Abstract
Yeast TGN resident proteins that frequently cycle between the TGN and endosomes are much more slowly transported to the prevacuolar/late endosomal compartment (PVC) than other proteins. However, TGN protein transport to the PVC is accelerated in mutants lacking function of Inp53p. Inp53p contains a SacI polyphosphoinositide phosphatase domain, a 5-phosphatase domain, and a proline-rich domain. Here we show that all three domains are required to mediate “slow delivery” of TGN proteins into the PVC. Although deletion of the proline-rich domain did not affect general membrane association, it caused localization to become less specific. The proline-rich domain was shown to bind to two proteins, including clathrin heavy chain, Chc1p. Unlike chc1 mutants, inp53 mutants do not mislocalize TGN proteins to the cell surface, consistent with the idea that Chc1p and Inp53p act at a common vesicular trafficking step but that Chc1p is used at other steps also. Like mutations in the AP-1 adaptor complex, mutations in INP53 exhibit synthetic growth and transport defects when combined with mutations in the GGA proteins. Taken together with other recent studies, our results suggest that Inp53p and AP-1/clathrin act together in a TGN-to-early endosome pathway distinct from the direct TGN-to-PVC pathway mediated by GGA/clathrin.
INTRODUCTION
Phosphoinositides are a minor class of membrane lipids known to regulate a variety of cellular processes including cell growth and apoptosis (Rameh and Cantley, 1999), cytoskeletal rearrangements (Caroni, 2001; Martin, 2001), and membrane trafficking (Simonsen et al., 2001). Interconversion between seven different phosphoinositide derivates is catalyzed by kinases and phosphatases that add or remove phosphates from the inositol ring. Phosphoinositides establish and maintain the identity of membrane domains and specify numerous membrane trafficking events. They play a role in membrane trafficking by acting as specific binding sites on the membrane bilayer for a wide array of proteins that mediate either vesicle formation or fusion.
Yeast resident trans-Golgi network (TGN) membrane proteins are known to continuously cycle between the TGN and the endosomal system and their trafficking appears to rely on phosphoinositides (Ha et al., 2001). Two of these TGN residents, dipeptidyl aminopeptidase (DPAP) A and Kex2p, are integral membrane proteins that undergo a relatively slow rate of transport to a prevacuolar endosomal compartment (PVC). They then undergo retrieval via a vesicle coat complex called the retromer that sorts TGN cargo proteins into PVC-derived vesicles for subsequent fusion with the TGN (Bryant and Stevens, 1997; Seaman et al., 1998; Nothwehr et al., 1999, 2000). Resident TGN proteins are thought to cycle through an early endosomal compartment as well as the PVC (Black and Pelham, 2000; Lewis et al., 2000); thus the slow rate of transport into the PVC presumably reflects retention in the TGN or early endosome. Other cargo proteins such as Vps10p appear to be transported directly from the TGN to PVC and thus exhibit a much more rapid delivery into the PVC (Bryant and Stevens, 1997; Costaguta et al., 2001; Ha et al., 2001). The DPAP A cytosolic domain contains a FXFXD motif that interacts with the retromer complex and is necessary for its retrieval from the PVC (Nothwehr et al., 1993, 2000). A second signal in the N-terminal region of the cytosolic domain is necessary for slow delivery into the PVC; mutation of this signal dramatically increases the rate of PVC delivery (Bryant and Stevens, 1997). A fusion protein (A-ALP) containing the cytosolic domain of DPAP A fused to the transmembrane and lumenal domains of ALP has served as a useful reporter protein for studying these pathways because its transport to compartments containing vacuolar proteolytic activity can be assayed by the specific proteolytic removal of its C-terminal propeptide (Nothwehr et al., 1993).
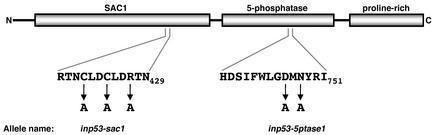
Recently, in a screen designed to identify yeast genes required for the slow delivery of A-ALP into the PVC, the INP53 gene encoding a synaptojanin-like protein was identified (Ha et al., 2001). Synaptojanin is a polyphosphoinositide phosphatase with a role in the vesicle uncoating step of the synaptic vesicle recycling pathway (Cremona et al., 1999; Harris et al., 2000). Synaptojanin is thought to mediate uncoating due to its hydrolysis of PtdIns(4,5)P2, a phosphoinositide that binds to clathrin coat components (McPherson et al., 1996; Chung et al., 1997; Martin, 2001). Synaptojanin contains three functional domains, two of which are catalytic: a domain that resembles the catalytic domain of the yeast polyphosphoinositide phosphatase SacIp, a phosphoinositide 5-phosphatase domain, and a C-terminal proline-rich domain (Figure 1). The SacI domain is capable of hydrolyzing PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 to PtdIns, whereas the 5-phosphatase domain selectively removes the phosphate group at the 5′ position of the inositol ring and uses PtdIns(4,5)P2 as its main physiological substrate (Guo et al., 1999).
Figure 1.
Inp53p mutant alleles generated to test the role of each enzymatic domain in TGN membrane protein trafficking. The domain structure of Inp53p is shown along with the two highly conserved regions thought to contain residues that participate in catalysis. The residues that were mutated to alanines are indicated as well as the names of the two resulting mutant alleles.
Yeast contains three synaptojanin-like proteins, Inp51p, Inp52p, and Inp53p (also called Sjl1p, Sjl2p, and Sjl3p), although the SacI domain present in Inp51p is noncatalytic (Guo et al., 1999). These proteins exhibit overlapping but not identical roles in membrane trafficking events, organelle dynamics, and actin cytoskeletal regulation. Although single and double null mutants of the three INP genes are viable, a triple knockout is lethal on standard medium (Stolz et al., 1998). The lethality of the triple knockout appears to be due to an accumulation of PtdIns(4,5)P2 because this strain can be rescued by a SacI domain-defective form of Inp52p (Stefan et al., 2002) or by a mammalian 5-phosphatase (O'Malley et al., 2001). INP single mutants and an inp51 inp53 double mutant exhibit relatively subtle and specific phenotypes. However, the inp51 inp52 and inp52 inp53 double mutants exhibit more severe and broad range phenotypes including slow growth, actin cytoskeletal defects, and defective vacuolar morphologies (Srinivasan et al., 1997; Stolz et al., 1998). Moreover, the inp51 inp52 but not the inp52 inp53 strain exhibited an endocytosis defect, suggesting that Inp51p and Inp52p but not Inp53 are involved in endocytosis (Singer-Krüger et al., 1998). In addition to the role of Inp53p in trafficking of the TGN protein A-ALP, INP53 genetically interacts with CHC1 (Bensen et al., 2000). Thus, Inp53p appears to be involved in clathrin-mediated trafficking within the TGN endosomal system.
In this study we investigated the relative contributions of the SacI, 5-phosphatase, and proline-rich domains of Inp53p to trafficking of A-ALP between the TGN and endosomes. We find that all three domains are necessary for Inp53p to mediate slow delivery of A-ALP into the PVC and show that the proline-rich domain binds to the clathrin heavy chain. Furthermore, our results suggest that Inp53p functions in trafficking between the TGN and early endosome rather than in a direct TGN-to-PVC pathway.
MATERIALS AND METHODS
General Methods and Antibodies
The production of minimal (synthetic dextrose) and rich (YPD) yeast medias, the genetic manipulation of yeast strains, and all general molecular biology methods were performed as described (Ausebel et al., 2000) or as otherwise noted. Rabbit polyclonal antibodies against alkaline phosphatase (ALP; Nothwehr et al., 1996) and Kex2p (Spelbrink and Nothwehr, 1999) have been previously described. A rabbit polyclonal antibody against pro-α-factor was a gift from G. Payne (University of California, Los Angeles, CA). Mouse anti-Chc1p was a gift from Sandra Lemmon (Case Western Reserve University, Cleveland, OH). Rabbit anti-HA antibodies were purchased from Covance (Richmond, CA) and mouse Vph1p antibodies were from Molecular Probes (Eugene, OR).
Plasmids and Mutagenesis
The wild-type INP53 gene was cloned into the LEU2 and URA3-based centromeric plasmids pRS315 and pRS316 (Sikorski and Hieter, 1989), respectively, by inserting a 4.4-kbp fragment from pSH12 (Ha et al., 2001) into the SpeI site of each vector. This resulted in pRS315 and pRS316-derived vectors pSH48 and pSH29, respectively. To generate the inp53-sac1 and inp53-5ptase1 alleles, regions of the INP53 gene contained in Bluescript KS+ (Stratagene, La Jolla, CA) were subjected to site-directed mutagenesis as described (Kunkel et al., 1987). The mutations were confirmed by DNA sequencing and fragments containing the desired mutations were subcloned into pSH48, resulting in inp53-sac1 and inp53-ptase1 plasmids pSH49 and pSH50, respectively. The same fragments were subcloned into pSH29 giving rise to the inp53-sac1 and inp53-ptase1 plasmids pSH31 and pSH32, respectively. To generate plasmids for integration of the inp53-sac1 and inp53-ptase1 alleles into the INP53 locus of yeast 4.4-kbp SpeI fragments from pSH31 and pSH32 were subcloned into the SpeI site of the URA3-based integration plasmid pRS306 (Sikorski and Hieter, 1989) giving rise to pSH38 and pSH39, respectively.
To introduce the inp53-ΔC::HA allele into the INP53 locus of yeast, three copies of the influenza hemagglutinin epitope were inserted at the 5′ end of the INP53 ORF in yeast strain TVY614 (Vida and Emr, 1995) as previously described (Schneider et al., 1995) to generate yeast strain SNY166. The INP53::HA allele was rescued by gap repair (Orr-Weaver et al., 1983) into plasmid pSH12 resulting in plasmid pJB17. A region encoding amino acids 910-1107 of Inp53p was deleted from pJB17 using a PCR approach resulting pSH47b. The 5.6-kbp XbaI fragment from pSH47b was then subcloned into the XbaI site of pRS306 to generate yeast integration vector pSH51. A vector for integration of the INP53::HA into yeast was made by inserting the 1.9-kbp EcoRI fragment from pSH47b into the EcoRI site of pRS306 giving rise to pSH56.
