Abstract
Little is known about sperm-binding proteins in the egg envelope of nonmammalian vertebrate species. We report here the molecular cloning and characterization of a recently identified sperm receptor (gp69/64) in the Xenopus laevis egg vitelline envelope. Our data indicate that the gp69 and gp64 glycoproteins are two glycoforms of the receptor and have the same number of N-linked oligosaccharide chains but differ in the extent of O-glycosylation. The amino acid sequence of the receptor is closely related to that of the mouse zona pellucida protein ZP2. Most of the sequence conservation, including a ZP domain, a potential furin cleavage site, and a putative transmembrane domain are located in the C-terminal half of the receptor. Proteolytic cleavage of the gp69/64 protein by a cortical granule protease during fertilization removes 27 amino acid residues from the N terminus of gp69/64 and results in loss of sperm binding to the activated eggs. Similarly, we find that treatment of eggs with type I collagenase removes 31 residues from the N terminus of gp69/64 and has the same effect on sperm binding. The isolated and purified N terminus-truncated receptor protein is inactive as an inhibitor of sperm–egg binding. Earlier studies on the effect of Pronase digestion on receptor activity suggest that this N-terminal peptide may contain an O-linked glycan that is involved in the binding process. Based on these results and the findings on the primary structure of the receptor, a pathway for the maturation and secretion of gp69/64, as well as its inactivation following fertilization, is proposed.
Fertilization in vertebrates requires that sperm recognize and bind to the extracellular coat of the egg and that they penetrate this coat so that they have access to the egg plasma membrane. These prerequisite steps have been most extensively studied in mice, and our current understanding of the function of egg coat glycoproteins is derived mostly from these studies (reviewed in ref. 1). Of the three major components that form the mouse zona pellucida (ZP1, ZP2, and ZP3), two glycoproteins exhibit sperm-receptor activity during fertilization (2–4). One of these (mouse ZP3) is considered to be the primary sperm receptor. It induces the acrosome reaction and sperm binding via its oligosaccharide chains. In a second step, acrosome-reacted sperm bind to ZP2, and this interaction is believed to facilitate sperm penetration through the zona pellucida (5). ZP3- and ZP2-like glycoproteins have been found in other mammals, including humans. However, the paradigm regarding the function of ZP glycoproteins in fertilization based on studies in mice may not hold true in all of the other mammalian species. In fact, in some mammalian species, ZP1 (rather than ZP3) homologs are believed to be the primary sperm receptor (6).
Much less information is available on the identity of sperm receptors in other, nonmammalian vertebrate species. Recently, a Xenopus laevis sperm receptor (gp69/64) in the egg vitelline envelope (VE) was identified (7, 8). It was shown that the purified gp69/64 proteins, as well as their antibodies, blocked sperm binding to unfertilized eggs or to beads coupled with gp69/64 proteins. It has been know for some time (9) that during fertilization, gp69/64 undergoes limited proteolysis. However, the molecular details of this cleavage have been unclear because the primary structure of gp69/64 was unknown. Here we report the molecular cloning and structure-function characterization of the Xenopus laevis sperm receptor gp69/64. We show that gp69/64 is a homolog to the mammalian sperm receptor ZP2. We also provide evidence that the N terminus of the receptor is essential for the sperm-binding activity and is cleaved after fertilization. Based on the full-length sequence of gp69/64 and the available information on its primary structure, a pathway for its maturation and inactivation on fertilization is proposed.
MATERIALS AND METHODS
Protein Purification and Analysis.
Individual egg envelope glycoproteins were purified as described (8). To determine the N-terminal sequence, protein samples were separated by 7.5% SDS/PAGE and electroblotted onto Immobilon-PSQ membranes (Millipore). Coomassie blue-stained bands (3 μg each) were cut out and used for microsequencing with a model 475A pulsed liquid protein sequencer (Applied Biosystems). gp69 and gp64 were treated with trifluoromethanesulfonic acid to remove both N and O-linked oligosaccharide chains. N-linked oligosaccharide chains were specifically removed by treatment with peptide N-glycosidase F (PNGase F, Boehringer Mannheim) using the following procedure: protein samples (0.5 mg protein per ml) were heated in 0.5% SDS and 1% 2-mercaptoethanol at 100°C for 5 min, and the reaction mixture containing 5 units/ml PNGase F, 10 mM EDTA, and 0.5% Nonidet P-40 in 50 mM Tris⋅HCl (pH 8.5) was added. After incubation at 37°C for 24 hr, the reaction was terminated by boiling at 100°C for 5 min.
