Abstract
Lantibiotics are ribosomally synthesized oligopeptide antibiotics that contain lanthionine bridges derived by the posttranslational modification of amino acid residues. Here, we describe the cinnamycin biosynthetic gene cluster (cin) from Streptomyces cinnamoneus cinnamoneus DSM 40005, the first, to our knowledge, lantibiotic gene cluster from a high G+C bacterium to be cloned and sequenced. The cin cluster contains many genes not found in lantibiotic clusters from low G+C Gram-positive bacteria, including a Streptomyces antibiotic regulatory protein regulatory gene, and lacks others found in such clusters, such as a LanT-type transporter and a LanP-type protease. Transfer of the cin cluster to Streptomyces lividans resulted in heterologous production of cinnamycin. Furthermore, modification of the cinnamycin structural gene (cinA) led to production of two naturally occurring lantibiotics, duramycin and duramycin B, closely resembling cinnamycin, whereas attempts to make a more widely diverged derivative, duramycin C, failed to generate biologically active material. These results provide a basis for future attempts to construct extensive libraries of cinnamycin variants.
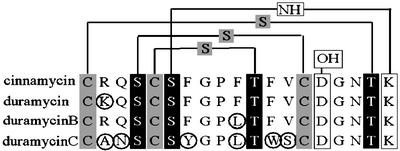
Cinnamycin (Fig. 1) is a peptide antibiotic produced by several Streptomyces strains, including Streptomyces cinnamoneus cinnamoneus DSM 40005. It is assumed to be made by posttranslational modification and processing of a larger ribosomally synthesized peptide. Modifications include the formation of lanthionine bridges, which are thio-ether bridges made when cysteine residues react with dehydroalanine or dehydrobutyrine moieties that have in turn been formed by dehydration of serine or threonine residues, respectively. The possession of lanthionine bridges defines cinnamycin as a lantibiotic. The lantibiotics studied so far are made as prepeptides, each consisting of a leader peptide and a propeptide. Modification of amino acid residues in the propeptide occurs before cleavage of the leader sequence, which releases the mature lantibiotic. In addition to lanthionine residues, lantibiotics may also contain other posttranslationally modified amino acid residues. Thus, cinnamycin contains a β-hydroxy-aspartate residue and a lysino-alanine bridge (apparently formed in a similar manner to a lanthionine bridge but with a lysine residue replacing cysteine; Fig. 1). Lantibiotic synthesis and classification have been reviewed (1).
Figure 1.
Sequence alignment of the propeptides of cinnamycin, duramycin, duramycin B, and duramycin C, highlighting the positions of posttranslational modifications present in the mature molecules. Lanthionine and methyllanthionine rings are indicated by thio-ether linkages, between cysteines (gray boxes) and dehydrated serines or threonines, respectively (black boxes). The hydroxylated aspartate residue, D15, and the lysinoalanine bridge between K19 and S6 are indicated by white boxes. Differences between the amino acid sequence of cinnamycin and the closely related family of duramycin variants are ringed.
There are two major subclasses of lantibiotics. Type A lantibiotics have been studied intensively and have a tertiary structure that is relatively flexible and rod-like. In contrast, the less-studied type B lantibiotics have a relatively globular and inflexible tertiary structure (2). Whereas type A lantibiotics form pores in the cell membrane, type B lantibiotics may inhibit enzymes involved in cell-wall biosynthesis (3). Cinnamycin is closely related to type B lantibiotics duramycin, duramycin B, duramycin C, and ancovenin. These compounds are all derived from 19-aa propeptides and have lanthionine residues in similar positions (Fig. 1). They are all produced by actinomycetes, the duramycins and cinnamycin exclusively by streptomycetes (1), although some of these species were formerly classified in the now defunct genus Streptoverticillium (4). Apart from their antimicrobial activities (5), these compounds also have other potentially useful pharmaceutical properties, including inhibition of angiotensin-converting enzyme, phospholipase A2, and prostaglandin, and leucotriene biosynthesis (1). The structural gene for cinnamycin from Streptoverticillium griseoverticillatum has previously been described (6).
Here we describe the cloning and sequence analysis of the complete cinnamycin biosynthetic gene cluster from S. cinnamoneus cinnamoneus DSM 40005 and demonstrate that the cluster can be manipulated to produce alternative lantibiotics (duramycin and duramycin B). These results suggest that it will be possible to generate novel cinnamycin-like lantibiotics by genetic manipulation of the cinnamycin biosynthetic gene cluster (cin).
Materials and Methods
Strains, Plasmids, and Culture Conditions.
S. cinnamoneus cinnamoneus strains DSM 40646 and DSM 40005 (duramycin and cinnamycin producers, respectively) were obtained from the German Collection of Microorganisms. Streptomyces lividans 1326 and Escherichia coli DH5α (Stratagene), and their culture and manipulation are described in refs. 7 and 8, respectively.