A vector for Escherichia coli expression of glutathione-S-transferase (GST) fused to the proline-rich domain of Inp53p was generated by PCR amplifying a ∼1-kbp fragment encoding amino acids 781-1107 and containing engineered BamHI and SalI sites at the upstream and downstream ends, respectively, from template pSH12. The PCR product was digested with BamHI/SalI and was inserted into the BamHI/SalI sites of pGEX-5X-1 (Pharmacia Biotec, Piscataway, NJ) to generate pAZ6. A 15-nucleotide deletion removing the LLDID919-encoding region of INP53 in pSH29 was introduced by PCR using the megaprimer method (Sarkar and Sommer, 1990) resulting in plasmid pSH54. An E. coli expression construct for GST-Inp53-CΔLLDID was constructed as described for pAZ6 (except template pSH54 was used) resulting in pSH57. Plasmids for E. coli expression of GST fused to full-length Inp53, Inp53-sac1, and Inp53-5ptase1 proteins were made by PCR amplifying the ORFs from the INP53, inp53-sac1, and inp53-5ptase1 alleles using primers that introduced BamHI and SalI sites at the 5′ and 3′ ends of the ORF, respectively. The PCR fragments were digested with BamHI and SalI and were subcloned into the BamHI/SalI sites of pGEX-5X-1, giving rise to plasmids pSH41, pSH44, and pSH43. Pfu DNA polymerase (Stratagene) was used for all PCR reactions.
Generation of Yeast Strains
Most of the yeast strains used in this study (Table 1) are based on SHY35, which is SNY36–9A (Nothwehr et al., 1995) mating type switched to α. To make strains SHY51, SHY57, SHY59, SHY63, SHY71, SHY72, SNY37, and SNY165 mutant alleles were integrated into yeast using the loop-in/loop-out procedure in which a pRS306-based constructs is linearized and targeted to a specific chromosomal site followed by growth on 5-fluoroorotic acid to select for strains that had excised the URA3-based construct. These strains were then screened for the desired mutation by PCR and, in some cases, analysis of PCR products by restriction enzyme analysis. A plasmid, pSL1699 (a gift from G. F. Sprague) was used to integrate the chc1ts allele into SNY17, giving rise to SNY37. Plasmid pLC1 (a gift from T. H. Stevens) was used to integrate the end3ts allele into SHY35 and SNY37, giving rise to SHY51 and SHY63, respectively. The inp53-sac1, inp53-5ptase1, INP53::HA, and inp53-CΔ::HA alleles were integrated into SHY35 using constructs pSH38, pSH39, pSH56, and pSH51 (described above) to generate strains SHY59, SHY57, SHY72, and SHY71, respectively. Finally, SNY165 was made by introducing the pho8-ΔX allele into YSC150 (Costaguta et al., 2001) using construct pSN111 (the pho8-ΔX allele in pRS306).
Table 1.
Saccharomyces cerevisiae strains used in this study
| Strain | Description | Source or reference |
|---|---|---|
| SHY35 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 | Ha et al. (2001) |
| SHY38 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53Δ::LEU2 | Ha et al. (2001) |
| SHY40 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53-1 | Ha et al. (2001) |
| SHY51 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53-1 end3ts | This study |
| SHY52 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53Δ::LEU2 pep4Δ::TRP1 | This study |
| SHY57 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53-5ptase1 | This study |
| SHY59 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53-sac1 | This study |
| SHY60-2A | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 (pho8Δ::ADE2 or pho8-ΔX) (LYS2 or lys2-801) inp53Δ::LEU2 gga1Δ::TRP1 | This study |
| SHY60-1A | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 (pho8Δ::ADE2 or pho8-ΔX) (LYS2 or lys2-801) inp53Δ::LEU2 gga2Δ::KanR | This study |
| SHY60-6C | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 (pho8Δ::ADE2 or pho8-ΔX) (LYS2 or lys2-801) inp53Δ::LEU2 gga1Δ::TRP1 gga2Δ::KanR | This study |
| SHY63 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 pho8Δ::LEU2 chc1-521(ts) end3ts | This study |
| SHY71 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 inp53-CΔ::HA | This study |
| SHY72 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 INP53::HA | This study |
| SNY17 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 pho8Δ::LEU2 | Nothwehr et al. (1995) |
| SNY37 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 lys2-801 pho8Δ::LEU2 chc1-521(ts) | This study |
| SNY94 | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 end3ts | Nothwehr et al. (1999) |
| SNY165 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ90 lys2-801 suc2-Δ9 pho8-ΔX gga1Δ::TRP1 gga2Δ::KanR | This study |
| SNY173-1B | MATaleu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 apl2Δ::KanR inp53Δ::LEU2 | This study |
| UFY2 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 suc2-Δ9 pho8Δ::ADE2 apl2Δ::KanR | This study |
| AAY143 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ90 lys2-801 suc2-Δ9 sac1Δ::TRP1 inp52Δ::HIS3 inp53Δ::TRP1 + pRS416-sac1-23 | Stefan et al. (2002) |
| YCS176 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 inp53Δ::hisG inp52Δ::HIS3 inp53Δ::TRP1 + pRS415-sjl2-8 | Stefan et al., (2002) |
Strains containing various combinations of inp53Δ, gga1Δ, and gga2Δ mutations were made by mating SNY165 with SHY34 (SNY36–9A made inp53Δ::LEU2). The resulting diploid was sporulated and dissected giving rise to SHY60-1A, SHY60-2A, and SHY60-6C. SHY52 was made by introducing the pep4Δ::TRP1 allele using gene-replacement construct pLS1-10 (Nothwehr lab collection) into SHY38. PCR-mediated gene disruption was used to replace the APL2 gene with a KanR cassette in SHY35 giving rise to UFY2. The inp53Δ apl2Δ double mutant was made by mating SHY64, a mating-type switched version of UFY2, with SHY38. The resulting diploid was sporulated and dissected giving rise to SNY173-1B.
Pulse/Chase Immunoprecipitation, Subcellular Fractionation, and Immunoblotting
Yeast strains were propagated at 30°C for all pulse-chase experiments. The procedure for immunoprecipitation of Kex2p and mutant A-ALP was performed as previously described (Nothwehr et al., 1993). Radioactively labeled proteins were quantified from gels using a Phosphorimager system (Fuji Photo Film Co., Tokyo, Japan). The half-time of Kex2p turnover was determined by calculating the percentage of protein remaining at a given time point compared with that present at time zero. Linear regression analysis was then carried out on plots of the log of the percentage protein remaining as a function of time. For calculation of the half-time of A(F→A)-ALP processing, the log of the percentage of A(F→A)-ALP that was unprocessed at each time point was plotted as a function of time and the plots were analyzed by linear regression analysis.
Yeast whole cell extracts were generated as previously described (Hill and Stevens, 1994) and 0.4 OD600 units of extract were loaded per lane. Subcellular fractionation was carried out by harvesting 50 OD600 units of yeast cells growing in midlog phase, washing them with 50 ml dH2O, and spheroplasting them in 14.5 ml of 1.4 M sorbitol, 50 mM Tris, pH 7.5, 2 mM MgCl2, and 10 mM NaN3 containing 0.16 mg oxalyticase (Enzogenetics, Corvallis, OR) for 45 min at 30°C. The spheroplasts were washed with 10 ml ice-cold 1.2 M sorbitol, 5 mM NaN3 and were resuspended in 10 ml of ice-cold lysis buffer (25 mM sodium phosphate, pH 7.4, 200 mM mannitol, 1 mM EGTA, and 5 mM MgCl2) containing freshly added protease inhibitors (0.5 mM PMSF, 1 μg/ml leupeptin, and 1 μg/ml pepstatin A). The resulting lysate was centrifuged at 460 × g for 12 min to pellet unlysed cells. The supernatant was then centrifuged at 15,000 × g for 15 min to create pellet (P15) and supernatant (S15) fractions. The S15 fraction was then centrifuged at 200,000 × g for 2 h to generate pellet (P200) and supernatant (S200) fractions. The P15, P200, and S200 fractions were separated by SDS-PAGE, blotted to nitrocellulose, and the blots probed with a rabbit anti-HA antibody. After subsequent incubation with an alkaline phosphatase–conjugated secondary antibody, the blots were developed using the Lumi-Phos substrate (Pierce, Rockford, IL). Films were scanned and imaged using Adobe Photoshop 5.5 software (Adobe Systems, San Jose, CA).
Immunoblots of secreted pro-α-factor were performed by applying freshly growing cells to the surface of a YEPD agar plate and immediately overlaying with a nitrocellulose filter. After incubating for 16 h at 30°C the cells were washed from the filter and the filter was blocked in 0.75% nonfat dry milk in TTBS (20 mM Tris, pH 7.5, 500 mM NaCl, 0.1% Triton X-100) and incubated with anti–α-factor serum. Detection was carried out using an anti-rabbit HRP secondary antibody and the Super Signal chemiluminescence substrate (Pierce, Rockford, IL).
Fluorescence Microscopy
The procedures for preparation of fixed spheroplasted yeast cells and attachment to microscope slides were previously described (Roberts et al., 1991). All secondary antibodies were diluted 1:500 before use. Simultaneous detection of A(F→A)-ALP and Vma2p was carried out by incubating with the following reagents followed by extensive washing: (a) rabbit anti-ALP and mouse anti-Vma2p antibodies, (b) biotin-conjugated goat anti-rabbit IgG (H+L), and (c) FITC-streptavidin and Texas Red–conjugated goat anti-mouse IgG (H+L).
Phosphoinositide Analysis and In Vitro Analysis of Inp53p Activity
The phosphoinositide analyses were performed with minor modifications of previously described procedures (Audhya et al., 2000; Ha et al., 2001). Five OD600 units of cells from log-phase cultures grown at 30°C in standard SD medium was harvested, washed, and resuspended in inositol-free medium. After a 10-min incubation at 38°C in this medium, [3H]myo-inositol (16 Ci/mmol; Amersham, Arlington Heights, IL) was added to the cells at 50 μCi/ml and labeling was carried out for 30 min at 38°C. Trichloroacetic acid was added (5% wt/vol final concentration) followed by incubation on ice for 1 h. Cells were washed with H2O and suspended in 0.5 ml of H2O. Lipids were extracted as described (Hanson and Lester, 1980) by combining the cells with 0.7 ml 95% ethanol/diethyl ether/pyridine/ammonium hydroxide (15:5:1:0.018), and extracting at 57°C for 30 min. Cell debris was removed by centrifugation and the supernatant was dried under N2.
Lipids were deacylated as previously described (Serunian et al., 1991) with minor modifications. Dried lipids were resuspended in 0.5 ml of methylamine reagent (42.8% of 25% methylamine, 45.7% of methanol, 11.4% of n-butanol) by bath sonication, incubated at 53°C for 50 min, and dried in vacuo. Deacylated lipids were suspended in 0.5 ml H2O and then extracted three times with 0.5 ml n-butanol/petroleum ether/ethyl formate (20:4:1). The aqueous phase was dried and suspended in a small volume of H2O for HPLC analysis.