Cloning of gp69/64 cDNA.
A unidirectional cDNA library in pBluescript SK(±) phagemid vector (Stratagene) was constructed by using poly(A)+ mRNA isolated from stage 1–3 Xenopus oocytes. To clone the cDNA encoding the gp69/64 protein, a degenerate PCR was performed in Perkin–Elmer GeneAmp PCR System 2400 by using frog oocyte phagemid library cDNA (0.1 μg) as template and a pair of primers; one was a forward, vector-specific T3 primer and the other a reverse, degenerate primer FE61AS (5′-GCXGCXACXGGDATYTCRTC-3′). The primer FE61AS was designed based on the N-terminal amino acid sequence determined for the gp66/61 protein isolated from the envelope of fertilized eggs. PCR conditions were: 30 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 2.5 min. The first cycle was preceded by a 5-min denaturation at 95°C, and the last cycle was followed by a 5-min extension at 72°C. PCR products were purified from a 1% agarose gel, subcloned into a TA cloning vector (Invitrogen), and sequenced. The correct PCR product was chosen by comparing the deduced amino acid sequence with the chemically determined N-terminal amino acid sequence of gp69/64 (see Results). The sequence of the remainder of gp69/64 cDNA was obtained by PCR using a gp69/64 cDNA-specific forward primer, SR-1S (5′-GGCGTCCTGCTATATCCCAA-3′), and a vector-specific T7 reverse primer under the same reaction conditions.
To obtain a full-length gp69/64 cDNA clone, 2 × 104 colonies of the oocyte cDNA library were screened by using a standard colony hybridization method with a 32P-labeled receptor cDNA fragment as a probe. Positive clones were selected after high-stringency (65°C) screening of the library. The full-length cDNA was sequenced in both directions manually with the dideoxynucleotide chain-termination method (10) and again by using an Applied Biosystems model 373A sequencer.
Polyclonal Antibodies.
A rabbit polyclonal antibody that reacted only with gp69 and gp64, but not with other VE proteins was prepared as described (7). Polyclonal anti-mouse ZP2 antiserum was a gift from P. Wassarman (Mount Sinai School of Medicine, New York). Western blot analysis was carried out by using the polyclonal anti-gp69/64 antibody (1:250 dilution) or anti-mZP2 antiserum (1:1,000 dilution) as primary antibody, and horseradish peroxidase-conjugated goat anti-rabbit IgG (Boehringer Mannheim) as secondary antibody. Reactivity was detected with LumiGLO (Kirkegaard and Perry Laboratories). To determine the reactivity of the polyclonal antibody on the surface of live, dejellied, unfertilized or fertilized eggs, the cells were incubated with the polyclonal anti-gp69/64 antibody (1:1 dilution) in MR (pH 6.5) for 30 min at 18–20°C, washed 3 times with MR, and fixed in 3% formaldehyde in MR for 2 hr at room temperature. Fixed eggs were then washed 3 × 20 minutes in MR and blocked for 1 hr in MR solution containing 5% BSA (Fraction V, Sigma). At the end of this period, fluorescein isothiocyanate-labeled goat anti-rabbit IgG secondary antibody (Sigma) was added to the blocking solution at a final concentration of 10 μg/ml. After another 30 min, the eggs were washed thoroughly with MR and viewed with a Nikon Diaphot fluorescence microscope with an ×4 objective.
Assays for Sperm Receptor Activity.
A quantitative sperm–egg binding competition assay was used to determine the inhibitory activity of the purified gp69/64 glycoproteins or their processed forms on sperm–egg binding (7).
RESULTS
gp69 and gp64 Are Two Glycoforms of a Single Gene Product.