Plasmid pCK51, containing the structural gene for cinnamycin (cinA) from Streptoverticillium griseoverticillatum (6), and sequence data for cinA and its flanking regions, were generously provided by Karl-Dieter Entian (Institut für Mikrobiologie, Frankfurt). Other plasmids used were pBluescript KS (Stratagene) and pOJ436 (9).
Cloning and Analysis of the Genes for Cinnamycin Production.
A ≈2.8-kb NdeI–BamHI restriction fragment was isolated from pCK51 by agarose gel electrophoresis by using a Qiagen gel extraction kit (Qiagen, Valencia, CA). This restriction fragment carries the 5′ part of a lanM-type gene encoding a protein that is probably involved in producing lanthionine residues. The restriction fragment was radiolabeled (Amersham Pharmacia Oligolabeling Kit) and used as a probe in Southern analyses and colony hybridizations.
Initial attempts to clone lantibiotic biosynthetic genes focused on S. cinnamoneus cinnamoneus DSM 40646 (a duramycin producer). By using the above probe, a ≈5-kb BglII restriction fragment was isolated from this strain and cloned into BamHI-cut pBluescriptII KS (Stratagene). The insert in this clone, pIJ10100, was sequenced, and one end of the fragment was found to encode the 3′ end of a lanM-like gene. A 294-bp PCR fragment corresponding to the presumed 3′ end of the lanM homologue was prepared by using primers BB3 (5′-GCC TAC GAG GAC CGG TAC GTC G-3′) and BB4 (5′-GGC GAA GCG CAG GAA GAG CTC G-3′) with pIJ10100 as template and was used to produce the radiolabeled probe-BB for use in Southern analyses, and subsequently E. coli colony hybridizations, of DSM 40646 DNA. Subsequent Southern analysis of the DNA flanking this region of the DSM 40646 genome indicated that the duramycin gene cluster had been disrupted by insertion of an integrating element (unpublished results). Consequently, further work focused on strain DSM 40005.
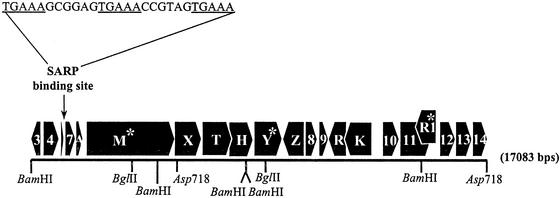
Probe BB was used to isolate and clone a BglII fragment of ≈5 kb from DSM 40005 in BamHI-cut pBluescriptII KS. Two clones, pIJ10101 and pIJ10102, were isolated with the 5-kb BglII fragment in opposite orientations. Sequencing of the insert in pIJ10101 revealed that the 4,981-bp BglII fragment contained the 3′ region of a lanM type gene, which had presumably hybridized to probe BB. The sequence from pIJ10101 was used to design primers to clone overlapping DNA fragments into pBluescriptIIKS. These fragments were sequenced and shown to correspond to a 17,083-bp fragment of the DSM 40005 genome (Fig. 2).
Figure 2.
The cinnamycin gene cluster of S. cinnamoneus cinnamoneus DSM 40005 showing the order and direction of transcription of all of the genes, plus restriction sites used in cloning the cluster. Genes containing a TTA codon are indicated by asterisks. The position and sequence of the potential CinR1 binding site are shown above the map. The individual clones that covered this region of the chromosome were as follows: pIJ10101 and pIJ10102, the central BglII fragment cloned in BamHI cut pBluescriptKSII in opposite orientations; pIJ10103, the BamHI fragment downstream of cinH cloned in BamHI cut pBluescriptIIKS; pIJ10104, the left-most BamHI fragment cloned in BamHI cut pBluescriptIIKS; pIJ10105, the right-most Asp718 fragment cloned into Asp718-cut pBluescriptIIKS; pIJ10108, the small BamHI–Asp718 fragment from pIJ10101 and the insert from pIJ10104 cloned, sequentially, in pOJ436 to give a fragment that corresponds to the left-most portion of the cluster up to the first Asp718 site.
The inserts of the plasmids described in the legend of Fig. 2 were cloned together in pOJ436, an E. coli replicon that can be mobilized into S. lividans, whereupon it integrates into the φ C31 att site (9). The inserts were cloned in a manner that restored the native organization of the constituent pieces in the DSM 40005 genome. The resulting plasmid, pIJ10109, contains a 17,083-bp region of the S. cinnamoneus cinnamoneus DSM 40005 chromosome, plus a small segment of the pBluescriptII KS polylinker. The Asp718 fragment from pIJ10105 was cloned in Asp718-cut pOJ436 to yield pIJ10110.
