x. [DOI] [PubMed] [Google Scholar]
- 64.Braun H, Koop R, Ertmer A, Nacht S, Suske G. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prives C, Manley J L. Cell. 2001;107:815–818. doi: 10.1016/s0092-8674(01)00619-5. [DOI] [PubMed] [Google Scholar]
- 66.Kuo M H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 67.Yoshida M, Kijima M, Akita M, Beppu T. J Biol Chem. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 68.Sternson S M, Wong J C, Grozinger C M, Schreiber S L. Org Lett. 2001;3:4239–4242. doi: 10.1021/ol016915f. [DOI] [PubMed] [Google Scholar]
- 69.Van Lint C, Emiliani S, Verdin E. Gene Expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 70.Chang Y-C, Illenye S, Heintz N H. Mol Cell Biol. 2001;21:1121–1131. doi: 10.1128/MCB.21.4.1121-1131.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Richon V M, Sandhoff T W, Rifkind R A, Marks P, A. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steffan J S, Kazantsev A, Spasic-Boskovic O, Greenwald M, Zhu Y Z, Gohler H, Wanker E E, Bates G P, Housman D E, Thompson L M. Proc Natl Acad Sci USA. 2000;97:6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schulz J B, Henshaw D R, MacGarvey U, Beal M F. Neurochem Int. 1996;29:167–171. doi: 10.1016/0197-0186(95)00122-0. [DOI] [PubMed] [Google Scholar]
- 74.Kim G W, Chan P H. J Cereb Blood Flow Metab. 2002;22:798–809. doi: 10.1097/00004647-200207000-00005. [DOI] [PubMed] [Google Scholar]
- 75.Zoghbi H Y, Orr H T. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]
- 76.McCampbell A, Fischbeck K H. Nat Med. 2001;7:528–530. doi: 10.1038/87842. [DOI] [PubMed] [Google Scholar]
- 77.Nucifora F C, Jr, Sasaki M, Peters M F, Huang H, Cooper J K, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson V L, et al. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 78.Arnold D B, Heintz N. Proc Natl Acad Sci USA. 1997;94:8842–8847. doi: 10.1073/pnas.94.16.8842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dressel U, Renkawitz R, Baniahmad A. Anticancer Res. 2000;20:1017–1022. [PubMed] [Google Scholar]
- 80.Takakura M, Kyo S, Sowa Y, Wang Z, Yatabe N, Maida Y, Tanaka M, Inoue M. Nucleic Acids Res. 2001;29:3006–3011. doi: 10.1093/nar/29.14.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Maehara K, Uekawa N, Isobe K. Biochem Biophys Res Commun. 2002;295:187–192. doi: 10.1016/S0006-291X(02)00646-0. [DOI] [PubMed] [Google Scholar]
- 82.Huang L, Sowa Y, Sakai T, Pardee A B. Oncogene. 2000;19:5712–5719. doi: 10.1038/sj.onc.1203963. [DOI] [PubMed] [Google Scholar]
- 83.McCampbell A, Taye A A, Whitty L, Penney E, Steffan J S, Fischbeck K H. Proc Natl Acad Sci USA. 2001;98:15179–15184. doi: 10.1073/pnas.261400698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes R E, Olson J M. Nat Med. 2001;7:419–423. doi: 10.1038/86486. [DOI] [PubMed] [Google Scholar]
- 85.Steffan J S, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol B L, Kazantsev A, Schmidt E, Zhu Y Z, Greenwald M, et al. Nature. 2001;413:739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 86.McGuinness M C, Lu J-F, Zhang H-P, Dong G-X, Heinzer A K, Watkins P A, Powers J, Smith K D. Mol Cell Biol. 2003;23:744–753. doi: 10.1128/MCB.23.2.744-753.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]