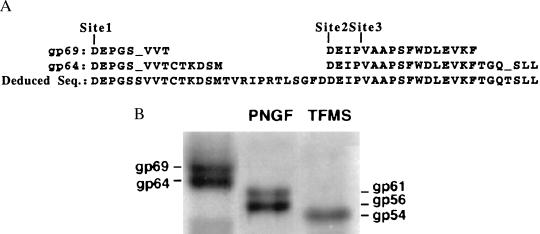
Three lines of evidence indicate that glycoproteins gp69 and gp64 share the same polypeptide chain. First, for each protein we determined the N-terminal amino acid sequence of the intact form of gp69 and gp64. In addition, the N-terminal sequences of the two proteolytically truncated forms of both proteins were determined (Fig. 1A). In all three cases, the peptide sequences obtained from the two glycoforms were found to be identical. Second, amino acid composition analysis of gp69 and gp64 also revealed that both polypeptides had identical compositions (data not shown). Third, to determine the basis for the difference in mass between gp69 and gp64, we carried out deglycosylation studies on the two glycoproteins. When gp69 and gp64 were treated with PNGase F to remove N-linked oligosaccharide chains, the apparent molecular masses of both glycoproteins were reduced by an equal extent, approximately 8 kDa, to ≈61 and ≈56 kDa, respectively, as detected by using Western blot analysis (Fig. 1B, lane 2). However, after deglycosylation with trifluoromethanesulfonic acid, which would be expected to remove all oligosaccharide chains, both proteins comigrated as a single band of ≈54 kDa (Fig. 1B, lane 3). These results indicated that both glycoproteins possess approximately the same mass of N-linked oligosaccharides (≈8 kDa) and different amounts of O-linked oligosaccharides (≈7 kDa in gp69 and ≈2 kDa in gp64).
Figure 1.
(A) Comparison of chemically determined amino acid sequences of gp69 and gp64 starting from three different sites in the polypeptide: site 1, N terminus of intact, mature gp69 and gp64; site 2, N terminus of the proteolytically processed forms of gp69 and gp64 (gp66 and gp61) after egg activation; and site 3, the 65- and 60-kDa forms generated by treatment of eggs with type I collagenase. (B) Deglycosylation of gp69 and gp64. Intact gp69/64, PNGase F-treated, and trifluoromethanesulfonic acid-treated gp69/64 were separated by using SDS/PAGE, transferred onto a membrane, and Western blotted with the specific anti-gp69/64 antibody.
Molecular Cloning of gp69/64 cDNA.
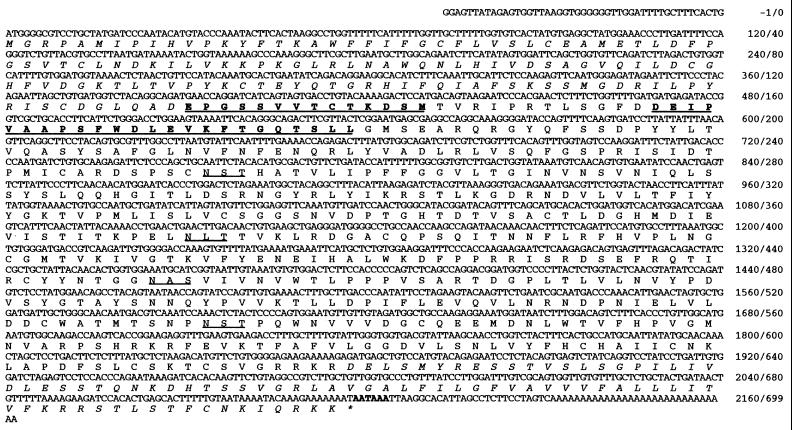
By using the degenerate PCR strategy outlined in Material and Methods, we first cloned a ≈540-bp cDNA fragment from the Xenopus oocyte cDNA library. The cDNA sequence encoded an ORF that contained the chemically determined peptide sequence at the N terminus of mature gp69/64 protein. The full-length gp69/64 cDNA clone was subsequently isolated from the same oocyte library by a high-stringency colony hybridization screen by using the 540-bp cDNA fragment as a probe. The 2,178-bp cDNA consisted of a 47-bp 5′ untranslated region, a 31-bp 3′ untranslated region, and a 2,100-bp ORF. A polyadenylation signal (AATAAA) was 27 nucleotides upstream of the poly(A) tail and overlapped the stop codon (see Fig. 2).