Plasmids pIJ10109, which contained the entire 17-kb cin cluster, and pOJ436, the vector, were introduced in S. lividans 1326 by transformation, and plasmids pIJ10108 and pIJ10110 (Fig. 2) that contained only part of the proposed cin cluster were introduced in S. lividans 1326 by conjugation.
Construction of a Plasmid for Producing Cinnamycin Variants.
Introduction of unique StuI and SpeI restriction sites within and downstream of cinA, respectively, generated a replaceable cassette that could be used along with the remainder of the cin cluster to produce variants of cinnamycin. cinA is part of the 4,872-bp BamHI fragment of pIJ10104. The StuI site was incorporated with an A1838G substitution in this BamHI fragment, which did not change the amino acid sequence of the predicted gene product. The StuI site lies four codons upstream of the first propeptide codon. The SpeI site was incorporated by making three changes (G1908C, G1910A, and C1912T) immediately downstream of the cinA stop codon. Thus, the whole propeptide-encoding region of cinA could be exchanged by replacement of the resulting StuI–SpeI fragment.
The 4,872-bp BamHI fragment of pIJ10104 was subcloned into pIJ10111, a pBluescriptII KS-based vector in which the XhoI and SpeI sites had been destroyed, to produce pIJ10112. Oligonucleotide primer pairs were made that introduced the StuI or the SpeI sites described above. These primers were as follows: FTP25, 5′-CTT CGT GTG CGA CGG CAA CAC C-3′; SpeI, 5′-GCA GCA ACT AGT TAC TTG GTG TTG CCG TCG-3′; Stu, 5′-CCA CGG AGG CCT TCG CCT GCC GCC AGA GCT GC-3′; StuI, 5′-GGC GAA GGC CTC CGT GGC GGC GAT GTC CTT GG-3′; Xho, 5′-TTC AGC AGT CCG TCG TGG ACG-3′; and Nde, 5′-GCG GAT ACG CGT TAC CCA TAC C-3′.
The StuI site was introduced first by producing PCR fragments by using pIJ10112 as template. The primer pairs Stu and Nde were used with Pfu polymerase to produce the fragment StuPCR1, whereas the primer pairs StuI and Xho were used with Taq polymerase to produce the fragment StuPCR2. The fragments were gel purified, mixed in approximately equimolar amounts, and subjected to PCR with Pfu polymerase and primer pairs Xho and Nde to produce fragment StuPCR3. This PCR product was used to replace the region between the XhoI and NdeI restriction sites of pIJ10112 to generate pIJ10113, which has a StuI site at the position described earlier.
The SpeI site was introduced next by producing PCR fragments by using pIJ10113 as template. The primer pairs FTP25 and Nde were used with Pfu polymerase to produce the fragment SpePCR1, whereas primer pairs SpeI and Xho were used with Pfu polymerase to produce SpePCR2. The fragments were gel purified, mixed in approximately equimolar amounts, and subjected to PCR with Pfu polymerase and primer pairs Xho and Nde to produce fragment SpePCR3. This PCR product was used to replace the region between the XhoI and NdeI restriction sites of pIJ10112 to generate pIJ10114, which possessed SpeI and StuI restriction sites in the positions described earlier. These changes were confirmed by sequencing; however, the sequence also revealed the unintended insertion of 3 bp downstream of the SpeI site. Because the insertion lies downstream of the cinA coding sequence, it was not expected to interfere with lantibiotic biosynthesis.
A further construct was made in which the cinA propeptide-encoding region was replaced with the tetracycline resistance gene (tetR) from pBR322. This was achieved by using PCR, with Taq polymerase, to produce a copy of tetR with a StuI site immediately downstream of the stop codon and a SpeI site 121 bp upstream of the start codon by using the following primers: tet3, 5′-GCG GCG AGG CCT CGC CGG CTT CCA TTC AGG; and tet4, 5′-GCG GCG ACT AGT AAT AGG CGT ATC ACG AGG. This was then used to replace the StuI–SpeI fragment of pIJ10114 encoding the propeptide region of cinA, yielding pIJ10115, which was used to make the cinnamycin derivatives.