Glycerophosphoinositol species were resolved using anion exchange chromatography with a Partisil 10 SAX (4.6 × 250 mm) column and a Beckman System Gold chromatograph. For each sample, equivalent counts were loaded (1 × 106 cpm). Fractions were collected every 20 s, mixed with 3 ml EcoLume (ICN), and counted in a liquid scintillation counter. Glycerophosphoinositol phosphate species eluted at identical times as previously chromatographed standards (Hama et al., 2000).
To measure in vitro activity of wild-type and mutant Inp53p proteins, GST, GST-Inp53, GST-Inp53-sac1, and GST-Inp53-5ptase1 proteins were expressed in E. coli from plasmids pGEX-5X-1, pSH41, pSH43, and pSH44, respectively, and were purified using glutathione agarose. Phosphoinositide phosphatase activity of 100-1000 ng of each purified protein was measured using a malachite green based chromogenic assay that measured the release of free phosphate from PdtIns(4)P and PtdIns(4,5)P2 substrates (Hess and Derr, 1975; Harder et al., 1994) as described previously (Marcus et al., 2001). Duplicate and triplicate values after 15 min (PtdIns(4,5)P2) or 30 min (PtdIns(4)P) were corrected for background (reaction lacking phosphoinositides) and converted to the amount of phosphate released (per amount of protein) using a standard curve made using varying amounts of sodium phosphate.
Affinity Chromatography with GST Fusion Proteins
GST, GST-Inp53-C, and GST-Inp53-CΔLLDID proteins were expressed from pGEX-5X-1, pAZ6, and pSH57, respectively, in E. coli strain BL21(DE3) (Novagen, Madison, WI) and were affinity purified using glutathione-agarose. To generate the yeast protein extract, a total of 1.75 × 1010 SHY52 cells were spheroplasted and resuspended in 7 ml of ice-cold lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.2 M sorbitol, 0.5 mM dithiothrietol [DTT], 0.6% Triton X-100, 0.5 mM PMSF, 1 μg/ml leupeptin, 1 μg/ml pepstatin A). The lysate was then subjected to 15 strokes with a dounce homogenizer and was centrifuged at 22,000 × g for 30 min. Two milliliters of supernatant was incubated with 50 μl bed volume of glutathione-agarose beads associated with either of the three proteins for 3 h at 4°C. The beads were washed four times with 1-ml volumes of lysis buffer before being denatured with SDS-PAGE sample buffer at 100°C for 5 min. Control samples were generated by denaturing 50 μl bed volumes of each of the three bead samples that had not been incubated with yeast protein extract. The eluted proteins were separated on SDS-PAGE and analyzed by coomassie staining and immunoblotting.
Identification of GST-Inp53-C Binding Proteins by Mass Spectrometry
Each Coomassie or silver-stained protein band was excised and transferred to a 0.5-ml microcentrifuge tube. The gel band was finely minced, dehydrated in 100 μl of CH3CN for 10 min, and centrifuged to remove the CH3CN, and then the step was repeated. The dehydrated gel pieces were dried in vacuo for 15 min and swelled in an equal volume of H2O for 15 min before another round of dehydration in CH3CN and drying was carried out.
For reduction and alkylation, 60 μl of 10 mM DTT in 100 mM NH4HCO3 was added and the gel pieces held at 56°C for 45 min. The solution was aspirated, an equal volume of 55 mM iodoacetamide in NH4HCO3 was added, and the gel pieces were incubated in darkness for 30 min. The solution was removed then 60 μl of 100 mM NH4HCO3 was added. After 5 min, the solution was removed and 100 μl of CH3CN was added. After 15 min the solution was aspirated, and the gel fragments were dried and reconstituted with 50 μl of 10 ng/μl modified trypsin in 50 mM NH4HCO3. After incubation on ice for 40 min they were topped with 50 mM NH4HCO3 and incubated at 37°C overnight. The gel pieces were then centrifuged and peptides from the supernatant were extracted once with 50 μl of milli-Q H2O and then three times with 50 μl 5% HCOOH and 50% CH3CN. All four supernatants were pooled, the solution was dried under vacuum to near dryness, and the peptides were reconstituted in 14 μl of 0.005% heptafluorobutyric acid, 0.4% acetic acid in H2O.
Analysis of peptides by microelectrospray liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed essentially as described (Gygi et al., 1999). Microelectrospray columns were constructed from 360 μm od × 75 μm id fused silica capillary with the column tip tapered to a 5–10-μm opening. The columns were packed with 200 Å, 5-μm C18 beads (Michrom BioResources Inc.), a reverse-phase packing material to a length of 10–12 cm. The flow through the column was split precolumn to achieve a flow rate of 300 nl/min. The mobile phase used for gradient elution consisted of (A) 0.4% acetic acid, 0.005% heptafluorobutyric acid, and 5% acetonitrile and (B) 0.4% acetic acid and 0.005% heptafluorobutyric acid in acetonitrile. The gradient was linear from 0.5 to 45% B in 35 min followed by 45–65% B in 5 min. Tandem mass spectra were recorded on a LCQ ion trap mass spectrometer (Thermoquest Corp., San Jose, CA) equipped with an in-house microelectrospray ionization source. Needle voltage was set at 1.6 kV. Ion signals above a predetermined threshold automatically triggered the instrument to switch from MS to MS/MS mode for generating collision-induced dissociation (CID) spectra (data-dependent MS/MS). The CID spectra were searched against a nonredundant yeast protein sequence database using the computer algorithm, SEQUEST (Yates et al., 1995).
RESULTS
Generation and Characterization of Inp53p Mutants Lacking Either SAC1 Or 5-Phosphatase Domain Activity
A complete loss of Inp53p function causes more rapid delivery of A-ALP and Kex2p into the PVC compared with wild type cells (Ha et al., 2001), suggesting that accumulation of one or more phosphoinositides on the membranes of TGN, endosomal, or transport intermediates is responsible for the defect. To determine which phosphoinositide(s) is/are responsible for the defect, and to analyze the structure and function of Inp53p, we generated Inp53p mutants in which either the SacI or 5-phosphatase activity was abolished (Figure 1). The highly conserved RTNCLDCLDRTN429 motif of the SacI domain is thought contain residues involved in catalysis (Hughes et al., 2000). The closely related Inp51p lacks SacI activity even though it contains a SacI-like domain (Guo et al., 1999). Within what would be the catalytic motif, however, Inp51p contains substitutions corresponding to the highly conserved C421, C424, and R427 positions of Inp53p. To inactivate the SacI domain we therefore substituted alanine for each of these three positions in Inp53p resulting in the inp53-sac1 allele.
The 5-phosphatase domain contains a motif (Figure 1) that is a defining feature of the 5-phosphatase family and mutation of D and N residues in 5-phosphatase II, corresponding to D746 and N748 of Inp53p, has been shown to cause a complete or near complete loss of activity (Jefferson and Majerus, 1996). Therefore, a mutant containing alanine substituations at D746 and N748 was also generated resulting in the inp53-5ptase1 allele. The steady state abundance and migration on SDS-PAGE of the inp53-sac1 and inp53-5ptase1 gene products was indistinguishable from that of wild-type INP53 (Ha and Nothwehr, unpublished data).
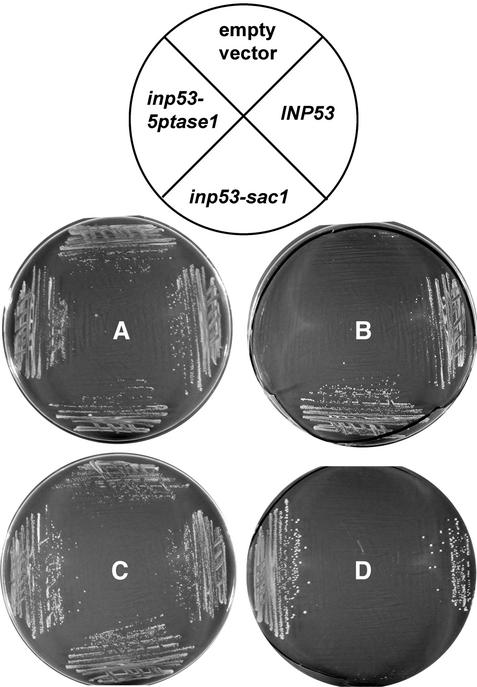
To assess whether the two mutant Inp53 proteins were inactive in their respective mutagenized domains and active in their nonmutagenized domains, we first determined whether each allele carried on a CEN plasmid could rescue an inp51Δ inp52ts inp53Δ strain that was inviable at a nonpermissive temperature due to a deficiency in 5-phosphatase activity (Stefan et al., 2002). The wild-type INP53 and inp53-sac1 alleles rescued growth of this strain at the nonpermissive temperature, whereas inp53-ptase1 and the empty vector did not (Figure 2B). These results suggested that the D746A and N748A mutations in the Inp53-5ptase1 protein inactivated function of the 5-phosphatase domain. Rescue of growth of this strain by the inp53-sac1 allele suggested that its protein product possesses 5-phosphatase activity. A related approach was to assess whether the alleles could rescue growth of an sac1ts-23 inp52Δ inp53Δ strain that is inviable at the nonpermissive temperature due to a deficiency in SacI activity. Inp53 (wild-type) and Inp53-5ptase1 proteins rescued the strain at nonpermissive temperature; however, the Inp53-sac1 mutant did not, indicating that it lacks SacI activity (Figure 2D).
Figure 2.
Growth of strains compromised for 5-phosphatase and SacI activity that carry various INP53 alleles on CEN plasmids. Strains were streaked onto minimal synthetic media lacking uracil and leucine and were grown for 3 d at 22°C (A and C) or 38°C (B and D). (A and B) The empty vector, INP53, inp53-sac1, and inp53-5ptase1 derivatives of parent strain YCS176 (inp51Δ inp52ts inp53Δ) consist of YCS176 transformed with pRS316, pSH29, pSH31, and pSH32, respectively. (C and D) The empty vector, INP53, inp53-sac1, and inp53-5ptase1 derivatives of parent strain AAY143 (sac1ts inp52Δ inp53Δ) consist of AAY143 transformed with pRS315, pSH48, pSH49, and pSH50, respectively.
