Abstract
Protein, mtDNA, and nuclear microsatellite DNA analyses have demonstrated that the Yellowstone grizzly bear has low levels of genetic variability compared with other Ursus arctos populations. Researchers have attributed this difference to inbreeding during a century of anthropogenic isolation and population size reduction. We test this hypothesis and assess the seriousness of genetic threats by generating microsatellite data for 110 museum specimens collected between 1912 and 1981. A loss of variability is detected, but it is much less severe than hypothesized. Variance in allele frequencies over time is used to estimate an effective population size of ≈80 across the 20th century and >100 currently. The viability of the population is unlikely to be substantially reduced by genetic factors in the next several generations. However, gene flow from outside populations will be beneficial in avoiding inbreeding and the erosion of genetic diversity in the future.
Managers of threatened populations face the challenge of balancing genetic and demographic concerns. The importance of genetic forces in determining the fate of small populations is a topic of extended debate (1–3). Recent studies demonstrate that inbreeding depression can affect demographic rates and thereby increase extinction probability (4–6). In many cases, however, the shifts in demographic rates that drive population decline have nongenetic origins such as habitat degradation or human-caused mortality (1). Genetic factors may simply hasten the extinction process once the population is small. To evaluate the relative importance of genetic factors, managers need to know the current and historical effective population size (Ne). By measuring the amount of the gene pool passed on to the next generation, Ne determines the rate that diversity declines and the degree of inbreeding (7). Unfortunately, estimating current Ne is difficult (8) and few estimates of historical Ne exist (e.g., refs. 9 and 10). Here we demonstrate how molecular genetic analysis of museum specimens can be combined with demographic data to estimate historical and current Ne and address genetic concerns for the Yellowstone grizzly bear.
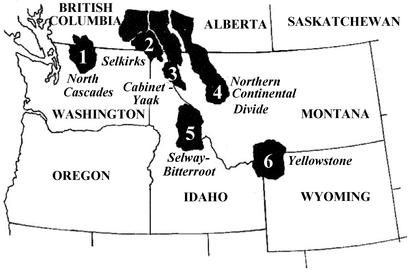
Since 1800, grizzly bears (a regional name for brown bears) have been extirpated from >99% of their historic range south of the Canadian border (11), which prompted their listing under the U.S. Endangered Species Act in 1975. Substantial numbers persist only in the Yellowstone Ecosystem (YE) population and the Northern Continental Divide Ecosystem (NCDE) population along with small numbers in several mountain ranges that extend south from Canada (ref. 12; Fig. 1). Unlike other grizzly populations in the lower 48 states, the one in the YE is and has been isolated for approximately a century. In the early 1990s the U.S. Fish and Wildlife Service began exploring options for addressing genetic concerns in the population. The importance of genetics was again highlighted in 1995 when a federal judge ruled that the U.S. Fish and Wildlife Service had failed to meet its Endangered Species Act recovery planning obligation by paying insufficient attention to the risk of genetic isolation.
Figure 1.
The six grizzly bear recovery zones in the lower 48 states. Approximate number of bears in each ecosystem: zone 1, ≈5; zone 2, ≈25–35; zone 3, ≈20–30; zone 4, ≈400–500; zone 5 = 0; zone 6, ≈280–610 (12, 18).
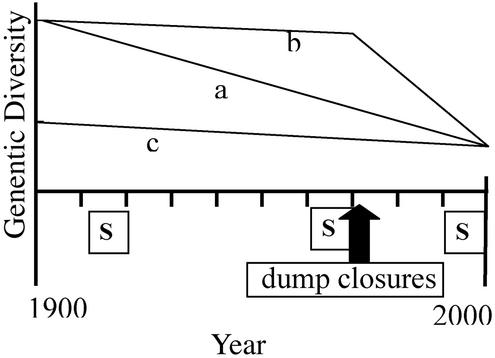
Recent genetic studies have further raised concerns by demonstrating that the YE has the lowest genetic diversity among continental brown bear populations in North America (13). Heterozygosity estimates at eight microsatellite loci are 55% in the YE, significantly lower than the 69% observed in the NCDE, only a few hundred kilometers to the north (13). Comparisons of heterozygosity at allozyme loci (0.8% vs. 1.4%; F. W. Allendorf, K. Knudson, and H. Reynolds, personal communication) and gene diversity at mtDNA (27% vs. 66%; ref. 14) are consistent but more extreme. With both populations embedded in the species' historic range (12), there is no reason to expect a priori that the YE population was historically less diverse (Fig. 2, hypothesis c). If the YE population was as diverse as the NCDE one before its isolation ≈10 generations ago and the NCDE population has not gained or lost substantial diversity since, the microsatellite heterozygosities would suggest a 2.3% rate of inbreeding per generation, or a harmonic mean Ne of 22 (based on Ht= Ho(1 − 1/2Ne)t; ref. 15; Fig. 2, hypothesis a).
Figure 2.