Large double-stranded oligonucleotides were made that varied in sequence from the pIJ10114 StuI–SpeI fragment (the cinA-propeptide-encoding region). The sequence changes would theoretically result in the modified cin cluster producing duramycin, duramycin B, or duramycin C instead of cinnamycin. The double-stranded fragments were constructed from 88-mer single-stranded oligonucleotides (duranA, duranB, and duranC, encoding the propeptide regions for duramycin, duramycin B, and duramycin C, respectively) that were annealed to a complementary 20-mer single-stranded oligonucleotide (endfill), which was then elongated with Pfu polymerase to produce 88-mer double-stranded oligonucleotides. The oligonucleotide sequences were as follows: duranA, 5′-GCG GCG AGG CCT TCG CCT GCA AGC AGA GCT GCA GCT TCG GCC CGT TCA CCT TCG TGT GCG ACG GCA ACA CCA AGT AAC TAG TCC GGC C-3′; duranB, 5′-GCG GCG AGG CCT TCG CCT GCC GCC AGA GCT GCA GCT TCG GCC CGC TCA CCT TCG TGT GCG ACG GCA ACA CCA AGT AAC TAG TCC GGC C-3′; duranC, 5′-GCG GCG AGG CCT TCG CCT GCG CGA ACA GCT GCA GCT ACG GCC CGC TCA CCT GGT CGT GCG ACG GCA ACA CCA AGT AAC TAG TCC GGC C-3′; and endfill, 5′-GGC CGG ACT AGT TAC TTG GT-3′.
The double-stranded oligonucleotides were digested with StuI and SpeI and used to replace the StuI–SpeI tetR gene fragment of pIJ10115, resulting in pIJ10116 (duranB), pIJ10117 (duranC), and pIJ10118 (duranA). The cinnamycin structural gene, cinA, had thus been replaced with a duramycin structural gene, a duramycin B structural gene, or a duramycin C structural gene. The plasmids were then used to reconstitute the duramycin equivalents of pIJ10109 in the same manner as pIJ10104. This resulted in pIJ10119, which carries a duramycin B structural gene; pIJ10120, which carries a duramycin C structural gene; and pIJ10121, which carries a duramycin structural gene. These plasmids were used to transform S. lividans 1326.
Bioassays for Production of Cinnamycin and Its Variants.
Streptomycetes were grown at 30°C for 5–7 days on R2YE medium (7). A total of 0.5 ml of an overnight culture of Bacillus subtilis EC1524 was used to inoculate 10 ml of LB medium, which was incubated at 37°C to OD600 0.3–0.4 (5–8 h). Samples (0.5 ml) of this culture were added to 5 ml of soft Oxoid nutrient agar (13 g of Oxoid nutrient broth/6 g of LabM agar per liter, held at 55°C), which was immediately poured over the R2YE plates containing the strains to be assayed. The assay plates were grown at 30°C overnight.
Detection of Cinnamycin by MS.
S. lividans strains were grown at 30°C for 5–7 days in 10 ml of TSB (7) and the mycelium removed by centrifugation. The supernatant was extracted twice with an equal volume of ethyl acetate, and the aqueous fraction was retained each time. The aqueous fraction was extracted twice with butanol equilibrated with water, and the two butanol fractions were pooled. The butanol fraction was extracted with 10× vol of 10% formic acid. The resulting material was subsequently used for analysis on a matrix-assisted laser desorption ionization–time-of-flight MS (Bruker ReflexII; Bruker, Billerica, MA).
Results and Discussion
Analysis of the Cinnamycin Biosynthetic Gene Cluster from S. cinnamoneus cinnamoneus DSM 40005.
Likely coding regions were identified by using the frame program (as implemented at http://watson.nih.go.jp/∼jun/cgi-bin/frameplot.pl; ref. 10), accepting the default values. Likely start codons were assigned by the presence of potential ribosome binding sites located 5–15 bp upstream (taking the sequence GGAGG as the ideal ribosome biding sites; ref. 7). The database search programs blast, based at the National Center for Biotechnology Information, and fasta, based at European Molecular Biology Laboratory, were used to identify proteins similar to those encoded by the cin cluster.
The 17,083-bp region of the S. cinnamoneus cinnamoneus DSM 40005 genome cloned here appears to contain 21 genes (Fig. 2). The two terminal genes are incomplete, and another, smallorf, a potential peptide of 29 aa, may be an artifact produced by a highly repetitive DNA sequence consisting of five direct repeats of a 15-mer nucleotide sequence (GAAGRCGGTGYTCCB).