The activity of the wild-type and mutant Inp53 proteins were assessed both in vivo and in vitro. The in vivo phosphoinositide levels in the inp51Δ inp52ts inp53Δ strain carrying the various INP53 alleles at the nonpermissive temperature were measured (Table 2). The inp51Δ inp52ts inp53Δ strain exhibits elevated levels of PtdIns(4,5)P2 (Stefan et al., 2002) because of its lack of 5-phosphatase activity. Expression of the wild-type INP53 allele in the inp51Δ inp52ts inp53Δ strain resulted in about sixfold lower PtdIns(4,5)P2 activity than expression of inp53-5ptase1, indicating that the Inp53-5ptase1 protein is highly impaired for 5-phosphatase activity. The strain carrying the inp53-sac1 allele also exhibited much lower PtdIns(4,5)P2 levels than the inp53-5ptase1 strain, although the levels were somewhat higher than that of wild-type (0.20 vs. 0.12%).
Table 2.
Phosphoinositide levels in strains compromised for 5-phosphatase or Sac1 activity that carry various INP53 alleles on CEN plasmids
| Strain | PtdIns levels (% of total 3H-labeled PtdIns)
|
|||
|---|---|---|---|---|
| PtdIns(3)P | PtdIns(4)P | PtdIns(3,5)P2 | PtdIns(4,5)P2 | |
| inp51Δ inp52ts inp53Δ + CEN-INP53 | 0.28 | 0.31 | 0.01 | 0.12a |
| inp51Δ inp52ts inp53Δ + CEN-inp53-sac1 | 0.36 | 0.51 | 0.01 | 0.20a |
| inp51Δ inp52ts inp53Δ + CEN-inp53-5ptase1 | 0.49 | 0.60 | 0.04 | 0.75a |
| sac1ts-23 inp52Δ inp53Δ + CEN-INP53 | 0.47 | 4.52a | 0.00 | 0.05 |
| sac1ts-23 inp52Δ inp53Δ + CEN-inp53-sac1 | 0.18 | 8.84a | 0.04 | 0.03 |
| sac1ts-23 inp52Δ inp53Δ + CEN-inp53-5ptase1 | 0.60 | 6.80a | 0.01 | 0.19 |
Boldface indicates increased levels (see text).
The expected trends were also observed when the alleles were carried in the sac1ts-23 inp52Δ inp53Δ strain. This strain background exhibits elevated PtdIns(4)P because of its lack of SacI activity (Stefan et al., 2002). Comparison of PtdIns(4)P levels in the three sac1ts-23 inp52Δ inp53Δ strains indicated that the strain expressing wild-type Inp53p had the lowest PtdIns(4)P level and the strain carrying inp53-sac1 had the highest. The sac1ts-23 inp52Δ inp53Δ derivative expressing inp53-ptase1 exhibited a PtdIns(4)P level that was intermediate between the other two strains.
Finally, in vitro phosphatase activity of the purified recombinant GST-Inp53, GST-Inp53-SacI, and GST-Inp53-5ptase1 proteins for hydrolysis of PtdIns(4)P and PtdIns(4,5)P2 was measured using a chromogenic assay with GST included as a control for background hydrolysis (Table 3). Consistent with the results discussed above, reactions containing GST-Inp53-sac1 exhibited only background levels of PtdIns(4)P hydrolysis. In contrast, GST-Inp53 and GST-Inp53-5ptase1 reactions exhibited PtdIns(4)P hydrolysis activity well above background although GST-Inp53-5ptase1 was somewhat less active than GST-Inp53. As expected, GST-Inp53-5ptase1 was completely devoid of PtdIns(4,5)P2 activity, whereas GST-Inp53-sac1 exhibited at least as much acivity as GST-Inp53. In summary, both analysis of cell growth and assessment of in vivo and in vitro activity indicates that for the Inp53-sac1 and Inp53-5ptase1 proteins the mutations have eliminated most or all of the activity of the domains that were mutated while leaving the nonmutagenized domains active.
Table 3.
In vitro phosphatase activity of wild type and mutant full-length Inp53 proteins fused to GST
| Relative amount of phosphate released
|
||
|---|---|---|
| PtdIns(4)P | PtdIns(4,5)P2 | |
| GST | 3.07 ± 0.04 | 63 ± 3 |
| GST-Inp53 | 4.29 ± 0.04 | 384 ± 16 |
| GST-Inp53-sac1 | 2.81 ± 0.05 | 566 ± 44 |
| GST-Inp53-5ptase1 | 3.55 ± 0.08 | 66 ± 10 |
Both the SAC1 and 5-phosphatase Domains of Inp53p Are Required for Slow Delivery of TGN Resident Proteins Into the PVC
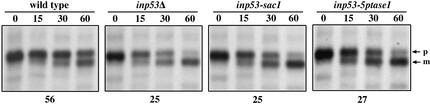
A mutant form of A-ALP, called A(F→A)-ALP, that lacks its PVC retrieval signal serves as a useful tool to measure the rate of TGN-to-PVC transport because upon reaching the PVC, it is promptly transported to and processed in the vacuole (Ha et al., 2001). Thus to compare TGN-to-PVC transport rates in wild-type and various inp53 mutant strains, cells were pulsed for 10 min with [35S]methionine/cysteine and chased for various times, and A(F→A)-ALP was immunprecipitated and analyzed (Figure 3). As previously observed (Ha et al., 2001), an inp53Δ strain exhibits a marked increase in the rate of A(F→A)-ALP processing compared with wild type (half-time of 25 vs. 56 min). Elimination of either the SacI domain activity or 5-phosphatase domain activity in the inp53-sac and inp53-5ptase1 strains caused a defect in the slow delivery mechanism (25 and 27 min, respectively) with similar severity as that observed in the null strain (25 min).
Figure 3.
Inactivation of the SacI and 5-phosphatase domains causes more rapid delivery of A(F→A)-ALP to the vacuole. Cells were pulsed with [35S]methione/cysteine for 10 min at 30°C and unlabeled amino acids were then added to initiate the chase. Samples were analyzed at the chase times (min) indicated above each panel. A(F→A)-ALP was immunoprecipitated from each strain and was analyzed by SDS-PAGE and fluorography. INP53 (SHY35), inp53Δ (SHY38), inp53-sac1 (SHY59), and inp53-5ptase1 (SHY57) yeast strains carrying a plasmid pSN100 expressing A(F→A)-ALP were analyzed. The half time of A(F→A)-ALP processing is indicated beneath the panel for each strain. The positions of A(F→A)-ALP precursor (p) and mature/processed (m) forms are indicated.
A loss of Inp53p function causes the endopeptidase Kex2p to be transported more rapidly into the PVC and to be also be mislocalized to the vacuole where it is turned over by vacuolar proteases (Ha et al., 2001). As a result of depletion of Kex2p from the TGN, MATα inp53 strains secrete unprocessed α-factor mating pheromone. As an alternative approach to assess the respective roles of SacI and 5-phosphatase activity in Inp53p, we measured pro-α-factor secretion and the rate of Kex2p turnover. Both the inp53-sac1 and inp53-5ptase1 mutants secreted similar amounts of pro-α-factor as the inp53Δ strain whereas little if any pro-α-factor was secreted by the wild-type strain (Figure 4A). Likewise, the inp53-sac1 and inp53-5ptase1 strains exhibited accelerated Kex2p turnover that was similar to that of the inp53Δ strain (Figure 4B). Taken together these results indicate that both domains are necessary for function of Inp53p in trafficking between the TGN and endosomes.
Figure 4.
Mutation of either of the enzymatic domains from Inp53p destablizes Kex2p. (A) Immunoblot analysis of pro-α-factor secretion was performed on strains SHY35 (INP53), SHY38 (inp53Δ), SHY59 (inp53-sac1), SHY57 (inp53-5ptase1), SHY72 (INP53:: HA), and SHY71 (inp53-CΔ:: HA). (B) Strains SHY35, SHY38, SHY59, and SHY57 were pulsed for 10 min with [35S]methionine/cysteine before unlabeled amino acids were added to initiate the chase. Kex2p was immunoprecipitated from the cells at the indicated chase times. The half-time of Kex2p turnover for each strain is indicated below each panel.
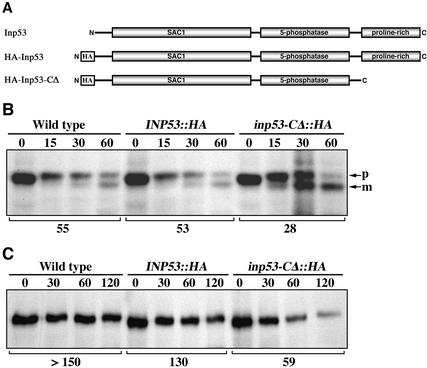
The C-terminal Proline-rich Domain of Inp53p Is Required for Function of Inp53p But Is Not Required for General Membrane Association
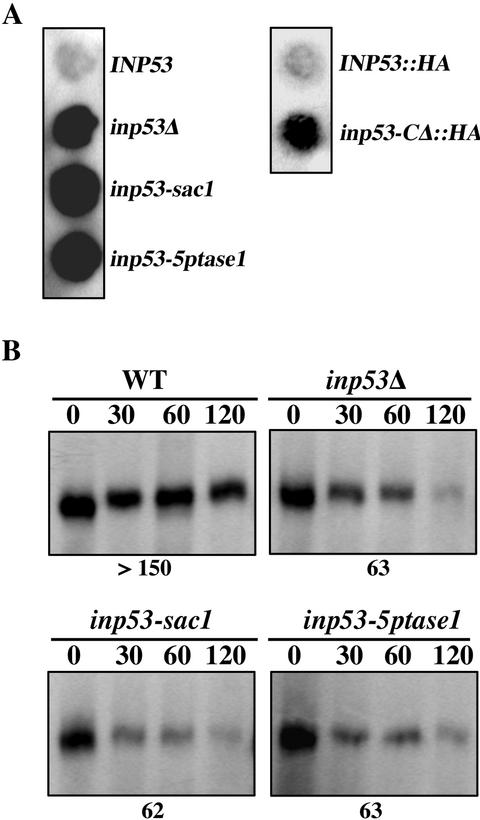
To determine whether the proline-rich domain is necessary for function of Inp53p, a mutant lacking this region (amino acids 910-1107) was generated (Figure 5A). The N-terminus of the deletion mutant and wild-type Inp53p were epitope-tagged with three copies of the influenza hemagglutinin (HA) epitope to facilitate detection. The HA-Inp53-ΔC protein was stable and its steady late level was similar to that of HA-Inp53 (Figure 6A). The ability of these proteins as well as untagged wild-type Inp53p to function in TGN/endosomal trafficking was assessed by A(F→A)-ALP processing kinetics and Kex2p turnover (Figure 5, B and C). Comparison of the phenotypes for wild-type with INP53::HA indicated that the HA tag caused little if any interference with Inp53p function. However, removal of the proline-rich domain caused both an increased rate of A(F→A)-ALP processing (28 min) over that of the full-length HA-Inp53 protein (53 min) and also more rapid Kex2p turnover (59 vs. 130 min). In accordance with the Kex2p turnover results, the inp53-CΔ::HA strain secreted substantial amounts of unprocessed α-factor compared with the wild-type tagged allele INP53::HA (Figure 4A). The severity of the phenotypes caused by removal of the proline-rich domain (Figure 5) rivaled that of the inp53Δ allele (Figures 3 and 4); thus the proline-rich domain is clearly essential for Inp53p function.