Three hypotheses (a–c) explaining low levels of genetic diversity in the YE and sample windows (S) used to resolve between them. a, Substantial but gradual decline in diversity and increase in inbreeding across century. b, Accelerated and substantial decline in diversity and increase in inbreeding after dump closures. c, Historically low diversity and slow inbreeding.
More troubling is the possibility that the YE underwent the majority of this hypothesized inbreeding in the last several decades. Garbage had become an important food resource by the early 1900s (16). Between 1968 and 1971 garbage dumps were phased out. Garbage-habituated bears were forced to disperse; many wound up near human habitation and were killed. Between 1967 and 1972, 220 known mortalities occurred (16). The population decline (refs. 16 and 17, but see ref. 18) may have been acute enough to precipitate a genetic bottleneck. If so, the recent rate of inbreeding could have been even more severe than suggested above (Fig. 2, hypothesis b).
These genetic and historic demographic data raise conservation concerns for several reasons. First, the 2.3% rate of inbreeding suggested by comparison with the NCDE is well above the maximum tolerable rate of 1% recommended by Franklin (7). Second, Picton et al. (19) found evidence of fluctuating asymmetry, a potential indicator of inbreeding depression, in post-dump closure YE bears. Third, inbreeding depression has been documented in captive brown bears (20), captive mammals (21), and wild mammal populations (22). Fourth, inbreeding and/or a loss of variability can increase a population's probability of going extinct (3–6). Fifth, the effects of inbreeding on both individual and population fitness are often exacerbated by stressful environments (22–24). Declines in important food resources [whitebark pine (Pinus albicaulis) because of blister rust (Cronartium rubicola) and cutthroat trout (Oncorhynchus clarki) because of non-native lake trout (Salvelinus namaychush)], global warming, continuing human growth, and habitat alteration will likely increase stress on bears in the YE (17). Lastly, if an island-type population that cannot shift geographic range is to persist, it must adapt to a changing environment. The rate at which it can adapt is directly proportional to its additive genetic variability (25).
The objective of this study is to assess the concern that the viability of the Yellowstone grizzly may be negatively affected by genetic factors. Using museum specimens to sample the population in the early 20th century and at the time of dump closures, we assess whether, when, and how rapidly diversity has been lost (Fig. 2). We estimate Ne across the century and obtain an estimate of the Ne/N ratio. Conservation management recommendations are drawn from these findings. We also use the dataset to examine the extent and nature of genotyping errors, a serious concern when working with low-quality/quantity DNA such as that from hair, feces, or museum specimens.
Materials and Methods
Major North American museum collections were sampled for historical YE specimens collected during two distinct windows of time: early in the century (1912–1920) and just before and during the dump closures (1959–1981; Fig. 2; post-dump closure samples were known to be alive before or during closures; see Table 2, which is published as supporting information on the PNAS web site, www.pnas.org, for a list of all historical specimens contributing data to this analysis). The contemporary YE and NCDE samples represent combinations of data from a previous study (13) and individual genotypes from hair samples collected for other studies (ref. 26; D. A. Roon, K. C. Kendall, and L.P.W., unpublished work). For historical samples, bone was sampled from the cranial cavity of skulls. Approximately 0.4 g of bone was ground to powder by using liquid nitrogen in a chlorine bleach-sterilized mortar and pestle. DNA was extracted by using the “silica” method (27, 28). Samples were typed at eight dinucleotide microsatellite loci (13). PCR was performed in 15 μl of total volumes by using 2–4 μl of DNA extract (of 200 μl total extract volume), 1.5 units of AmpliTaq gold polymerase, 1× AmpliTaq buffer, 2.5 μM MgCl2, 0.1 μM of each of the four dNTPs, and primer concentrations of 0.2 μM (G1A, G10C), 0.3 μM (G10B, G1D, G10L, G10P, G10X), and 0.5 μM (G10M). A total of 45–55 cycles were conducted as follows: 10 min at 95°C; 45–55 cycles of 95°C for 30 s, 52°C (for G10M and G10P) or 56°C (for other loci) for 30 s, 72°C for 40 s; finish at 72°C for 2 min. Loci were generally amplified in the following multiplexes: G1A+G10C+G10L, G10B+G1D, G10M+G10P, G10X. Allele size was measured by separation on 6% polyacrylamide gels in an ABI 377 automated sequencer (Perkin–Elmer) followed by analysis with genescan and genotyper software (Perkin–Elmer).
Authenticating genetic data is a serious concern when working with ancient or historical samples (29, 30). To authenticate ancient DNA sequence data it is common practice to extract and sequence samples in two independent laboratories (30). However, with several hundred extractions, >5,000 PCRs, and limited bone available from museums, it was financially and logistically infeasible to replicate this study. Thus extensive steps were taken to ensure that the historical dataset under analysis is accurate. There are three sources of error: (i) contamination, (ii) human error, and (iii) enzymatic errors such as allelic dropout and false alleles. Several precautions were taken to guard against contamination. Extractions and PCR setup occurred in a separate building where no bear DNA had ever been present and no materials (clothing included) were allowed to move from the PCR/gel building back to the extraction/setup facility. One of every 10 extractions was a negative control, and one of every 16 PCRs was a negative control. Approximately one-third of the samples were extracted a second time. Genotypes were replicated extensively (on average 3.5 times per locus per sample). The use of negative controls and replication at the extraction and genotyping levels also functioned to check for human errors.