Most lantibiotic gene clusters contain the following genes: lanA, the lantibiotic prepeptide structural gene; lanBC or lanM, the products of which are responsible for the formation of dehydroalanine, dehydrobutyrine, and/or lanthionine residues; lanT, which encodes a single subunit ATP-binding cassette (ABC) transporter for export of the lantibiotic; lanP, which encodes a peptidase that releases the finished lantibiotic from its leader peptide; lanRK, which encodes a two-component regulatory system; and lanFEG, encoding an ABC transporter responsible for immunity to the mature lantibiotic. Where possible, these conventions have been retained when interpreting the cin gene cluster. Proceeding from left to right of the cluster as represented in Fig. 2, the genes are as follows.
cinorf3 (346 bp and incomplete) encodes the N-terminal portion of a protein with a putative N-terminal signal peptide with 78% amino acid sequence identity to SCO1313, a putative integral membrane protein of Streptomyces coelicolor (11).
cinorf4 (573 bp) probably encodes a regulatory protein with a helix–turn–helix DNA-binding motif similar to the pfam TETR family; Cinorf4 shows 67% identity to SCO1312 of S. coelicolor (11).
smallorf (89 bp) was described earlier.
cinorf7 has a length of 360 bp. The encoded protein has no significant similarities to other proteins.
cinA (236 bp) encodes the cinnamycin prepropeptide. The nucleotide sequence is almost identical (>99%) to the gene previously cloned from Streptomyces griseoverticillatus (formerly Streptoverticillium griseoverticillatum; ref. 6). The leader peptide is unlike those of other lantibiotics and resembles the signal peptides of proteins that are exported via the general secretory (Sec) pathway (for example, the three amino acid residues before the cleavage site of mature cinnamycin, AFA, conform to the AXA motif required for cleavage by type I signal peptidases; ref. 12).
cinM (3,276 bp) encodes a member of the LanM family of proteins, which are responsible for the dehydration of serine and threonine residues in propeptides and the subsequent formation of lanthionine bridges. The most similar homologue is MrsM, the putative modification enzyme for the type B lantibiotic mersacidin (13). cinM contains the rare streptomycete leucine codon TTA, translation of which requires the tRNA encoded by bldA in S. coelicolor, where it is required for morphological development and antibiotic production (14). A putative stem–loop structure lies between cinA and cinM that may reflect an attenuator or a transcript processing site.
cinX has a length of 978 bp. The encoded protein has no significant similarities to other proteins.
cinT and cinH (930 and 873 bp, respectively) encode two translationally coupled subunits of an ABC transporter. CinT appears to be the ATP-binding subunit and is a member of the pfam family of ABC transporters; it shows 36% identity to MrsF (which is part of a putative self-protection transporter found in the mersacidin cluster; ref. 6). CinH appears to be an integral membrane subunit of the pfam family of ABC-2 type transporters commonly involved in drug efflux and resistance, or carbohydrate export.
cinY (1,023 bp) encodes a product that may possess an N-terminal signal peptide, suggesting that it may be exported; no other significant similarities to known proteins. cinY contains a TTA codon within the sequence encoding the putative signal peptide.
cinZ has a length of 627 bp. The encoded protein is 79% identical to SCO1302 of S. coelicolor (11). CinZ shares ≈40% identity with PepQ, a proline dipeptidase from E. coli (GenBank accession no. CAA38501) and appears to be a member of a family of proteins (pfam UPF0029) of unknown function.
cinorf8 (420 bp) encodes a protein that is 75% identical to SCO1304 of S. coelicolor (11). Cinorf8 appears to be a member of the pfam CoA-binding family of proteins.
cinorf9 has a length of 321 bp. The encoded protein may possess a possible N-terminal signal peptide, suggesting that it may be exported; otherwise, no significant similarities to known proteins.
cinR and cinK (651 and 1,065 bp, respectively) overlap by 4 bp, and encode a two-component regulatory system. CinR and CinK are homologues of DegU (a sensory histidine kinase) and DegS (a response regulator) of B. subtilis, which are adjacent to a homologue (yviA) of cinZ.
cinorf10 has a length of 498 bp. The encoded protein has no significant similarities to other proteins.
cinorf11 (1,191 bp) encodes a protein that possesses a domain similar to the pfam family DedA. This family of domains, none of which have been functionally characterized, occurs in proteins with multiple membrane-spanning helices. The start codon of cinorf11 overlaps with the stop codon of cinorf10. cinorf11 appears to overlap the downstream convergently transcribed gene (cinR1) by 570 base pairs.
cinR1 (786 bp) encodes a protein that belongs to the Streptomyces antibiotic regulatory protein (SARP) family of antibiotic pathway-specific regulatory proteins. SARPs possess an OmpR-like DNA-binding motif, which recognizes distinctive sequences in the promoter regions of regulated genes. These sequences are composed of 5- to 6-bp repeats at intervals of about 11 bp, placing them on the same face of the DNA helix (15). A likely SARP-binding site, a 5-bp motif (TGAAA) repeated three times with a 6-bp spacer, lies 83 bp upstream of the cinorf7 start codon. Like several other SARP genes, cinR1 contains a TTA codon (16). This is the first example of a lantibiotic gene cluster that is apparently regulated by a SARP.
cinorf12, cinorf13, and cinorf14 have lengths of 492, 585, and 535 bp, respectively. The encoded proteins have no significant similarities to other proteins. cinorf14 is incomplete.
pIJ10109, Containing the cin Cluster, Confers the Ability to Produce Cinnamycin on S. lividans 1326.