Figure 5.
The proline-rich domain of Inp53p is required for normal trafficking between the TGN and endosomes. (A) Graphical illustration of wild-type Inp53, HA-Inp53, and HA-Inp53-ΔC proteins. (B) Processing kinetics of A(F→A)-ALP expressed from pSN100 in strains containing the INP53 (SHY35), INP53:: HA (SHY72), and inp53:: HA-CΔ (SHY71) alleles expressing the proteins shown in A. (C) Kinetics of Kex2p turnover in the same strains analyzed in B. The half-times of A(F→A)-ALP processing and Kex2p turnover are indicated below each panel. The experiments shown in B and C were performed as described in the legend to Figures 3 and 4, respectively.
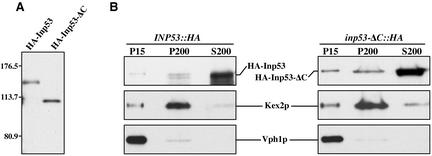
Figure 6.
Removal of the C-terminal proline-rich domain of Inp53p affects specificity of membrane association. (A) Whole cell extracts of INP53:: HA (SHY72) and inp53-CΔ:: HA (SHY71) strains were analyzed by SDS-PAGE and immunoblotted using a mouse mAb directed against the HA tag. (B) The same strains were spheroplasted, lysed in hypotonic buffer, and centrifuged at 450 × g to pellet unlysed cells. The supernatant was subjected to centrifugation at 15,000 × g to generate pellet (P15) and supernatant fractions. The supernatant was then centrifuged at 200,000 × g to generate pellet (P200) and supernatant (S200) fractions. HA-Inp53, HA-Inp53-ΔC, Kex2p, and Vph1p were then detected in the three fractions by immunblot analysis.
One possible function of the proline-rich domain of Inp53p could be to target the protein to appropriate membranes where the enzymatic domains would act on their phosphoinositide substrates. To test the role of this domain in membrane association, lysates from strains expressing full-length HA-Inp53 and the HA-Inp53-CΔ mutant were centrifuged at 15,000 × g to generate pellet (P15) and supernatant (S15) fractions. The S15 fraction was then centrifuged at 200,000 × g to generate pellet (P200) and supernatant (S200) fractions. In this type of fractionation scheme the P15 contains vacuoles, ER, and plasma membranes while the P200 contains Golgi, endosomes, and vesicles (Marcusson et al., 1994; Ha et al., 2001) and indeed we observe the TGN marker Kex2p predominantly in the P200 fraction and the vacuolar marker Vph1p in the P15 fraction. As previously observed for untagged Inp53p (Ha et al., 2001), HA-Inp53 predominantly fractionated in the cytosolic S200 fraction with a small pool in the P200 fraction (Figure 6B). The fractionation pattern of HA-Inp53-CΔ was similar to wild-type except a small pool of the truncated protein was also found in the P15, suggesting that this protein is more promiscuous in the way it associates with membranes. Thus the C-terminal domain is not required for general membrane association but it may function in specifying which organelle Inp53p associates with.
The C-terminal Proline-rich Domain of Inp53p Associates With Clathrin Heavy Chain
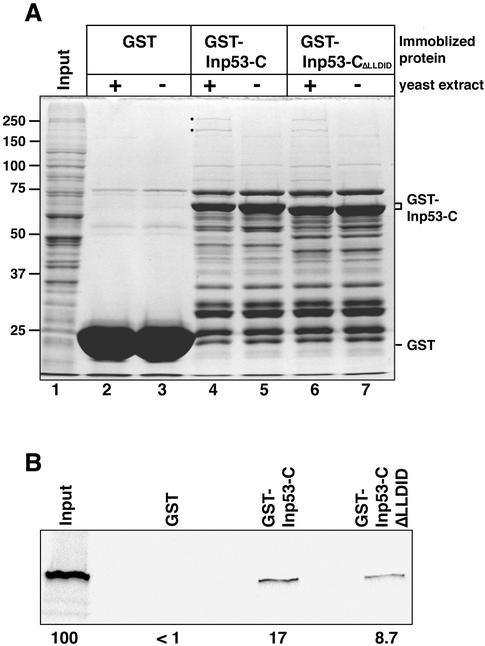
Inp53p may, like its mammalian cousin synaptojanin, associate with proteins involved in vesicular trafficking through its C-terminal proline-rich domain. To identify yeast proteins that bind to the proline-rich domain of Inp53p, we expressed amino acids 781-1107 of Inp53p in E. coli as a GST fusion protein. This region included a small portion of the 5-phosphatase domain and the entire proline-rich domain. The GST-Inp53-C protein and GST alone were purified from E. coli on gluthathione agarose beads and were incubated with a crude protein extract made from Triton X-100–solublized yeast spheroplasts. After washing extensively, the proteins associating with the beads were denatured and analyzed by SDS-PAGE and Coomassie stain detection (Figure 7A). Control samples included immobilized proteins that were not incubated with the yeast protein extract. Two proteins of 254 and 189 kDa reproducibly associated with the GST-Inp53-C beads (lane 4) but were not associated with either the GST beads (lane 2) or the GST-Inp53-C beads not incubated with yeast proteins (lane 5). The 189-kDa band was shown by mass spectrometry to be clathrin heavy chain (Chc1p), whereas unequivocal identification of the 254-kDa band was not successful.
Figure 7.
The C-terminal proline-rich domain of Inp53p interacts with Chc1p. A C-terminal region of Inp53p (amino acids 781-1107) fused to GST (GST-Inp53-C), the same fusion containing a deletion of the LLDID919 motif (GST-Inp53-CΔLLDID), or GST were bound to glutathione agarose beads, incubated with an extract from yeast SHY52, and were extensively washed. (A) Bead samples (+) were then incubated with yeast extract, washed, denatured, and analyzed by SDS-PAGE. Proteins were visualized by coomassie staining. Control bead samples (−) were not incubated with yeast extract. The input lane represents 1.67% of the extract that was incubated with with each immobilized protein. 257 and 189 kDa yeast proteins that associated with GST-Inp53-C, but not with GST alone, are indicated (●). The 189-kDa protein was identified as Chc1p by mass spectrometry analysis. (B) Immoblized proteins incubated with yeast extract (samples loaded in lanes 2, 4, and 6 of part A) were analyzed by SDS-PAGE and immunoblotted with an anti-Chc1p antibody. The control input lane represents 100% of the yeast extract that was incubated with the beads. The percentage of Chc1p in the input yeast extract that was pulled down by each immobilized protein is indicated below the panel.
To confirm the identification of the 189-kDa protein and to assess the efficiency of its binding to GST-Inp53-C, the samples loaded in lanes 2 and 4 of Figure 7A along with 100% of the input yeast extract were separated by SDS-PAGE and immunoblotted with an antibody against Chc1p. In accordance with mass spectrometry analysis 17% of the input Chc1p associated with the GST-Inp53-C beads but no Chc1p associated with GST alone.
A five-amino acid consensus motif for binding to the terminal domain of clathrin heavy chain has been identified: L(L/I)(D/E/N)(L/F)(D/E) (Dell'Angelica et al., 1998; ter Haar et al., 2000; Drake and Traub, 2001). Examination of the Inp53p sequence revealed a similar motif in the proline-rich domain: LLDID919. To determine if this sequence mediates binding to clathrin, the pull-down experiment described above was also performed with beads coated with a mutant GST-Inp53-C fusion in which the LLDID919 motif was deleted. The amount of GST-Inp53-CΔLLDID fusion present on the beads was indistinguishable from that of the wild-type fusion (compare lanes 4 and 5 with 6 and 7 of Figure 7A). Although some binding of GST-Inp53-CΔLLDID to Chc1p was observed (Figure 7B), the extent of binding was reduced to about half that of the wild-type fusion. These results suggest that clathrin association with Inp53p may be mediated in part by the LLDID919 motif but that other structural features also play a role in clathrin interaction.
inp53 and chc1 Mutants Affect A(F→A)-ALP Trafficking In Distinct Ways
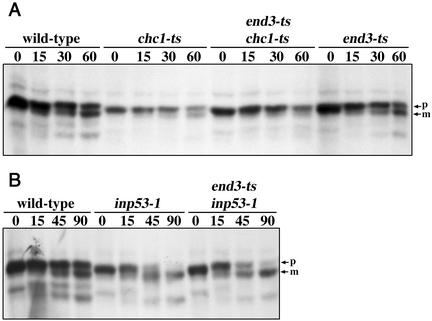
The identification of Chc1p as a binding partner for Inp53p complements other studies demonstrating a genetic interaction between inp53 and chc1 (Bensen et al., 2000) as well as interaction between mammalian synaptojanin and clathrin (Haffner et al., 2000). However, in chc1 mutants the TGN resident protein DPAP A is mislocalized to the plasma membrane (Seeger and Payne, 1992), raising the question of whether A(F→A)-ALP in both chc1 and inp53 mutants is initially mislocalized to the plasma membrane before being transported to the PVC.
At the nonpermissive temperature the end3tsallele blocks the internalization step of endocytosis (Benedetti et al., 1994). Thus to determine whether A(F→A)-ALP in inp53 and chc1 strains is transported to the plasma membrane and then uses the endocytic pathway to access the PVC, we introduced the end3ts allele into these strains. The rate of processing of A(F→A)-ALP in the chc1ts mutant at the nonpermissive temperature was somewhat variable and in some experiments was more rapid than wild-type while in others it was similar to wild-type as shown in Figure 8A. Processing was blocked in the chc1ts end3ts strain consistent with trafficking of A(F→A)-ALP via the plasma membrane upon a loss of clathrin function. As expected, the end3ts single mutation did not block processing in strains with functional clathrin confirming that A(F→A)-ALP did not reach the PVC and vacuole via the plasma membrane. In contrast to the results with chc1ts, no block in processing was observed in a strain lacking both Inp53p and End3p function (Figure 8B). Thus, in inp53 mutants, as in wild-type, A(F→A)-ALP reaches the PVC/vacuole independent of the plasma membrane.