Figure 2.
cDNA sequence and translated single-letter amino acid sequence of gp69/64 protein. The N-terminal pre-pro-peptide sequence and the C-terminal peptide sequence after the putative furin cleavage site (RRKR) is italicized. The chemically determined peptide sequences are underlined and in boldface. The four putative N-glycosylation sites are underlined. The polyadenylation signal (AATAAA) is in boldface.
The ORF encoded a polypeptide chain of 699 amino acid residues with a calculated molecular mass of 77,867 Da. All three chemically determined peptide sequences (two of which overlapped each other) were found at the predicted positions in the deduced sequence of the clone (Fig. 2). The calculated mass is significantly larger than the apparent molecular mass (≈54 kDa) of the mature, deglycosylated gp69/64 protein, suggesting that posttranslational processings may occur to the nascent polypeptide chain. A cleavable signal-peptide sequence was predicted to be present at the N terminus (residues 1–33) by published methods (11, 12). Hydropathy analysis of the deduced polypeptide with the Kyte–Doolittle algorithm predicted a highly hydrophobic region with a core of 17 continuous hydrophobic residues (633–679) followed by several positively charged residues (KRR). These features are characteristic of a typical transmembrane domain. Four potential N-glycosylation sites were found in the proposed mature form of the receptor. Of course, numerous potential O-linked glycosylation sites (Ser or Thr) also were found in the sequence.
Homology Between gp69/64 and Mammalian ZP2.
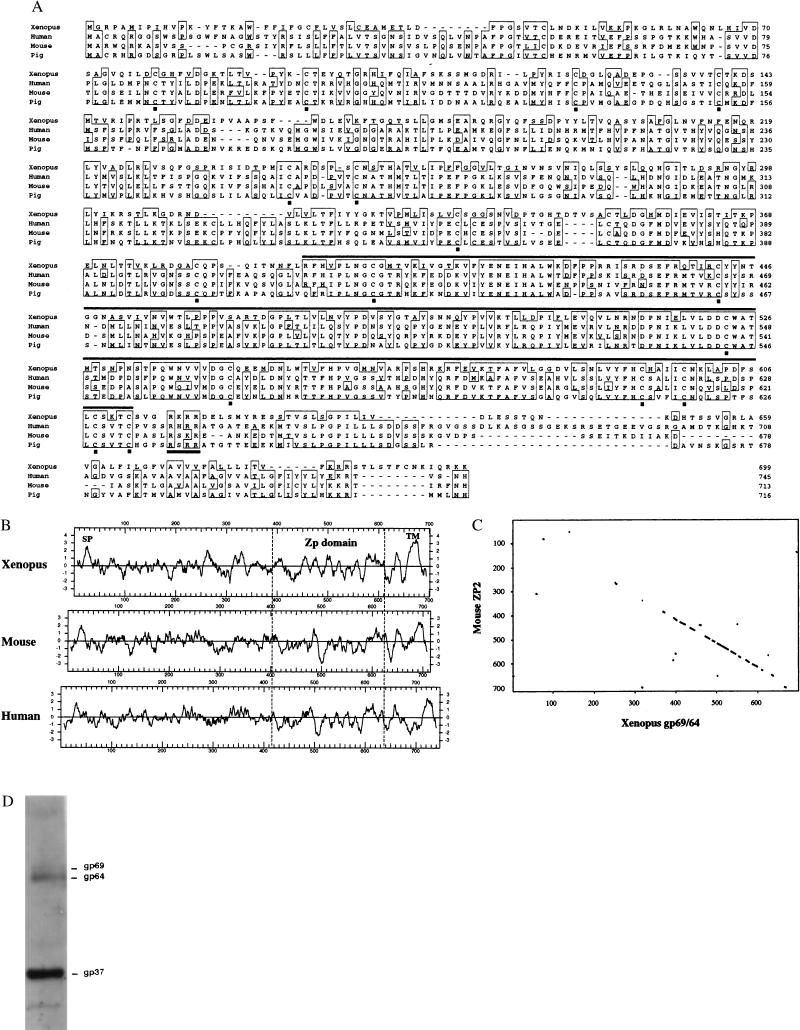
A search of the GenBank database revealed that the primary sequence of Xenopus gp69/64 is homologous to the mammalian zona pellucida glycoprotein ZP2 (ZPA) family (Fig. 3A). The level of sequence identity was found to be highly significant, with the probability of a random match between the Xenopus gp69/64 and various mammalian ZP2 family members between 2 × 10−132 and 1 × 10−106. Pairwise sequence alignments revealed 34–37% overall sequence identity between the Xenopus gp69/64 and the human, mouse, and pig ZP2 proteins. In addition, these polypeptide chains were similar in length, and all of them possessed an N-terminal signal sequence and a putative C-terminal transmembrane domain as predicted by hydropathy plots (Fig. 3B). All of the cysteine residues (16 of 16) in gp69/64 were conserved among the mammalian ZP2s (Fig. 3A), suggesting conserved overall structures among Xenopus and mammalian ZP2s. We also found that most of the conserved domains