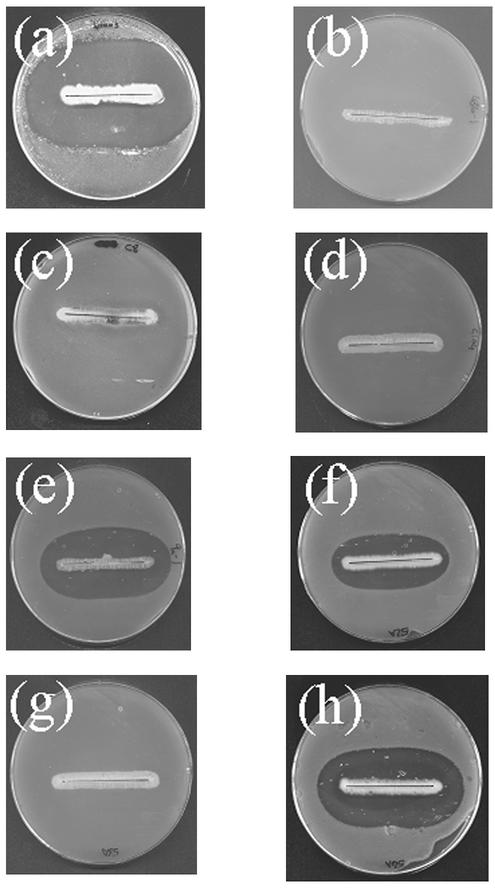
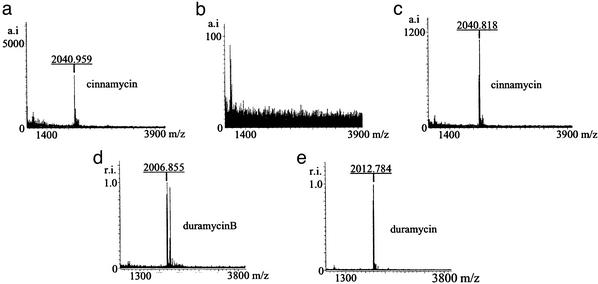
S. cinnamoneus cinnamoneus DSM 40005 inhibited the growth of B. subtilis EC1524 in overlay assays (Fig. 3a) and produced a compound with a Mr of 2,040 (Fig. 4a), identical to that of cinnamycin. S. lividans/pOJ436, S. lividans/pIJ10108 (containing cinA and cinM), and S. lividans/pIJ10110 (containing the Asp-718 fragment that occurs downstream of cinM) lacked both of these properties. S. lividans/pIJ10109, containing the cloned putative cinnamycin pathway, inhibited B. subtilis EC1524 and produced a compound of Mr 2040 (Fig. 4c). These results suggest that the 17,083-bp segment contains all of the genes needed for cinnamycin production in S. lividans.
Figure 3.
Undersides of plates streaked with various Streptomyces strains overlaid with B. subtilis EC1524. Lantibiotic production is indicated by a zone of inhibition of B. subtilis EC1524 around the Streptomyces culture. The strains are (a) S. cinnamoneus cinnamoneus DSM 40005 (cinnamycin) and S. lividans derivatives carrying the following plasmids; (b) pOJ436; (c) pIJ10108; (d) pIJ10110; (e) pIJ10109 (cinnamycin); (f) pIJ10119 (duramycin B); (g) pIJ10120 (duramycin C); (h) pIJ10121 (duramycin). The lantibiotic predicted to be made by the strain is given in parentheses.
Figure 4.
Matrix-assisted laser desorption ionization time-of-flight mass spectra of extracts of media in which the following strains were grown: (a) S. cinnamoneus cinnamoneus DSM 40005 (cinnamycin); (b) S. lividans/pOJ436; (c) S. lividans/pIJ10109 (cinnamycin); (d) S. lividans/pIJ10119 (duramycin B); (e) S. lividans/pIJ10121 (duramycin). The lantibiotic predicted to be made by the strain is given in parentheses. The masses shown are 1 Da in excess of actual masses because of the addition of a proton during ionization. Masses quoted elsewhere have been rounded up and the mass of the extra proton has been subtracted. The larger peaks to the right of the peak for duramycin B in d correspond to masses predicted for the unmodified duramycin B propeptide and its oxidized form.
Because the biosynthetic gene cluster for nisin (17, 18) comprises only 11 genes, and that for the type B lantibiotic mersacidin (13, 19) contains only 10, it is unlikely that all 21 of the genes described here are involved in the production of cinnamycin.