Figure 8.
Comparison of the role of Inp53p and Chc1p in trafficking of A(F→A)-ALP. Cells were propagated at 22°C overnight and shifted to 37°C for 15 min before labeling with [35S]methione/cysteine for 10 min followed by a chase for times (min) indicated above each panel. At each time point, A(F→A)-ALP was immunoprecipitated and analyzed by SDS-PAGE and fluorography. Wild-type (SHY35), inp53-1 (SHY40), end3-ts (SNY94), inp53-1 end3ts (SHY51), chc1-ts (SNY37), and chc1-ts end3ts (SHY63) yeast strains carrying a plasmid expressing A(F→A)-ALP (pSN100) were analyzed.
Inp53p Acts in A Pathway Distinct from the GGA-mediated TGN-to-PVC Pathway
Recently two clathrin-mediated pathways for transport between the yeast TGN and endosomes have been described (Black and Pelham, 2000; Costaguta et al., 2001). One of these appears to lead from the TGN to the PVC independent of the early endososome and is mediated by the GGA proteins and clathrin. Yeast contain two functionally redundant GGA genes, GGA1 and GGA2, that encode ARF-interacting adapator-like proteins that function with clathrin at the TGN (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000). The other pathway appears to lead to the early endosome and involves clathrin and the AP-1 adaptor complex although the issue of whether clathrin and AP-1 act at the TGN or at the early endosome itself has not been fully resolved (Black and Pelham, 2000; Valdivia et al., 2002). It was previously shown that blocking both pathways by simultaneously inactivating both Apl2p, the β subunit of AP-1, and the GGA proteins caused a dramatic growth defect (Costaguta et al., 2001), suggesting a degree of functional redundancy between the two pathways. The interaction between Inp53p and clathrin and the phenotype of inp53 mutants suggest that Inp53p acts in one or more of these pathways.
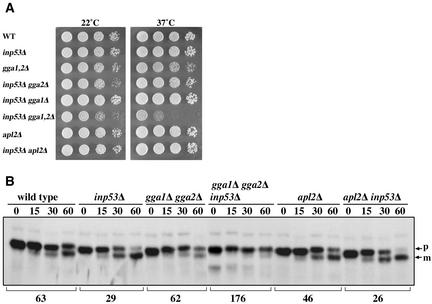
To determine which pathway Inp53p acts in, we combined the inp53Δ mutation with apl2Δ and with the gga1Δ gga2Δ mutations and assessed yeast growth at 22° and 37°C (Figure 9). Although no synthetic growth defect was observed when inp53Δ was combined with apl2Δ, a dramatic synthetic growth defect was observed upon combining inp53Δ with gga1Δ gga2Δ, particularly at 37°C (Figure 9A).
Figure 9.
Combination of inp53 with the gga1Δ gga2Δ mutations causes synthetic phenotypes. (A) Growth of the indicated mutant strains (from top to bottom: SHY35, SHY38, SNY165, SHY60-1A, SHY60-2A, SHY60-6C, UFY2, and SNY173-1B) was assessed by spotting 10-fold serial dilutions onto YEPD plates and growing for 3 d at the indicated temperature. (B) A subset of the strains from (A) carrying plasmid pSN100 to direct expression of A(F→A)-ALP were analyzed for A(F→A)-ALP processing kinetics as described in the legend to Figure 3.
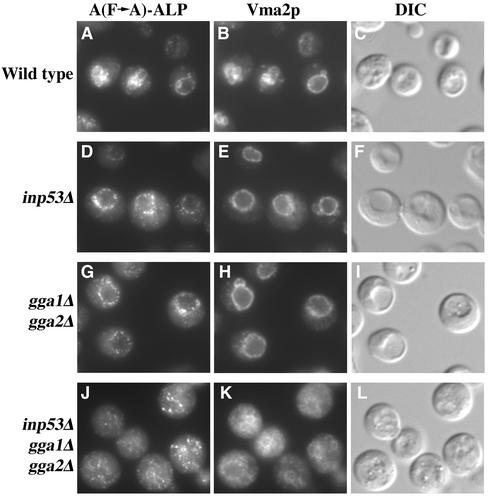
Several key observations were made upon analysis of A(F→A)-ALP processing in a subset of the strains shown in Figure 9A. First, the apl2Δ strain exhibited faster processing of A(F→A)-ALP compared with wild-type (46 vs. 63 min), although processing was not as fast as inp53Δ (29 min). The inp53Δ apl2Δ double mutant exhibited a similar rate of processing to the inp53Δ single mutant. In contrast to the apl2Δ and inp53Δ mutants, the gga1Δ gga2Δ mutant exhibited essentially no increase in the rate of processing compared with wild type. Strikingly, the growth-defective inp53Δ gga1Δ gga2Δ triple mutant exhibited a dramatic delay in processing (176 min) compared with the inp53Δ single mutant (29 min) and the gga1Δ gga2Δ double mutant (62 min). Vacuolar processing of ALP in the inp53Δ gga1Δ gga2Δ triple mutant was delayed by only ∼10 min (half-time of 5 and 15 min in wild-type and triple mutant strains, respectively; data not shown). Thus the delay in A(F→A)-ALP processing in the triple mutant is not accounted for by a lack of vacuolar proteolytic activity and instead must be due to a transport defect. Analysis of the inp53Δ gga1Δ gga2Δ strain by indirect immunofluorescence microscopy revealed that A(F→A)-ALP primarily localized to nonvacuolar small punctate structures, which could represent fragmented TGN or endosomes (Figure 10). In contrast, A(F→A)-ALP was primarily vacuolar in wild-type, inp53Δ, and gga1Δ gga2Δ strains. A substantial vacuolar morphology defect was also apparent in the inp53Δ gga1Δ gga2Δ strain that was not apparent in the control strains (compare panel K to B, E, and H), consistent with the idea that transport of cargo to the vacuole via multiple pathways may be blocked in this mutant.
Figure 10.
A(F→A)-ALP localization in inp53Δ strains also containing apl2Δ and gga mutations. Wild-type, inp53Δ, gga1Δ gga2Δ, and inp53Δ gga1Δ gga2Δ strains (see legend to Figure 9 for names) carrying plasmid pSN100 to direct expression of A(F→A)-ALP were fixed, spheroplasted, and costained for A(F→A)-ALP and Vma2p, a vacuolar marker. After subsequent treatment with fluorochrome-conjugated secondary antibodies, the cells were viewed by differential interference contrast optics and by epifluorescence through filters specific for FITC and Texas Red.
DISCUSSION
In this study we have investigated the function of the three domains of Inp53p in its role in mediating trafficking between the TGN and endosomes and have addressed which TGN-to-endosome pathway Inp53p functions in. We show that both enzymatic domains of Inp53p are required for mediating slow transport of the TGN protein A-ALP from the TGN to the PVC. The C-terminal proline-rich domain was found to not influence the overall extent of membrane association but was shown to associate with two other proteins, one of which is clathrin heavy chain. Interestingly, chc1 and inp53 mutants are both defective in trafficking of TGN resident proteins, but the phenotypes are different in that chc1 mutants mislocalize TGN proteins to the cell surface, whereas inp53 mutants do not. Combining an inp53Δ allele with the gga1Δ gga2Δ mutations resulted in dramatic synthetic defects in growth and in transport of A(F→A)-ALP trafficking to the PVC, whereas no synthetic defects were observed when inp53Δ was combined with a mutation in the AP-1 adaptor complex. Taken together, these results indicate that Inp53p functions in collaboration with clathrin and AP-1 in mediating trafficking between the TGN and endosomes.
Slow Transport of TGN Proteins into the PVC Requires Both Enzymatic Domains of Inp53p
Loss of function of all three synaptojanin-like proteins in yeast is lethal under normal growth conditions but this lethality can be rescued by providing only a functional 5-phosphatase domain (Stefan et al., 2002), suggesting that an accumulation of PtdIns(4,5)P2 causes the lethality but accumulation of other phosphoinositides do not. The growth requirement for a functional 5-phosphatase domain from a synaptojanin family member likely reflects the role of these proteins in mediating the level of PtdIns(4,5)P2 at the plasma membrane. PtdIns(4,5)P2 is known to mediate actin dynamics at the plasma membrane (Janmey, 1994; Martin, 2001) and yeast cell wall synthesis appears to rely on actin cytoskeletal components (Mulholland et al., 1997; Tang et al., 2000). Accordingly, the yeast synaptojanins are required for proper cell wall synthesis (Srinivasan et al., 1997; Stolz et al., 1998; Stefan et al., 2002) and Inp52p and Inp53p have been shown to localize to actin cortical patches under osmotic stress conditions (Ooms et al., 2000). Finally, the inp51 inp52 inp53 mutant strain can be partially rescued by including an osmotic stabilizer in the media (Stolz et al., 1998), suggesting that defective cell wall structure is a critical factor in lethality of the triple knockout.
In contrast, our observation that both the SacI and 5-phosphatase domains are required for mediating slow transport into the PVC indicates that an accumulation of other phosphoinositides in addition to PtdIns(4,5)P2 is deleterious for this process. Accumulation of phosphoinositides in inp53 mutants could affect vesicular transport between the TGN and endosomes in various ways. Mammalian synaptojanin has been proposed to act in uncoating of clathrin-coated vesicles by hydrolyzing PtdIns(4,5)P2 to PtdIns(4)P (Cremona et al., 1999), which in turn would promote dissociation of components of the clathrin coat that bind to PtdIns(4,5)P2, such as AP-1 (Crottet et al., 2002) and AP-2 (Collins et al., 2002; Rohde et al., 2002). In yeast, it is possible that Inp53p could be involved in uncoating TGN-derived clathrin-coated vesicles via its 5-phosphatase domain, especially given the interaction between Inp53p and Chc1p (Figure 7). By analogy with synaptic vesicle recycling, a lack of uncoating of such vesicles could block transport by preventing fusion with the acceptor organelle or by preventing recycling of components necessary for vesicular formation such as AP-1 and clathrin.