were located in the C-terminal half of the ZP2s, such as the so-called ZP domain (13), the putative transmembrane domain, and a furin-like protease cleavage site (RXR/KR) immediately after the ZP domain. The sequence identity between Xenopus and mammalian ZP2s in the C-terminal ZP domain reached 54–60% and dropped to 22–27% in the N-terminal half preceding the ZP domain. This is clearly revealed in the protein matrix plot (Fig. 3C). Among the mammalian ZP2s, the sequence identity differs in these two regions, but to a lesser extent (71–73% in the C-terminal ZP domain vs. 45–61% in the N-terminal half).
Figure 3.
Protein homology of Xenopus gp69/64 VE protein with mammalian ZP2. (A) The sequence of Xenopus gp69/64 was compared with the sequence of ZP2 from human, mouse, and pig (20–22). At each position, residues that are identical in 3 of 4 sequences are boxed. The ZP domain is overlined. The 16 conserved cysteines are indicated by closed squares. The putative furin-cleavage site is underlined. (B) Comparison of hydropathy plots of Xenopus gp69/64 and mouse and human ZP2s. Hydropathy plots were made by using DNA Strider and the Kyte–Doolittle algorithm. Plots were aligned at the conserved ZP domain. A putative signal peptide (SP) and a C-terminal hydrophobic domain (TM) are present in all three sequences. (C) Protein-matrix plots (pam250 matrix) of mouse ZP2 protein vs. the Xenopus gp69/64. Parameters: Window size = 8, Minimum % score = 60; Hash value = 2. (D) Western blot of total VE proteins with polyclonal antiserum against mouse ZP2 protein.
The homology between Xenopus gp69/64 and mammalian ZP2s predicted that these glycoproteins may share some common antigenic properties. As shown in Fig. 3D, polyclonal anti-mouse ZP2 antiserum crossreacted with Xenopus gp69/64 as well as with the very abundant Xenopus VE protein, gp37, but not with the other VE proteins. The faint bands shown are artifacts because they do not correspond with any of the VE proteins. The polyclonal anti-gp69/64 antibody could recognize isolated mouse ZP2 as well (data not shown).
The relationship between gp69/64 and other cloned VE proteins also was examined. Pairwise sequence alignments revealed 32% sequence identity between gp69/64 and gp37. The identity increased to 39% in the ZP domain, and blocks of continuous identical amino acid residues found in this region could explain why gp37 was reactive with polyclonal anti-mZP2 antiserum. The homology between gp69/64 and gp41 was insignificant (13% identity, 24% similarity).
The N terminus of gp69/64 Is Required for Sperm Binding.