Allelic dropout, amplification of only one of the two alleles in a heterozygote, was addressed by replicating genotypes until their estimated reliabilities met or exceeded 95% under the model of Miller et al. (29). A subsequent analysis was conducted to examine factors potentially affecting allelic dropout under the assumption that the accepted genotypes were correct. A logistic regression was performed with sample identity and locus as dummy-coded categorical predictor variables, distance between allele lengths as a continuous predictor variable and dropout/no-dropout as the response variable. No interaction terms were considered. All eight possible models comprised of combinations of these three predictor variables (including the intercept-only model) were evaluated by using Akaike information criteria (AIC) corrected for small sample size (AICc; ref. 31). Models with an AICc score two or more poorer than the best were rejected (31). Among models within two AICc units of the best, the model with fewest parameters was favored. A second analysis was performed to determine whether the shorter or longer allele is more likely to drop out and whether this differs between loci. Dropout events across all individuals within each locus were pooled (sample sizes were too small to consider individual effects). The likelihood of the data was calculated under three binomial models of increasing complexity: (i) shorter and longer allele equally likely to dropout across all loci, (ii) one estimable probability of the shorter allele dropping out across all loci, and (iii) separate estimable probabilities of the shorter allele dropping out at each locus. The models were again evaluated by using AICc.
For data used in estimating changes in genetic diversity and Ne, the potential for false alleles, an enzymatic error in the PCR yielding an allele not present in the true genotype, was addressed by requiring that every allele be observed twice (29). When four or more replicates failed to yield a second observation of an allele (e.g., ab, a, a, a, a), the locus was scored as half a diploid genotype (n rather than 2n) for the allele observed multiple times (a rather than aa or ab in the example). With the number of replicates rarely exceeding six, it was impossible in these circumstances to be certain of the true diploid genotype. We therefore did not perform a logistic regression for causes of false alleles. Instead we simply made best guesses of the true genotypes (based on the principle of parsimony), pooled replicates from all samples, and calculated the incidence of false alleles at each locus. For example, the above result (ab, a, a, a, a) would be putatively typed aa with one false allele rather than as ab with four dropout events. Not included in this analysis of false alleles were cases where three or more alleles appeared in a replicate (e.g., the result abc was excluded). We estimated the distance between the true and false alleles by restricting the analysis to cases where the “true” genotype was a homozygote and tallying across samples and loci.
Tests for Hardy–Weinberg equilibrium were carried out by locus, by sample, and across all loci and samples by using GENEPOP 3.1b (32). Genotypic linkage equilibrium was tested by using GENEPOP 3.1b both within each sample and across samples. Within each test (e.g., Hardy–Weinberg test by locus) a Bonferroni correction for multiple tests was applied (33). Unbiased expected heterozygosity (He) was calculated for each sample along with allelic richness (A = mean number of alleles per locus). Confidence intervals on He were estimated by bootstrap resampling alleles at each locus. Tests for differences in He were performed by using a paired t test on arcsine-transformed heterozygosities (13). A decline in He was also tested by comparing observed He with equilibrium He calculated by assuming an upper bound on Ne (see below), a conservatively high mutation rate of u = 0.001, and either an infinite or stepwise mutation model by using the equations He(eq) = 4Neu/(4Neu + 1) and He(eq) = 1 −/(8 Neu + 1)0.5, respectively (15). To test for differences in A between samples of different sizes, 1,000 subsamples were drawn at random and without replacement at each locus from a larger, more recent sample at smaller historical sample size. For within YE comparisons, the P value was estimated as the proportion of the 1,000 subsamples where A was ≥A in the historic sample. For YE–NCDE comparisons, the P value was estimated as the proportion of the 1,000 subsamples where NCDE A was ≤A in the YE sample. The trend in A was also estimated by using a sample coverage method (34). FST was calculated between each YE sample and the current NCDE sample by using GENEPOP 3.1b. To test for a pre-1910 bottleneck, a Wilcoxon one-tailed test for heterozygosity excess (35) was conducted on the 1910 YE sample under the two-phase model (variance = 30, 70% stepwise mutational model, 1,000 iterations) by using the program bottleneck (www.ensam.inra.fr/URLB/bottleneck/bottleneck.html).