Database searches have enabled us to propose roles for several of the genes contained in the cloned segment. Likely functions are readily assigned to the lanA-type gene, cinA, and the lanM-type gene, cinM. The two-component regulatory system encoded by cinR and cinK may play a role functionally analogous to that of lanR and lanK. In addition to these two regulatory genes, there is also a SARP (15) gene cinR1. In some other streptomycete antibiotic gene clusters, for example those for undecylprodigiosin (20) and daunorubicin (21), expression of a SARP is controlled by another regulatory gene. Consequently, we propose that CinK and CinR regulate transcription of cinR1. cinT and cinH encode an ABC transporter that does not closely resemble either a typical lantibiotic export ABC transporter (LanT) or a typical lantibiotic resistance ABC transporter (LanFEG). Although cinT does have some similarity to mrsF, a lanF-type gene, cinH does not show close similarity to any lanE- or lanG-type genes. However, CinH appears to be a member of a family of proteins that includes many antibiotic resistance transporters. Therefore, we propose that cinT and cinH are involved in cinnamycin self-protection.
These assignments still leave several functions necessary for the production of cinnamycin unaccounted for. In addition to the characteristic lanthionine residues, cinnamycin has a lysino-alanine ring and a hydroxy-aspartate residue, potentially accounting for some of the remaining genes. If CinT and CinH are involved in self-protection, then an export mechanism will be required. This function is performed by a single subunit ABC transporter (LanT) in most other lantibiotic clusters. There is no such gene in the cin cluster, although it is conceivable that the CinTH transporter is responsible for both export and self-protection. A protease activity is also required to cleave the modified propeptide from its leader peptide and thus release cinnamycin. In other lantibiotic clusters, this activity is performed either by a single subunit protease (LanP) or by a protease domain incorporated into the LanT protein. This cin cluster does not contain an obvious candidate for a protease or a protease domain on either of the CinTH subunits.
Because the functions of Cinorf7 and CinX are unclear, they could represent novel LanT or LanP proteins. Alternatively, they could be responsible for the additional modifications required for cinnamycin production. Intriguingly, the CinA prepeptide has a much longer leader peptide than other lantibiotics (1). The leader peptide weakly resembles a signal peptide and has a potential signal peptidase cleavage site just before the N-terminal cysteine of the cinnamycin propeptide. Therefore, cinnamycin may be secreted by a more general export mechanism, such as the general secretory (Sec) pathway.
Even if the last hypothesis is false, it is unlikely that modification, export, and cleavage account for all of the remaining 15 genes in the 17,083-bp fragment of pIJ10109. If the SARP and its potential binding site delineate the ends of the cluster, this still encompasses 15 genes, of which 8 cannot yet be assigned functions.
There must be at least five promoters in the cluster. Four of these are needed to express the following divergently transcribed gene pairs; cinorf3 + cinorf4, cinZ + cinorf8, cinK + cinorf10, and cinR1 + cinorf12. The putative SARP-binding site is highly indicative of a fifth promoter, which is likely to allow transcription of cinorf7, cinA, cinM, and cinX as part of an operon; cinT, cinH, and cinY may also be cotranscribed with these genes.
Modification of the cinA Structural Gene Results in Production of Cinnamycin Variants.
Introduction of StuI and SpeI sites on either side of the cinnamycin propeptide-encoding region of cinA allowed replacement of that region with oligonucleotides encoding three different lantibiotics: duramycin, duramycin B, and duramycin C. S. lividans/pIJ10121 produced a substance with the expected mass of duramycin (Mr 2,012; Fig. 4e) and inhibited B. subtilis EC1524 (Fig. 3h). Similar observations were made for S. lividans/pIJ10119, which produced a compound with the expected mass of duramycin B (Mr 2,006; Figs. 3f and 4d; a prominent peak that corresponds to the calculated mass for duramycin B propeptide can also be seen in Fig. 4d). Less prominent peaks of similar masses are present in the other S. lividans lantibiotic-producing strains, indicative of a small amount of the respective unmodified propeptide. However, S. lividans/pIJ10120 did not inhibit B. subtilis, nor did it produce a compound with a mass that corresponded to that of duramycin C (Mr 1,949; Fig. 3g).
We have shown that the cinnamycin gene cluster can be used to produce related lantibiotics. We were able to produce duramycin and duramycin B by using this cluster, but not duramycin C. Duramycin and duramycin B differ from cinnamycin by only one amino acid residue each (Fig. 1). For duramycin, this residue is R2K, and for duramycin B, it is F10L. These changes are conservative, but they show that there is some flexibility in the cinnamycin production machinery that will tolerate at least some changes in unmodified amino acid residues. In contrast, there are six differences between cinnamycin and duramycin C: R2A, Q3N, F7Y, F10L, F12W, and V13S. The cinnamycin biosynthetic machinery does not appear to be able to cope with all of these changes. Of the six differences between cinnamycin and duramycin C, the most detrimental are likely to be the nonconservative changes R2A and V13S.