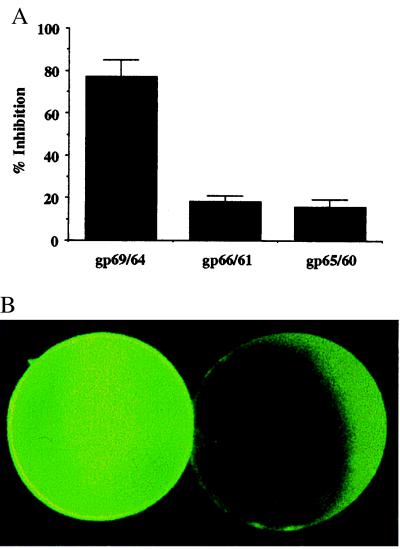
As shown in Fig. 1A, N-terminal peptide sequencing indicated that gp69 and gp64 were proteolytically cleaved between D156 and D157 during fertilization and between P160 and V161 by type I collagenase treatment. The two cleavage sites were separated by only 4 residues and would be expected to produce 27 and 31 amino acid residue peptides, respectively. We tested the activity of the N-terminal truncated receptors to inhibit sperm–egg binding by using an in vitro binding competition assay. We found that neither the purified gp66/61 from fertilized eggs nor the gp65/60 from collagenase-treated VE could inhibit sperm binding (Fig. 4A). As a positive control in this assay, purified, intact gp69/64 showed positive inhibition activity at the same protein concentration. The 27-aa fragment predicted to be produced on fertilization was produced as a glutathione S-transferase fusion protein in Escherichia coli. The purified fusion protein exhibited no inhibition of sperm binding in the competition assay. This finding is consistent with earlier findings (7) indicating that a glycan chain is involved in sperm binding. In this context, it is noteworthy that the chemically determined sequence of part of the 27-aa peptide failed to yield an amino acid residue corresponding to Ser-6 in the deduced sequence (see Fig. 1A). This may be indicative of an O-linked glycan at this position. We also examined the surface accessibility of the sperm receptor in the VE after type I collagenase treatment. As shown in Fig. 4B, the intensity of oocyte surface staining by anti-gp69/64 was greatly reduced after the treatment, suggesting that an exposed epitope on gp69/64 had been removed by proteolysis.
Figure 4.
The N terminus of gp69/64 is essential for sperm binding. (A) Comparison of the inhibitory activity of gp69/64, gp66/61, and gp65/60 on sperm–egg binding. Each protein (5 μg/ml) was used as competitor in the sperm-binding competition assays. (B) Comparison of anti-gp69/64 antibody staining of the surface of a dejellied unfertilized egg before (Left) and after (Right) treatment with type I collagenase. The apparent “staining” seen after collagenase on the vegetal hemisphere is an artifact caused by autofluorescence.
DISCUSSION
In earlier studies we established that one pair of glycoproteins in the VE of Xenopus mediates sperm binding. A major structural element in these glycoproteins involved in this binding is their oligosaccharide chains, although the possibility that the polypeptide chain also is involved cannot be excluded. In the current study, we have established that both of these glycoproteins are different glycoforms of the same polypeptide. In addition, we have cloned and sequenced the cDNA that encodes this polypeptide. Isolation and N-terminal sequencing of the form of this protein present in the coat of the fertilized egg revealed that it was proteolytically truncated by 27 amino acids. Similarly, isolation of the protein after crude collagenase treatment indicated a truncation of 31 amino acids. These three N-terminal sequences were found to be present in the deduced sequence of the cDNA.
Analysis of the deduced sequence of the frog sperm receptor in the egg VE reveals a close relationship with the ZP2 family of mammalian zona pellucida glycoproteins and suggests the existence of a common evolutionary origin for this family of glycoproteins. Among the conserved sequence features are the ZP domain, the putative cleavage site by a furin-like protease (RXK/RR), the putative C-terminal transmembrane domain, and all 16 cysteine residues at conserved positions. The hydropathy plots of these proteins are also similar. These conserved characteristics suggest that the overall structure of gp69/64 may resemble that of ZP2 and also strongly argues that the gp69/64 is a homolog to the mammalian ZP2, which in some mammalian species functions as a sperm receptor. In fact, the homology is largely restricted to the ZP domain in the C-terminal half of the protein. The ZP-domain is a ≈260 residue domain common to proteins of apparently diverse function and tissue specificity (13). Neither the structure nor the functional role of this domain has been determined. In Xenopus, in addition to gp69/64, two abundant VE glycoproteins, gp37 and gp41, also have identifiable ZP-domains in their C-terminal half; however, the domain in gp69/64 is more similar to gp37 than to gp41. It has been suggested that proteins with a ZP domain may share a common tertiary structure (13). Therefore, we suspect that this domain may primarily play a structural role in generation of the highly organized coat of the egg and therefore is present in several VE proteins.