Ne in YE between the 1910s, 1960s, and 1990s was estimated from changes in allele frequencies by using three maximum-likelihood estimators (36–38) and a moment estimator (8). For the moment estimator (8), the equation for plan I was used setting N1910s–60s = 350, N1910s–90s = 320, and N1960s–90s = 280. The ratio of effective to census population size (Ne/N) was estimated by combining the estimate of Ne between the 1960s and 1990s with an estimate of N over the same interval. Estimates of Ne from temporally spaced samples represent the harmonic mean of Ne over the interval. Assuming Ne/N is constant, the ratio is equivalent to the harmonic mean Ne divided by the harmonic mean N (39). Low, point, and high estimates of N between 1965 and 1995 (Table 1) were obtained from published studies on the YE (18, 40–45). The best estimate of Ne/N derives from the ratio of the best estimates of Ne over the best estimate of N. The lower bound on Ne/N comes from the lower bound on Ne divided by the upper bound on N whereas the upper bound represents the upper bound on Ne divided by the lower bound on N. A lower bound on the current size of the Yellowstone population was obtained in two different ways: (i) by dividing the lower bound on the 1996–1998 estimated 3-year sum of females with cubs (46) by the estimated proportion of the population composed of females with cubs of the year (0.274; ref. 18) and (ii) by using the 1998 and 1999 lower bounds on N from radio mark-aerial resight data (47).
Table 1.
Lower bound, point, and upper bound estimates of N in YE between 1965 and 1995 with source/justification
| Estimate | Year | N̂ | Source/justification |
|---|---|---|---|
| Lower bound | 1965 | 229 | Estimated 1960–1965 mean from National Academy of Sciences (40) |
| 1970 | 182 | Midpoint between N̂1965 and N̂1975 | |
| 1975 | 136 | From Craighead et al. (41) | |
| 1980 | 136 | Analyses (18, 42–44) suggest YE ∼ stable 1975–1980 | |
| 1985 | 163 | 1985–1995: N1995 lower bound of 90% confidence interval (18) | |
| 1990 | 220 | N1985 and N1990 assume exponential growth between 1980 and 1995 (i.e. r = 0.036) | |
| 1995 | 280 | ||
| Harmonic mean | 180 | ||
| Point | 1965 | 312 | Best estimate for 1959–1970 by McCullough (45) |
| 1970 | 277 | Midpoint between N1965 and N1975 | |
| 1975 | 243 | A 22% decline from N1965 is ∼ the decline in 3-yr sums of females with cubs between 1965 and 1975 (42) and ½ the extreme 44.5% decline suggested by Craighead et al. (16) | |
| 1980 | 243 | Analyses (18, 42–44) suggest YE ∼ stable 1975–1980 | |
| 1985 | 265 | 1985–1995: N1995 is best estimate of Eberhardt and Knight (18) | |
| 1990 | 306 | Ref. 18; N1985 and N1990 assume exponential growth | |
| 1995 | 344 | Ref. 18; N1985 and N1990 assume exponential growth between 1980 and 1995 (i.e., r = 0.017) | |
| Harmonic mean | 280 | ||
| Upper bound | 1965 | 412 | McCullough's estimate (45) of 312 +100 backcountry bears suggested by Cole (as cited in ref. 40) |
| 1970 | 412 | 1970–1980; although no decline across this period is unlikely, Eberhardt and Knight (18) suggest decline may have been slight | |
| 1975 | 412 | ||
| 1980 | 412 | ||
| 1985 | 454 | N1995 is upper 90% confidence interval from Eberhardt and Knight (18); N1985 and N1990 assume exponential growth between 1980 and 1995 (i.e., r = 0.020) | |
| 1990 | 535 | ||
| 1995 | 610 | ||
| Harmonic mean | 454 |
The number of migrants (Nm) from the NCDE into the YE needed to maintain genetic diversity was calculated by using two approaches. First, we consider an island (YE)–continent (NCDE) model in which equilibrium He in the YE is given by HYE(eq) = HNCDE[4Nm/(4Nm + 1)] (15). Second, we calculated Nm per generation needed to maintain the level of divergence observed between the NCDE and the 1910s YE by using the equation Nm = (1 − FST)/4FST (15).
Results
Thirty-eight specimens collected between 1912 and 1920 contributed one or more loci of data to the 1910s sample, yielding a weighted mean collection year of 1917 (see Table 2 for individual museum numbers and genotypes). The 1960s sample came from 72 individuals collected between 1959 and 1981 with a weighted mean collection year of 1969. The 1990s sample came from 136 individuals sampled between 1992 and 1999 with a mean collection year of 1996.
The mean number of independent positive PCRs per sample per locus was 3.5. The mean estimated dropout rate across individuals was 14% with 96% of the estimates ≤50%. Assuming all dropouts were detected, the mean observed dropout rate was 16%. In the allelic dropout logistic regression, the model with individual identity and locus was more than two AICc units better than the next best model. The odds ratios for the eight loci provide estimates of the relative dropout probabilities after accounting for individual sample effects. They are (by locus): 1.7 (A), 0.5 (B), 0.3 (C), 5.4 (D), 1.5 (L), 5.8 (M), 0.6 (P), and 1 (X). The count of short alleles to long alleles dropping out (by locus) were 18:7 (A), 12:8 (B), 6:10 (C), 20:13 (D), 10:8 (L), 10:12 (M), 14:8 (P), and 6:5 (X). The zero parameter model with an equal dropout probability for short and long alleles across all loci [P(short) = 0.5; AICc = 37.8] was similar to the model with one free rate [P̂(short) = 0.575 (≈95% confidence interval: 0.495–0.650); AICc = 36.1] whereas the eight free rates (AICc = 43.3) model performed more poorly. The incidence of false alleles (with counts; by locus) was 3.6% (7/196; A), 3.1% (6/194; B), 2.4% (5/211; C), 1.2% (2/172; D), 2.9% (6/205; L), 2.7% (4/148; M), 1.3% (3/225; P), and 5.5% (12/217; X). Among the 21 false alleles observed at putatively homozygous loci, 14 (67%) were one repeat less than the true allele, with the other seven differing from the true allele by +6, +1, −2, −3, −3, −6, and −11 repeats.
