Abstract
The Zeste-White 10 (ZW10) and Rough Deal (ROD) proteins are part of a complex necessary for accurate chromosome segregation. This complex recruits cytoplasmic dynein to the kinetochore and participates in the spindle checkpoint. We used immunoaffinity chromatography and mass spectroscopy to identify the Drosophila proteins in this complex. We found that the complex contains an additional protein we name Zwilch. Zwilch localizes to kinetochores and kinetochore microtubules in a manner identical to ZW10 and ROD. We have also isolated a zwilch mutant, which exhibits the same mitotic phenotypes associated with zw10 and rod mutations: lagging chromosomes at anaphase and precocious sister chromatid separation upon activation of the spindle checkpoint. Zwilch's role within the context of this complex is evolutionarily conserved. The human Zwilch protein (hZwilch) coimmunoprecipitates with hZW10 and hROD from HeLa cell extracts and localizes to the kinetochores at prometaphase. Finally, we discuss immunoaffinity chromatography results that suggest the existence of a weak interaction between the ZW10/ROD/Zwilch complex and the kinesin-like kinetochore component CENP-meta.
INTRODUCTION
Eukaryotic organisms require extraordinary fidelity in chromosome segregation during meiosis and mitosis because mistakes in chromosome distribution can be catastrophic to an organism or its progeny. At least one of the macromolecular assemblies that ensure the fidelity of chromosome distribution seems to have evolved exclusively in multicellular eukaryotes. We have characterized a complex containing at least two proteins, Zeste-White 10 (ZW10) and Rough Deal (ROD), that is required for proper chromosome segregation and that is evolutionarily conserved in plants and animals (Williams et al., 1992, 1996; Williams and Goldberg, 1994; Starr et al., 1997, 1998; Scaerou et al., 1999, 2001). ZW10 and ROD proteins have not yet been found in any unicellular eukaryote, and the genome of Saccharomyces cerevisiae does not contain genes with detectable homologies to zw10 or rod. Mutations in the Drosophila zw10 or rod genes cause similar defects, most noticeably lagging chromatids that remain at the metaphase plate late in anaphase, leading to high levels of aneuploidy among daughter cells.
ZW10 and ROD proteins display remarkable changes in their intracellular location during cell division (Williams and Goldberg, 1994; Scaerou et al., 1999; Wojcik et al., 2001). Both proteins accumulate strongly at the outer kinetochore plates during prometaphase. At metaphase, ZW10 and ROD migrate off the kinetochores onto the kinetochore microtubules (kMTs). During anaphase, the proteins are no longer found on kMTs and instead localize exclusively to the kinetochores of the separating chromosomes. The association of ZW10 and ROD with kMTs during metaphase suggests that the movement of these proteins from kinetochores to kMTs might require bipolar tension across the chromosomes exerted by the spindle. This expectation seems to be valid. In Drosophila spermatocytes at meiotic metaphase I, ZW10 and ROD remain on the kinetochores of univalents (homologous chromosomes attached to a single centromere). In the same cells, these proteins migrate off the kinetochores of the bivalents (normally paired homologous chromosomes, each with its own centromere) onto kMTs (Williams et al., 1996; Scaerou et al., 1999).
ZW10 and ROD have two functions critical to the accuracy of chromosome segregation. First, they help target the microtubule motor cytoplasmic dynein to kinetochores (Starr et al., 1998; Chan et al., 2000; Wojcik et al., 2001). This is mediated at least in part by direct contacts between ZW10 and dynamitin, one of the subunits of the dynein-associated dynactin complex (Echeverri et al., 1996; Starr et al., 1998). Presumably, the strongly reduced velocity at which chromosomes move toward the poles in Drosophila spermatocytes mutant for zw10 or rod is due to the loss of dynein from the kinetochore (Savoian et al., 2000). A recent finding indicates that dynein returns the favor and helps direct the movement of ROD (and perhaps ZW10) from kinetochores to kMTs during metaphase (Wojcik et al., 2001).