A lack of SacI domain activity in yeast synaptojanins primarily causes accumulation of PtdIns(4)P and to a lesser extent PtdIns(3,5)P2 (Stefan et al., 2002; Table 2). PtdIns(4)P pools are also regulated by the PtdIns(4)P kinases Pik1p, Stt4p, and Lsb6p. Stt4p and Lsb6p appear restricted to regulation of PtdIns(4)P pools at the plasma membrane and, possibly, vacuolar membrane (Audhya and Emr, 2002; Han et al., 2002). In contrast, Pik1p is localized to the TGN and nucleus and appears to have roles in secretory vesicle budding from the TGN, maintenance of Golgi structure and in cytokinesis (Garcia-Bustos et al., 1994; Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). Thus it is possible that an accumulation of PtdIns(4)P at the TGN in the inp53-sac1 could affect trafficking of TGN resident proteins to endosomes by affecting Golgi integrity or via a previously unrecognized role in budding of vesicles bound for endosomes.
The Inp53p C-terminal Proline-rich Domain Associates with Clathrin Heavy Chain
Using an in vitro pull down assay we showed that the C-terminal proline-rich domain of Inp53p associated with clathrin heavy chain and an unidentified 254-kDa protein. Mutation of a putative clathrin-binding motif in the proline-rich domain of Inp53p (LLDID919) cut the extent of binding in half. Other sequences exist in the proline-rich domain that loosely resemble the canonical clathrin box [L(L/I)(D/E/N)(L/F)(D/E)] such as LLSLD898, therefore, it is possible that one or more motifs other than LLDID919 may contribute to clathrin interaction. We cannot completely exclude the possibility that LLDID919 is not a true clathrin-binding motif and that by deleting this sequence the structure of the proline-rich domain was altered to interfere with the structure of the true clathrin-binding motif. Nevertheless, it is interesting that sequence alignments reveal an LIDLD948 sequence in Inp52p that aligns with the LLDID919 motif of Inp53p (S. Nothwehr, unpublished data), suggesting that this sequence may mediate association of the proline-rich domain of Inp52p with Chc1p. It is likely that Inp53p may function similarly to mammalian synaptojanin in clathrin-coated vesicle vesicle uncoating (discussed above), because mammalian synaptojanin has also been shown to associate with clathrin heavy chain and with the AP-2 adaptor complex (Haffner et al., 2000). Mammalian synaptojanin and clathrin have been shown to directly associate (Haffner et al., 2000), suggesting that this is probably also the case for the yeast Inp53p/Chclp interaction; however, we cannot at this time rule out the possibility that the interaction is mediated by a third protein.
A minor pool of Inp53p associates with a P200 fraction that contains Golgi, endosomes, or vesicles. Association of Inp53p with this fraction is not affected by deletion of the C-terminal proline-rich domain, although deletion of this domain increased association with membranes fractionating in the P15 fraction. Thus a domain distinct from the proline-rich domain, such as one of the catalytic domains, must be involved in general membrane association. The proline-rich domain may then specify binding to specific membrane domains via its interaction with other proteins such as clathrin. Binding of the proline-rich domain to other proteins could also regulate the activity and/or specificity of the catalytic domains.
Inp53p Appears to Function in an AP-1/Clathrin-mediated Transport Step
Multiple pathways exist in yeast for trafficking of proteins from the TGN to endosomes. For example, Vps10p and Cps1p reach the PVC very rapidly (∼15 min) from the TGN (Bryant and Stevens, 1997; Ha et al., 2001) and entry of Cps1p into the PVC is blocked in strains lacking GGA protein function (Costaguta et al., 2001). In contrast trafficking of A(F→A)-ALP into the PVC occurs much more slowly (∼60 min; Ha et al., 2001) and is not blocked in the gga1Δ gga2Δ strain (Figure 9). A t-SNARE of the PVC, Pep12p, reaches the PVC via the GGA pathway, and its sorting into the GGA pathway relies on a cytosolic sorting signal (Black and Pelham, 2000). If the signal is mutated or if GGA function is lost, Pep12p is instead transported to early endosomes.
Recent evidence suggests that the AP-1 adaptor complex in collaboration with clathrin appear to be involved in a TGN-to-endosome pathway distinct from the GGA/clathrin pathway. Severe growth defects have been observed in cells lacking the function of the β subunit of AP-1, Apl2p, and the GGA proteins (Costaguta et al., 2001), suggesting that AP-1 and the GGAs represent distinct clathrin-associated adaptors that function in separate but partially redundant pathways. In addition, clathrin genetically and physically interacts with both AP-1 and the GGA proteins (Pishvaee and Payne, 1998; Yeung et al., 1999; Costaguta et al., 2001). Cells lacking clathrin function are severely compromised for growth and mislocalize TGN resident membrane proteins to the cell surface (Lemmon and Jones, 1987; Payne et al., 1987; Seeger and Payne, 1992), consistent with clathrin being required for both pathways. The difference in the A(F→A)-ALP trafficking phenotype between inp53Δ and chc1ts strains can be explained by Chc1p being needed in two pathways, whereas inp53Δ is needed only in one. The AP-1/clathrin coat complex likely functions in transport between the TGN and early endosome because trafficking of both Chs3p, a protein that maintains an intracellular localization via transport between the TGN and early endosome, and Tlg1p, an early endosome/TGN t-SNARE, is affected in AP-1 mutants (Valdivia et al., 2002).
The observation that synthetic growth and transport defects occurred upon combining the inp53Δ mutation with gga1Δ gga2Δ (Figure 9) suggests that Inp53p functions in AP-1/clathrin–mediated transport between the TGN and early endosome. We observe more rapid transport of A-ALP into the PVC upon inactivation of either Inp53p or AP-1 (Ha et al., 2001; Figure 9), suggesting that if transport between the TGN and early endosome is disrupted, then A-ALP is transported into the PVC more rapidly. Given the likely role of Inp53p in vesicle uncoating (see above), we favor the idea that AP-1/clathrin and Inp53p act in the same transport step. More rapid transport into the PVC could then occur because of a lack of formation of vesicles from the TGN bound for the early endosome that could cause default entry of A-ALP into the GGA/clathrin pathway. Alternatively, AP-1/clathrin may act in retrieval from the early endosome (see Valdivia et al., 2002) and if this were the case, a lack of retrieval could result in early endosome-to-PVC transport by default. However, a failure to retrieve A(F→A)-ALP from the early endosome would not be expected to cause a lengthy delay in transport to the PVC as is observed when both Inp53p and the GGAs are inactivated. Thus, our data seem most consistent with AP-1/clathrin and Inp53p being needed for formation of vesicles from the TGN for delivery to early endosomes. In this case both pathways would be blocked, thus preventing TGN proteins from reaching the PVC. If AP-1/clathrin and Inp53p mediate the return pathway from the early endosome then in the inp53Δ gga1Δ gga2Δ mutant traffic from the early endosome to the PVC must also be blocked. We have observed a delay in trafficking of the lipophilic endocytic tracer dye FM4–64 from the PM to the vacuole in inp53Δ gga1Δ gga2Δ cells compared with wild type (S. Nothwehr, unpublished results). However, the magnitude of the delay is much less than that observed for A(F→A)-ALP processing in the inp53Δ gga1Δ gga2Δ strain, suggesting that a general block in trafficking through the endocytic pathway is probably not responsible for the block in trafficking of A(F→A)-ALP to the PVC.
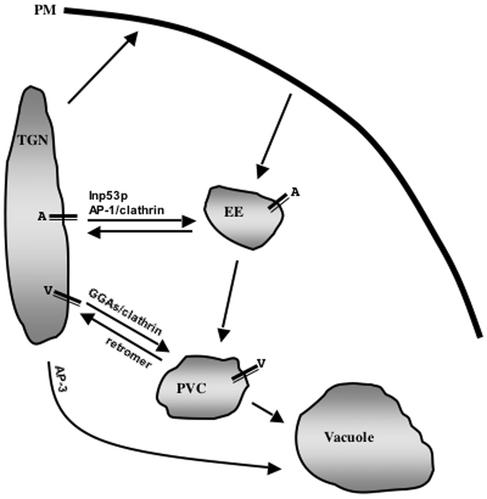
A working model of the role of Inp53p in trafficking of A-ALP to the PVC is depicted in Figure 11. We propose that the slow delivery of A-ALP into the PVC occurs because A-ALP frequently cycles between the TGN and early endosome and is less frequently transported to the PVC. The infrequent passage of A-ALP into the PVC could occur via inefficient retrieval from the early endosome and direct early endosome-to-PVC transport, or by inefficient entry into the TGN-to-early endosome pathway resulting in TGN-to-PVC direct transport. Addressing the role of the slow delivery signal in the A-ALP cytosolic domain in TGN/early endosome cycling will be a subject of future study.
Figure 11.
A model for the role of Inp53p and other machinery in trafficking of TGN resident proteins between the TGN and endosomes. In this model the AP-1 adaptor complex, clathrin, and Inp53p are required for anterograde TGN-to-early endosome transport while clathrin and the GGA proteins are required for TGN-to-PVC transport. TGN proteins such as A-ALP (A) tend to cycle between the TGN and early endosome and infrequently enter the PVC perhaps because of inefficient sorting at the TGN or early endosome. Inp53p acts in concert with the AP-1/clathrin perhaps in vesicle uncoating. In contrast, proteins such as Vps10p and Cps1p (V) are transported directly to from the TGN to the PVC via the GGA/clathrin pathway.
ACKNOWLEDGMENTS
We acknowledge Sandra Lemmon, Scott Emr, and Greg Payne for providing antibodies and strains. This work was supported by grants from the National Institutes of Health, GM-53449 and RR-11823, awarded to S.F.N. and R.A., respectively, and from the American Cancer Society awarded to D.B.D.
Abbreviations used:
- ALP
alkaline phosphatase
- CEN
yeast centromere
- CPY
carboxypeptidase Y
- DPAP
dipeptidyl aminopeptidase
- ER
endoplasmic reticulum
- GST
glutathione-S-transferase
- ORF
open reading frame
- PtdIns
phosphatidylinositol
- PtdIns(3)P
phosphatidylinositol 3-phosphate
- PtdIns(4)P
phosphatidylinositol 4-phosphate
- PtdIns(3,5)P2
phosphatidylinositol (3,5)-bisphosphate
- PtdIns(4,5)P2
phosphatidylinositol (4,5)-bisphosphate
- PVC
prevacuolar/endosomal compartment
- TGN
trans-Golgi network
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0686. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–10–0686.