The model and replication strategy used to filter allelic dropout assumes an even dropout rate across loci and no bias in dropout probability among alleles. The logistic regression indicates that there is considerable heterogeneity in dropout rates among loci. This finding suggests that a more complex model is needed to identify dropouts (29) and that the dropout rate was likely underestimated at some loci and a small number of dropout errors may have gone undetected. Fortunately, the focus here is on allele counts and not multilocus genotypes. As long as dropout events are random with respect to allele identities, our allele counts (Table 3, which is published as supporting information on the PNAS web site) should provide unbiased estimates of the population allele frequencies. This statement is supported by the fact that the shorter and longer alleles were approximately equally likely to drop out and that the logistic regression found no effect of distance between alleles on the dropout rate. These findings do not support previous speculations that the longer allele is more likely to drop out (48). False alleles appear to be a less serious source of error, being observed in <3% of the total PCRs. Requiring that alleles be observed at least twice should severely limit the number of false alleles in the data set (e.g., 0.032 = 0.0009).
The mean number (and range) of observations per locus (2n) for the 1910s, 1960s, and 1990s are: 44 (35–54), 99 (76–123), and 224 (114–272), respectively. Although samples were taken over time and in an opportunistic rather than a random manner, hypothesis tests revealed no evidence of deviations from Hardy–Weinberg or linkage equilibrium.
Estimated He in the YE has declined slightly across the century from 0.580 (95% confidence interval: 0.537–0.618) to 0.579 (0.553–0.602) to 0.560 (0.538–0.572), although none of the paired comparisons are significant (P1910–60 = 0.49; P1960–90 = 0.13; P1910–90 = 0.31). He in the 1990s NCDE population is significantly greater than the YE in the 1910s (PNCDE-YE1910 = 0.028) whereas comparisons with the 1960s and 1990s are suggestive, but not statistically significant (PNCDE-YE1960 = 0.057, PNCDE-YE1990 = 0.074). Using Ne = 250 (see below), equilibrium He in the YE under the infinite allele and stepwise mutational models is 0.5 and 0.42, respectively, less than observed He. Observed allelic richness in the YE shows an increasing trend from 4.50 in the 1910s to 4.63 in the 1960s to 4.88 in the 1990s. However, observed A depends strongly on sample size. The results from subsampling at the smaller sample size indicates that YE1910s was significantly more diverse than YE1960s (P = 0.041) and YE1990s (P = 0.004) whereas there is little evidence of a difference between YE1960s and YE1990s (P = 0.123). The NCDE is more diverse than either YE1910s (P < 0.001) or YE1960s (P < 0.001). Concordantly, the estimated allelic richness by using the sample coverage method shows a declining trend: 5.875 in the 1910s, 5.624 in the 1960s, and 5.500 in the 1990s. The test for heterozygosity excess revealed no evidence for a bottleneck in the YE 1910 sample (P = 0.37). Pairwise FST values between the current NCDE and the 1910s, 1960s, and 1990s YE populations are 0.122, 0.129, and 0.123, respectively.
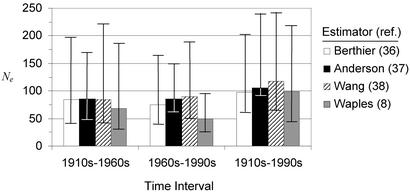
All three maximum-likelihood estimates of Ne between the 1910s and the 1960s are ≈85 (Fig. 3). Similarly, likelihood estimates between the 1960s and the 1990s are between 75 and 89. The estimated harmonic mean N for the interval 1965–1995 is 280 with lower and upper bounds of 180 and 454 (Fig. 3). Combining the (Berthier, cited in ref. 38) maximum-likelihood estimates of Ne (see Fig. 3) with these estimates of N yields an estimate of Ne/N = 0.27 with lower and upper bounds of 0.09 and 0.92. Using other likelihood estimates of Ne yields similar estimates of Ne/N [Anderson (cited in ref. 39): 0.31 (0.15–0.94); Wang (cited in ref. 40): 0.32 (0.11–1.04)]. The lower bound on N in the YE between 1996 and 1998 based on females with young is 430 (118.5/0.274), whereas the lower bounds on the 1998 and 1999 mark-resight estimates are 614 and 627, respectively. The estimated number of effective migrants per generation needed to maintain genetic diversity in the YE is 1.0. The number needed to maintain divergence between the NCDE and the YE is 1.8.
Figure 3.