Compared with the C-terminal half that contains the highly conserved ZP domain, the N-terminal half of the receptor is much less conserved. This also is true among the mammalian ZP2 proteins. Perhaps, at least in the case of the frog, this is indicative of the functional role of the N-terminal domain in species-specific gamete recognition. Indeed, both in vivo and in vitro data indicated that the N terminus of gp69/64 is essential for its binding to sperm. Cleavage of the N-terminal 27 residues during fertilization or of the 31 residues by type I collagenase abolishes sperm binding to the egg surface. Moreover, the isolated purified N-terminally truncated receptors no longer inhibit sperm–egg binding in competition assays. However, as mentioned above, it remains to be determined whether such epitopes include polypeptide sequences as well as putative oligosaccharide chains. Based on the above findings, we propose that the function of cleavage of the N terminus of the sperm receptor is to abolish sperm–egg interaction after egg activation (see ref. 8 for more evidence). Whether this cleavage also induces a conformational change of the VE or causes envelope “hardening” is not obvious from our study. The mouse homolog mZP2 also is proteolytically modified during fertilization, and its apparent Mr is reduced from 120,000 to 90,000 (ZP2f) (5, 14). However, the molecular details of this modification are not clear. In view of the close structural similarities between the mouse ZP2 and the Xenopus gp69/64, it is possible that the mouse ZP2 also is cleaved at a site close to its N terminus and that this cleavage blocks mouse sperm–egg binding. Indeed, there is evidence that postfertilization cleavage occurs at or near this same site in the pig ZP2 homolog to gp69/64 (15).
Several lines of evidence suggest that both ends of the nascent sperm receptor polypeptide are posttranslationally processed to generate the mature form present in the oocyte envelope. N-terminal peptide sequencing of the mature gp69/64 revealed that a pre-pro-sequence is cleaved just preceding Asp-130. C-terminal processing is proposed, based on several observations. First, without such a cleavage in the C terminus, the calculated molecular mass of the uncleaved polypeptide (residues 130–699) would be 63,365 Da, approximately 9 kDa larger than the apparent molecular mass of deglycosylated gp69/64 (≈54–55 kDa), as determined by SDS/PAGE. Second, the two strong hydrophobic domains at the C-terminus do not seem to be present in the mature receptor. When extracted by Triton X-114, the purified gp69 and gp64 glycoproteins remained in the aqueous phase, not in the Triton phase (unpublished data). Third, a potential furin cleavage site [RXK/RR (16), first reported in ZPs by Yurewicz et al. (17)] is found between the ZP domain and the putative transmembrane domain. This is a good candidate site for the cleavage, because a cleavage at or around this site would generate a mature polypeptide of ≈54.6 kDa, which would agree with the experimentally determined apparent molecular mass of the deglycosylated gp69/64. This cleavage site also is found in gp41 and gp37 (18, 19) at approximately the same relative position. It seems likely that these envelope glycoproteins are synthesized and processed at approximately the same time and together form the VE. The furin cleavage site also is conserved in the mammalian ZP2, suggesting that the biosynthetic pathway of the egg-coat glycoproteins may be conserved in mammals. However, the C-terminal sequence of the mature Xenopus receptor and the mechanism of cleavage remain to be experimentally determined.
Acknowledgments
We thank Dr. Paul M. Wassarman for sending the anti-mZP2 antiserum and purified mouse ZP2 protein, Dr. Jurrien Dean for helpful conversations, Lorraine Conroy for help in preparation of this manuscript, and Dr. Gerald H. Thomsen for scientific advice. We thank Dr. Anu Leinonen at the University of Kansas Medical Center for pointing out some sequence differences that we confirmed on further analysis. This work was supported in part by a Predoctoral Fellowship Award to J.T. from the Institute for Cell and Developmental Biology and by National Institutes of Health Grant HD18590 and National Science Foundation Grant IBN9728656 to W.J.L.