Acknowledgments
We are grateful to Prof. Karl-Dieter Entian for donating plasmid pCK51 and sequence data, to Prof. David Hopwood and Dr. Mark Buttner for advice on the preparation of this manuscript, and to Dr. Mike Naldrett for assistance with MS. This work was funded by Biotechnology and Biological Sciences Research Council Grant 208/P08242.
Abbreviations
- cin
cinnamycin biosynthetic gene cluster
- SARP
Streptomyces antibiotic regulatory protein
- ABC
ATP-binding cassette
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ536588).
References
- 1.Sahl H-G, Bierbaum B. Annu Rev Microbiol. 1998;52:41–79. doi: 10.1146/annurev.micro.52.1.41. [DOI] [PubMed] [Google Scholar]
- 2.Jung G. In: Nisin and Novel Lantibiotics. Jung G, Sahl H A, editors. Leiden, The Netherlands: ESCOM; 1991. pp. 1–34. [Google Scholar]
- 3.Brotz H, Josten M, Wiedemann I, Schneider U, Gotz F, Bierbaum G, Sahl H G. Mol Microbiol. 1998;30:317–327. doi: 10.1046/j.1365-2958.1998.01065.x. [DOI] [PubMed] [Google Scholar]
- 4.Witt D, Stackebrandt E. Syst Appl Microbiol. 1990;13:361–371. [Google Scholar]
- 5.Choung S Y, Kobayashi T, Takemoto K, Ishitsuka H, Inoue K. Biochim Biophys Acta. 1998;940:180–187. doi: 10.1016/0005-2736(88)90193-9. [DOI] [PubMed] [Google Scholar]
- 6.Kaletta C, Entian K-D, Jung G. Eur J Biochem. 1991;199:411–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- 7.Kieser T, Bibb M J, Buttner M J, Chater K F, Hopwood D A. Practical Streptomyces Genetics. Norfolk, U.K.: The John Innes Foundation; 2000. [Google Scholar]
- 8.Sambrooke J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Bierman M, Logan R, O'Brien K, Seno E T, Rao R N, Schoner B E. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 10.Ishikawa J, Hotta K. FEMS Microbiol Lett. 1999;174:251–253. doi: 10.1111/j.1574-6968.1999.tb13576.x. [DOI] [PubMed] [Google Scholar]
- 11.Bentley S D, Chater K F, Cerdeño-Tárraga A-M, Challis G L, Thomson N R, James K D, Harris D E, Quail M A, Kieser H, Harper D, et al. Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 12.Carlos J L, Paetzel M, Brubaker G, Karla A, Ashwell C M, Lively M O, Cao G, Bullinger P, Dalbey R E. J Biol Chem. 2000;275:38813–38822. doi: 10.1074/jbc.M007093200. [DOI] [PubMed] [Google Scholar]
- 13.Altena K, Guder A, Cramer C, Bierbaum G. Appl Environ Microbiol. 2000;66:2565–2571. doi: 10.1128/aem.66.6.2565-2571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leskiw B K, Lawlor E J, Fernandez-Abalos J M, Chater K F. Proc Natl Acad Sci USA. 1991;88:2461–2465. doi: 10.1073/pnas.88.6.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wietzorrek A, Bibb M. Mol Microbiol. 1997;43:1177–1184. doi: 10.1046/j.1365-2958.1997.5421903.x. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Moreno M A, Caballero J L, Hopwood D A, Malpartida F. Cell. 1991;66:769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- 17.Kuipers O P, Bierbaum G, Ottenwalder B, Dodd H M, Horn N, Metzger J, Kupke T, Gnau V, Bongers R, van den Bogaard P, et al. Antonie Leeuwenhoek. 1996;69:161–169. doi: 10.1007/BF00399421. [DOI] [PubMed] [Google Scholar]
- 18.Entian K D, deVos W M. Antonie Leeuwenhoek. 1996;69:109–117. doi: 10.1007/BF00399416. [DOI] [PubMed] [Google Scholar]
- 19.Chatterjee S, Chatterjee D K, Jani R H, Blumbach J, Ganguli B N, Klesel N, Limbert M, Seibert G. J Antibiot. 1992;45:839–845. doi: 10.7164/antibiotics.45.839. [DOI] [PubMed] [Google Scholar]
- 20.White J, Bibb M. J Bacteriol. 1997;179:627–633. doi: 10.1128/jb.179.3.627-633.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuya K, Hutchinson C R. J Bacteriol. 1996;178:6310–6318. doi: 10.1128/jb.178.21.6310-6318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]