Estimates of variance effective population size (and 95% confidence interval) in the Yellowstone grizzly bear population for three time intervals from four different estimators that use between-generation changes in microsatellite allele frequencies.
Discussion
Genetic diversity has declined slightly in the Yellowstone grizzly bear population since the early 20th century as evidenced by a significant drop in allelic diversity. The decline is also implied by an Ne <250, a value too small to maintain heterozygosity at or above the observed 55–60%. The similarity of Ne estimates in the two intervals suggests a gradual decline of diversity over the century, not an acute drop after dump closure. More importantly, the loss of diversity and the rate of inbreeding appear to have been much less severe than originally hypothesized (ref. 13; hypothesis c in Fig. 2). These observations may be explained by the long generation time of brown bears (≈10 years) and that either the population spent little to no time below a few hundred in the last century or Ne/N is well above our lower bound of ≈0.1 (or both). Interestingly, the 1910s YE population was not as diverse as the NCDE's and there is no evidence of a bottleneck in the 1910s sample. It is likely that diversity was historically low in the YE. The reason for this is not clear, but a FST ≈0.12 between the NCDE samples and all three YE samples suggests that gene flow into the YE from the north was historically restricted.
The data illustrate that the detection of low levels of genetic diversity in an extant population may not be not strong evidence of a recent bottleneck (35, 49). Alternate hypotheses should also be considered when using genetic data to make decisions for conservation and management. Historical specimens afford an ideal opportunity to evaluate competing hypotheses. The use of historical specimens for genetic study also demonstrates how museums and museum collections continue to play an important role in conservation and management, despite an atmosphere of budget cuts and a misconceived reputation for being obsolete (50).
The three maximum-likelihood estimates of Ne from the 1910–1960s and 1960–1990s are similar with point estimates ≈80, lower bounds ≈50, and upper bounds ≈150–200. Although the moment estimates for the same intervals are smaller than the likelihood estimates, we favor the likelihood estimates because simulation studies have shown their improved accuracy over the moment estimator (36, 38). To estimate Ne/N it was necessary to obtain an estimate of the harmonic mean N over the same interval. Estimating the size of grizzly bear populations in forested landscapes is notoriously difficult (42) and published estimates from the YE have been criticized for making unverified and/or unrealistic assumptions (18). We therefore caution that our estimates are only approximate. Nonetheless, the point estimate (≈0.3) and lower bound (≈0.1) are broadly consistent with two previous estimates of Ne/N in Ursus arctos, one demographic (0.20–0.38; ref. 51) and one genetic (0.04–0.19; ref. 13).
The minimum effective size to avoid the negative short-term effects of inbreeding is not known and probably varies between species. Based on domestic animal breeding, Franklin (7) suggests Ne should remain >50. Studies of wild (4–6) and laboratory (24, 52) populations have confirmed that viability can be significantly depressed at and below this effective size. Trend data suggest that the YE population is larger now than it has been in the past three decades (12, 18, 46), implying that Ne is probably >50. If recent evidence that N is at least 400 is accurate, then Ne is likely to be near or >100 (0.27 × 400 = 108). In our opinion, it is unlikely that genetic factors will have a substantial effect on the viability of the Yellowstone grizzly over the next several decades.
It has been argued that Ne should be at least 500–5,000 to maintain long-term evolutionary potential in the form of additive genetic variance (7, 53, 54). However, Ne in an isolated Yellowstone population is unlikely to ever approach or exceed even 500 (if Ne/n = 0.27, N would need to be ≥1,850). Furthermore, there is no guarantee that N will not decline. Genetic variability can be maintained over longtime spans only through gene flow. Thus we evaluated the potential long-term genetic impacts of gene flow from the NCDE. The NCDE is a good candidate source population because it is the nearest geographical population to the YE; it has high levels of diversity and bears historically occupied intervening regions. If Ne in the NCDE is and remains large, one effective migrant per generation into the YE will maintain the current levels of diversity while the current level of divergence between the two populations will be maintained by ≈two migrants per generation. We therefore argue that one to two effective migrants per generation from the NCDE to the YE is an appropriate level of gene flow.
Migration could either take the form of artificial transplantation or natural movements. Transplantation could occur at any time. Natural gene flow is, at the least, a generation or two away. Natural connectivity will require a concerted and cooperative effort on the part of federal and state agencies, private landowners, industry, political leaders, and the general public. As illustrated by the ongoing political controversy surrounding the U.S. Fish and Wildlife Service proposal to reintroduce bears to the Selway–Bitterroot Ecosystem in Idaho (ref. 55; Fig. 1), the obstacles to achieving natural connectivity are substantial. Because the need for gene flow into the YE is not urgent, we argue that concentrating current efforts on establishing intermediate populations and protecting and restoring intervening habitat are justified. If gene flow does not occur naturally within several decades, however, we argue that translocation should be conducted.