ABBREVIATIONS
- VE
vitelline envelope
- PNGase F
peptide N-glycosidase F
- ZP
zona pellucida
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF038151).
References
- 1.Wassarman P M. Development (Cambridge, UK) 1990;108:1–17. doi: 10.1242/dev.108.Supplement.1. [DOI] [PubMed] [Google Scholar]
- 2.Bleil J D, Wassarman P M. Cell. 1980;20:873–882. doi: 10.1016/0092-8674(80)90334-7. [DOI] [PubMed] [Google Scholar]
- 3.Bleil J D, Wassarman P M. J Cell Biol. 1986;102:1363–1371. doi: 10.1083/jcb.102.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimizu S, Tsuji M, Dean J. J Biol Chem. 1983;258:5858–5863. [PubMed] [Google Scholar]
- 5.Bleil J D, Beall C F, Wassarman P M. Dev Biol. 1981;86:189–197. doi: 10.1016/0012-1606(81)90329-8. [DOI] [PubMed] [Google Scholar]
- 6.Topfer-Petersen E, Mann K, Calvete J J. Biol Chem Hoppe-Seyler. 1993;374:411–417. doi: 10.1515/bchm3.1993.374.7-12.411. [DOI] [PubMed] [Google Scholar]
- 7.Tian J-D, Gong H, Thomsen G H, Lennarz W J. J Cell Biol. 1997;136:1099–1108. doi: 10.1083/jcb.136.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tian J-D, Gong H, Thomsen G H, Lennarz W J. Dev Biol. 1997;187:143–153. doi: 10.1006/dbio.1997.8607. [DOI] [PubMed] [Google Scholar]
- 9.Gerton G L, Hedrick J L. Dev Biol. 1986;11:1–7. doi: 10.1016/0012-1606(86)90036-9. [DOI] [PubMed] [Google Scholar]
- 10.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGeoch D J. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 12.Von Heijne G. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bork P, Sander C. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- 14.Moller C C, Wassarman P M. Dev Biol. 1989;132:103–112. doi: 10.1016/0012-1606(89)90209-1. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa A, Koyama K, Okazaki Y, Sugimoto M, Isojima S. J Reprod Fertil. 1994;100:245–255. doi: 10.1530/jrf.0.1000245. [DOI] [PubMed] [Google Scholar]
- 16.Hosaka M, Nagahama M, Kim W S, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. J Biol Chem. 1991;266:12127–12130. [PubMed] [Google Scholar]
- 17.Yurewicz E C, Hibler D, Fontenot G K, Sacco A G, Harris J. Biochim Biophys Acta. 1993;1174:211–214. doi: 10.1016/0167-4781(93)90119-x. [DOI] [PubMed] [Google Scholar]
- 18.Yang J C, Hedrick J L. Dev Growth Differ. 1997;39:457–467. doi: 10.1046/j.1440-169x.1997.t01-3-00007.x. [DOI] [PubMed] [Google Scholar]
- 19.Kubo H, Kawano T, Tsubuki S, Kawashima S, Katagiri C, Suzuki A. Dev Growth Differ. 1997;39:405–417. doi: 10.1046/j.1440-169x.1997.t01-3-00001.x. [DOI] [PubMed] [Google Scholar]
- 20.Liang L, Dean J. Dev Biol. 1993;156:399–408. doi: 10.1006/dbio.1993.1087. [DOI] [PubMed] [Google Scholar]
- 21.Liang L, Chamow S M, Dean J. Mol Cell Biol. 1990;10:1507–1515. doi: 10.1128/mcb.10.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taya T, Yamasaki N, Tsubamoto H, Hasegawa A, Koyama K. Biochem Biophys Res Commun. 1995;207:790–799. doi: 10.1006/bbrc.1995.1256. [DOI] [PubMed] [Google Scholar]