REFERENCES
- Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausebel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K, Smith JA. Current protocols in molecular biology. New York: John Wiley & Sons, Inc; 2000. [Google Scholar]
- Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensen ES, Costaguta G, Payne GS. Synthetic genetic interactions with temperature-sensitive clathrin Saccharomyces cerevisiae: roles for synaptojanin-like Inp53p and dynamin-related Vps1p in clathrin-dependent protein sorting at the trans-Golgi network. Genetics. 2000;154:83–97. doi: 10.1093/genetics/154.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black MW, Pelham HRB. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman AL, Zhang C, Zhu X, Kahn RA. A family of ADP-ribosylation factor effectors that can alter membrane transport through the. trans-Golgi. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Two separate signals act independently to localize a yeast late Golgi membrane protein through a combination of retrieval and retention. J Cell Biol. 1997;136:287–297. doi: 10.1083/jcb.136.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caroni P. Actin cytoskeleton regulation through modulation of PI(4,5)P-2 rafts. EMBO J. 2001;20:4332–4336. doi: 10.1093/emboj/20.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JK, Sekiya F, Kang HS, Lee C, Han JS, Kim SR, Bae YS, Morris AJ, Rhee SG. Synaptojanin inhibition of phospholipase D activity by hydrolysis of phoshpatidylinositol 4,5-bisphosphate. J Biol Chem. 1997;272:15980–15985. doi: 10.1074/jbc.272.25.15980. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Costaguta G, Stefan CJ, Bensen ES, Emr SD, Payne GS. Yeast Gga coat proteins function with clathrin in Golgi to endosome transport. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremona O, et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell. 1999;99:179–188. doi: 10.1016/s0092-8674(00)81649-9. [DOI] [PubMed] [Google Scholar]
- Crottet P, Meyer DM, Rohrer J, Spiess M. ARF1-GTP, tryrosine-based signals, and phoshatidylinositol 4,5-bisphosphate constitute a minimal machinery to recruit the AP-1 clathrin adaptor to membranes. Mol Biol Cell. 2002;13:3672–3682. doi: 10.1091/mbc.E02-05-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC, Klumperman J, Stoorvogel W, Bonifacino JS. Association of the AP-3 adaptor complex with clathrin. Science. 1998;280:431–434. doi: 10.1126/science.280.5362.431. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica EC, Puertollano R, Mullins C, Aguilar RC, Vargas JD, Hartnell LM, Bonifacino JS. GGAs. A family of ADP ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J Cell Biol. 2000;149:81–93. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem. 2001;276:28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- Garcia-Bustos JF, Marini F, Stevenson I, Frei C, Hall MN. PIK1, an essential phosphatidylinositol 4-kinase associated with the yeast nucleus. EMBO J. 1994;13:2352–2361. doi: 10.1002/j.1460-2075.1994.tb06519.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SL, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- Ha S-H, Bunch JT, Hama H, DeWald DB, Nothwehr SF. A novel mechanism for localizing membrane proteins to the yeast trans-Golgi network requires function of a synaptojanin-like protein. Mol Biol Cell. 2001;12:3175–3190. doi: 10.1091/mbc.12.10.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffner C, Di Paolo G, Rosenthal JA, De Camilli P. Direct interaction of the 170 kDa isoform of synaptojanin 1 with clathrin and with the clathrin adaptor AP-2. Curr Biol. 2000;10:471–474. doi: 10.1016/s0960-9822(00)00446-2. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hama H, Takemoto JY, DeWald DB. Analysis of phosphoinositides in protein trafficking. Methods: A Companion to Methods in Enzymology. 2000;20:465–473. doi: 10.1006/meth.2000.0959. [DOI] [PubMed] [Google Scholar]
- Han GS, Audhya A, Markley DJ, Emr SD, Carman GM. The Saccharomyces cerevisiae LSB6 gene encodes PI 4-kinase activity. J Biol Chem. 2002;277:47709–47718. doi: 10.1074/jbc.M207996200. [DOI] [PubMed] [Google Scholar]
- Hanson BA, Lester RL. The extraction of inositol-containing phospholipids and phosphatidylcholine from Saccharomyces cerevisiae and Neurospora crassa. J Lipid Res. 1980;21:309–315. [PubMed] [Google Scholar]
- Harder KW, Owen P, Wong LK, Aebersold R, Clark-Lewis I, Jirik FR. Characterization and kinetic analysis of the intracellular domain of human protein tyrosine phosphatase beta (HPTP beta) using synthetic phosphopeptides. Biochem J. 1994;298:395–401. doi: 10.1042/bj2980395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris TW, Hartwieg E, Horvitz RH, Jorgensen EM. Mutations in synaptojanin disrupt synaptic vesicle recycling. J Cell Biol. 2000;150:589–599. doi: 10.1083/jcb.150.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess HH, Derr JE. Assay of inorganic and organic phosphorus in the 0.1–5 nanomole range. Anal Biochem. 1975;63:607–613. doi: 10.1016/0003-2697(75)90388-7. [DOI] [PubMed] [Google Scholar]
- Hill KJ, Stevens TH. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol Biol Cell. 1994;5:1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WWY, Bright NA, Totty N, Seaman MNJ, Robinson MS. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–79. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WE, Cooke FT, Parker PJ. Sac phosphatase domain proteins. Biochem J. 2000;350:337–352. [PMC free article] [PubMed] [Google Scholar]
- Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56:169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- Jefferson AB, Majerus PW. Mutation of the conserved domains of two inositol polyphosphate 5-phosphatases. Biochemistry. 1996;35:7890–7894. doi: 10.1021/bi9602627. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lemmon SK, Jones EW. Clathrin requirement for normal growth of yeast. Science. 1987;238:504–509. doi: 10.1126/science.3116672. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HRB. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus SL, Wenk MR, Steele-Mortimer O, Finlay BB. A synaptojanin-homologous region of Salmonella typhimurium SigD is essential for inositol phosphatase activity and Akt activation. FEBS Lett. 2001;494:201–207. doi: 10.1016/s0014-5793(01)02356-0. [DOI] [PubMed] [Google Scholar]
- Marcusson EG, Horazdovsky BF, Cereghino JL, Gharakhanian E, Emr SD. The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell. 1994;77:579–586. doi: 10.1016/0092-8674(94)90219-4. [DOI] [PubMed] [Google Scholar]
- Martin TFJ. PI(4,5)P-2 regulation of surface membrane traffic. Curr Opin Cell Biol. 2001;13:493–499. doi: 10.1016/s0955-0674(00)00241-6. [DOI] [PubMed] [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang XM, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, Decamilli P. A presynaptic inositol-5-phosphatase. Nature. 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Mulholland J, Wesp A, Riezman H, Botstein D. Yeast actin cytoskeleton mutants accumulate a new class of Golgi-derived secretory vesicle. Mol Biol Cell. 1997;8:1481–1499. doi: 10.1091/mbc.8.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bruinsma P, Strawn LS. Distinct domains within Vps35p mediate the retrieval of two different cargo proteins from the yeast prevacuolar/endosomal compartment. Mol Biol Cell. 1999;10:875–890. doi: 10.1091/mbc.10.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Bryant NJ, Stevens TH. The newly identified yeast GRD genes are required for retention of late-Golgi membrane proteins. Mol Cell Biol. 1996;16:2700–2707. doi: 10.1128/mcb.16.6.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Conibear E, Stevens TH. Golgi and vacuolar membrane proteins reach the vacuole in vps1 mutant yeast cells via the plasma membrane. J Cell Biol. 1995;129:35–46. doi: 10.1083/jcb.129.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Ha S-A, Bruinsma P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J Cell Biol. 2000;151:297–309. doi: 10.1083/jcb.151.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley CJ, McColl BK, Kong AM, Ellis SL, Wijayaratnam APW, Sambrook J, Mitchell CA. Mammalian inositol polyphosphate 5-phosphatase II can compensate for the absence of all three yeast. SacI-like-domain-containing 5-phosphatases. Biochem J. 2001;355:805–817. doi: 10.1042/bj3550805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms LM, McColl BK, Wiradjaja F, Wijayaratnam APW, Gleeson P, Gething MJ, Sambrook J, Mitchell CA. The yeast inositol polyphosphate 5-phosphatases Inp52p and Inp53p translocate to actin patches following hyperosmotic stress: mechanism for regulating phosphatidylinositol 4,5-bisphosphate at plasma membrane invaginations. Mol Cell Biol. 2000;20:9376–9390. doi: 10.1128/mcb.20.24.9376-9390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Payne GS, Hasson TB, Hasson MS, Schekman R. Genetic and biochemical characterization of clathrin-deficient Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:3888–3898. doi: 10.1128/mcb.7.11.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pishvaee B, Payne GS. Clathing coats—threads laid bare. Cell. 1998;95:443–446. doi: 10.1016/s0092-8674(00)81611-6. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Rohde G, Wenzel D, Haucke V. A phosphatidylinositol (4,5)-bisphosphate binding site within μ2-adaptin regulates clathrin-mediated endocytosis. J Cell Biol. 2002;158:209–204. doi: 10.1083/jcb.200203103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G, Sommer SS. The “megaprimer”method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, McCaffery JM, Emr SD. A membrane coat complex essential for endosome-to-Golgi retrograde transport in yeast. J Cell Biol. 1998;142:665–681. doi: 10.1083/jcb.142.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serunian LA, Auger KR, Cantley LC. Identification and quantification of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Wurmser AE, Emr SD, Stenmark H. The role of phosphoinositides in membrane transport. Curr Opin Cell Biol. 2001;13:485–492. doi: 10.1016/s0955-0674(00)00240-4. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger B, Nemoto Y, Daniell L, Ferro-Novick S, De Camilli P. Synaptojanin family members are implicated in endocytic membrane traffic in yeast. J Cell Sci. 1998;111:3347–3356. doi: 10.1242/jcs.111.22.3347. [DOI] [PubMed] [Google Scholar]
- Spelbrink RG, Nothwehr SF. The yeast GRD20 gene is required for protein sorting in the trans-Golgi network/endosomal system and for polarization of the actin cytoskeleton. Mol Biol Cell. 1999;10:4263–4281. doi: 10.1091/mbc.10.12.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, Decamilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stefan CJ, Audhya A, Emr SD. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol Biol Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Huynh C, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP53, INP52, and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H-Y, Xu J, Cai M. Pan1p, End3p, and Sla1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the b-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Baggot D, Chuang JS, Schekman R. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Yates JRD, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67:1426–1436. doi: 10.1021/ac00104a020. [DOI] [PubMed] [Google Scholar]
- Yeung BG, Phan HL, Payne GS. Adaptor complex-independent clathrin function in yeast. Mol Biol Cell. 1999;10:3643–3659. doi: 10.1091/mbc.10.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]