An alternative course of management would be to monitor genetic diversity in the YE and facilitate gene flow only if a significant decline in diversity is detected. We do not support this approach for two reasons. First, power analyses of bottleneck tests have shown that small to moderate declines in diversity have a small probability of detection (56). Second, there is no question whether diversity will decline in an isolated population with an Ne in the vicinity of 100–200. A good estimate of Ne provides more information about the decline in variation than measuring it directly will. Resources would be better allocated obtaining a good estimate of N, from which a better estimate of Ne could be made.
The viability of the Yellowstone grizzly bear population is unlikely to be compromised by genetic factors in the near future as we hypothesized based on modern samples (13). Rather, the genetic consequences of inbreeding and isolation are likely to transpire over longer time periods (decades and centuries). The more immediate threats to the Yellowstone grizzly (and nearly all other U. arctos populations) are habitat loss and human-caused mortality. We argue that management should therefore focus on maintaining the YE and NCDE populations at or above their current sizes and encouraging range expansion through natural dispersal and/or reintroduction. Success in these regards will improve the demographic security of the grizzly bear south of the Canadian border as well as address long-term genetic concerns.
Supplementary Material
Acknowledgments
We acknowledge Fred Allendorf, Paul Joyce, J. Michael Scott, David Roon, Alan Cooper, and the numerous museums and curators for their contributions to this article. Funding was provided by National Science Foundation Experimental Program to Stimulate Competitive Research Grant 9720634 and National Science Foundation Grants 0080935 and 9871024.
Abbreviations
- Ne
effective population size
- YE
Yellowstone Ecosystem
- NCDE
Northern Continental Divide Ecosystem
- AIC
Akaike information criteria
- He
expected heterozygosity
- Nm
number of migrants
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Lande R. Science. 1988;241:1455–1460. doi: 10.1126/science.3420403. [DOI] [PubMed] [Google Scholar]
- 2.Caro T M, Laurenson M K. Science. 1994;263:485–486. doi: 10.1126/science.8290956. [DOI] [PubMed] [Google Scholar]
- 3.Soulé M E, Mills L S. Science. 1998;282:1658–1659. [Google Scholar]
- 4.Westemeier R L, Brawn J D, Simpson S A, Esker T L, Jansen R W, Walk J W, Kershner E L, Bouzat J L, Paige K N. Science. 1998;282:1695–1698. doi: 10.1126/science.282.5394.1695. [DOI] [PubMed] [Google Scholar]
- 5.Madsen T, Shine R, Olsson M, Wittzell H. Nature. 1999;402:34–35. [Google Scholar]
- 6.Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I. Nature. 1998;392:491–494. [Google Scholar]
- 7.Franklin I R. In: Conservation Biology: An Evolutionary-Ecological Perspective. Soulé M E, Wilcox B A, editors. Sunderland, MA: Sinauer; 1980. pp. 135–149. [Google Scholar]
- 8.Waples R S. Genetics. 1989;121:379–391. doi: 10.1093/genetics/121.2.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller L M, Kapuscinski A R. Genetics. 1997;147:1249–1258. doi: 10.1093/genetics/147.3.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Queney G, Ferrand N, Marchandeau S, Azevedo M, Mougel F, Branco M, Monnerot M. Mol Ecol. 2000;9:1253–1264. doi: 10.1046/j.1365-294x.2000.01003.x. [DOI] [PubMed] [Google Scholar]
- 11.Allendorf F W, Servheen C. Trends Ecol Evol. 1986;1:88–89. [Google Scholar]
- 12.Servheen C. In: Bears: Status Survey and Conservation Action Plan. Servheen C, Herrero S, Peyton B, editors. Cambridge, U.K.: International Union for the Conservation of Nature; 1999. pp. 50–54. [Google Scholar]
- 13.Paetkau D, Waits L P, Clarkson P L, Craighead L, Vyse E, Wark R, Strobeck C. Conserv Biol. 1998;12:418–429. [Google Scholar]
- 14.Waits L P, Talbot S L, Ward R H, Shields G F. Conserv Biol. 1998;12:408–417. [Google Scholar]
- 15.Hedrick P W. Genetics of Populations. Sudbury, MA: Jones and Bartlett; 2000. [Google Scholar]
- 16.Craighead J J, Summner J S, Mitchell J A. The Grizzly Bears of Yellowstone: Their Ecology in the Yellowstone Ecosystem, 1959–1992. Washington, DC: Island Press; 1995. [Google Scholar]
- 17.Matson D J, Reid M M. Conserv Biol. 1991;5:364–372. [Google Scholar]
- 18.Eberhardt L L, Knight R R. J Wildl Manage. 1996;60:416–421. [Google Scholar]
- 19.Picton H D, Palmisciano D, Nelson G. Intl Con Bear Res Manage. 1990;8:421–424. [Google Scholar]
- 20.Laikre L, Andren R, Larsson H-O, Ryman N. Biol Conserv. 1996;76:69–72. [Google Scholar]
- 21.Ralls K, Ballou J D, Templeton A. Conserv Biol. 1988;2:185–193. [Google Scholar]
- 22.Crnokrak P, Roff D A. Heredity. 1999;83:260–270. doi: 10.1038/sj.hdy.6885530. [DOI] [PubMed] [Google Scholar]
- 23.Coltman D W, Pilkington J G, Smith J A, Pemberton J M. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- 24.Bijlsma R, Bundgaard J, Boerema A C. J Evol Biol. 2000;13:502–514. [Google Scholar]
- 25.Lande R, Shannon S. Evolution. 1996;50:434–437. doi: 10.1111/j.1558-5646.1996.tb04504.x. [DOI] [PubMed] [Google Scholar]
- 26. Haroldson, M., Gunther, K., Reinhart, D. P., Podrozny, S. R., Cegelski, C. & Waits, L. (2003) Ursus, in press.
- 27.Boom R, Sol C J A, Salimans M M M, Jansen C L, Wertheim-van Dillen P M E, van der Noordaa J. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Höss M, Pääbo S. Nucleic Acids Res. 1993;21:3913–3914. doi: 10.1093/nar/21.16.3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller C R, Joyce P, Waits L P. Genetics. 2002;160:357–366. doi: 10.1093/genetics/160.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Austin J J, Smith A B, Thomas R H. Trends Ecol Evol. 1997;12:303–309. doi: 10.1016/S0169-5347(97)01102-6. [DOI] [PubMed] [Google Scholar]
- 31.Burnham K P, Anderson D R. Model Selection and Inference: A Practical Information-Theoretic Approach. New York: Springer; 1998. [Google Scholar]
- 32.Raymond M, Rousset F. Heredity. 1995;86:248–249. [Google Scholar]
- 33.Rice W R. Evolution. 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 34.Huang S, Weir B S. Genetics. 2001;159:1365–1373. doi: 10.1093/genetics/159.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cornuet J M, Luikart G. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berthier P, Beaumont M, Cornuet J-M, Luikart G. Genetics. 2002;160:741–751. doi: 10.1093/genetics/160.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson E C, Williamson E G, Thompson E A. Genetics. 2000;156:2109–2118. doi: 10.1093/genetics/156.4.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J. Gen Res. 2001;78:243–257. doi: 10.1017/s0016672301005286. [DOI] [PubMed] [Google Scholar]
- 39.Kalinowski S T, Waples R S. Conserv Biol. 2002;16:129–136. doi: 10.1046/j.1523-1739.2002.00134.x. [DOI] [PubMed] [Google Scholar]
- 40.Cowan I M, Chapman D G, Hoffmann R S, McCullough D R, Swanson G A, Weeden R B. Report of Committee on the Yellowstone Grizzlies. Washington, DC: Natl. Acad. Sci. Press; 1974. [Google Scholar]
- 41.Craighead J J, Varney J R, Craighead F C., Jr . A Population Analysis of the Yellowstone Grizzly Bears. Univ. of Montana, Missoula: Montana Forest and Conservation Experiment Station; 1974. [Google Scholar]
- 42.Eberhardt L L, Knight R R, Blanchard B M. J Wildl Manage. 1986;50:613–618. [Google Scholar]
- 43.Knight R R, Eberhardt L L. Ecology. 1985;66:323–334. [Google Scholar]
- 44.Pease C M, Mattson D J. Ecology. 1999;80:957–975. [Google Scholar]
- 45.McCullough D R. In: Dynamics of Large Mammal Populations. Fowler C W, Smith T D, editors. New York: Wiley; 1981. pp. 173–193. [Google Scholar]
- 46.Boyce M, MacKenzie D, Manly B, Haroldson M, Moody D. J Wildl Manage. 2001;65:498–509. [Google Scholar]
- 47.Schwartz C C. In: Yellowstone Grizzly Bear Investigations: Annual Report of the Interagency Grizzly Bear Study Team. Schwartz C C, Haroldson M A, editors. Bozeman, MT: U.S. Geologic Survey; 1998/1999. [Google Scholar]
- 48.Gerloff U, Schlötterer C, Rassmann K, Rambold I, Hohmann G, Fruth B, Tautz D. Mol Ecol. 1995;4:515–518. [Google Scholar]
- 49.Matocq M D, Villablanca F X. Biol Conserv. 2001;98:61–68. [Google Scholar]
- 50.Pennisi E. Science. 2001;293:194–198. doi: 10.1126/science.293.5528.194. [DOI] [PubMed] [Google Scholar]
- 51.Allendorf F W, Harris R B, Metzgar L H. In: The Unity of Evolutionary Biology. Dudley E C, editor. Univ. of Maryland, College Park: Bioscorides Press; 1990. pp. 650–654. [Google Scholar]
- 52.Reed D H, Bryant E H. Anim Conserv. 2000;3:7–14. [Google Scholar]
- 53.Franklin I R, Frankham R. Anim Conserv. 1998;1:69–70. [Google Scholar]
- 54.Lynch M, Lande R. Anim Conserv. 1998;1:70–72. [Google Scholar]
- 55. Jehl, D. (June 21, 2001) N.Y. Times, Section A, p. 14.
- 56.Luikart G, Sherwin W B, Steele B M, Allendorf F W. Mol Ecol. 1998;7:963–974. doi: 10.1046/j.1365-294x.1998.00414.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.