The chromosome segregation defects in zw10 and rod mutants might also involve a second activity of ZW10 and ROD: they are required for the function of the spindle assembly (metaphase) checkpoint. This checkpoint normally blocks anaphase onset until all chromosomes are aligned properly on the metaphase plate (reviewed by Amon, 1999). The checkpoint produces an inhibitory signal that prevents the anaphase promoting complex from ubiquitinating substrates whose degradation is a prerequisite for sister chromatid separation and mitotic exit (Zachariae and Nasmyth, 1999). Unlike wild-type cells, which arrest in response to spindle damage caused by microtubule poisons, cells mutant for zw10 or rod instead separate sister chromatids, degrade cyclin B, and exit mitosis in the presence of these drugs (Basto et al., 2000; Chan et al., 2000; Savoian et al., 2000). This phenotype is also seen in mutants for the BUBR1, BUB3, MAD1, and MAD2 proteins known to participate in the spindle checkpoint in yeast and higher organisms (Hoyt et al., 1991; Li and Murray, 1991; Basu et al., 1998, 1999).
Similarities in the intracellular localization of ZW10 and ROD with that of the BUB and MAD checkpoint components suggest the possibility of a mechanistically important interaction. Just as ZW10 and ROD, the BUB and MAD proteins accumulate preferentially on the kinetochores of chromosomes that have not yet congressed to the metaphase plate (Li and Benezra, 1996; Jablonski et al., 1998; Taylor et al., 1998; Waters et al., 1998; Martinez-Exposito et al., 1999). More recently, MAD2 has been found to migrate along kMTs at metaphase from the kinetochores of properly oriented chromosomes (Howell et al., 2000). But despite these parallels, the relationship of ZW10 and ROD with the canonical spindle checkpoint components is unclear. In both Drosophila and human cells, the two sets of proteins are mutually independent at the level of kinetochore targeting. Fly Bub1 and Bub3 accumulate at the kinetochores in zw10 or rod mutant cells, whereas ZW10 associates with kinetochores in bub1 and bub3 mutants (Basu et al., 1998, 1999; our unpublished data). Likewise, kinetochores depleted of human ZW10 and ROD retain hBUBR1, hBUB1, Mad1, and Mad2, whereas these checkpoint proteins can selectively bind to unattached kinetochores despite the absence of dynein, hZW10, and hROD (Chan et al., 2000). These results do not of course preclude the existence of more subtle interactions between these molecules.
Together with the laboratory of Dr. R. Karess (Centre National de la Recherche Scientifique, Gif-sur-Yvette, France), we have previously shown that ZW10 and ROD associate with each other as part of a large protein complex (Scaerou et al., 2001). This result explains why mutations in both genes cause identical phenotypes, why the two proteins colocalize throughout mitosis and meiosis, and why each protein depends on the other for its localization to kinetochores and kMTs. The multiple functions of ZW10/ROD suggest that the complex may contain additional components. In fact, the complex elutes from sizing columns with an apparent molecular mass of 700–900 kDa, considerably larger than the sum of the masses of ZW10 (85 kDa) and ROD (240 kDa). This article reports our efforts to characterize the complex by immunoaffinity chromatography of Drosophila embryo extracts on a column containing immobilized anti-ZW10 antibodies.
MATERIALS AND METHODS
Immunoaffinity Chromatography and Mass Spectrometry
To make the column resins, 2 mg of affinity-purified anti-ZW10 IgG or preimmune serum (Williams et al., 1992) were bound to 1 ml of Affigel-protein A beads (Bio-Rad, Hercules, CA) in phosphate-buffered saline (PBS). Antibodies were cross-linked to protein A by using dimethylpimidiliate (Sigma-Aldrich, St. Louis, MO) according to Harlow and Lane (2000) and stored at 4°C in PBS + 0.01% sodium azide. Protein extraction from 0- to 4-h embryos (Drs. William Sullivan and Kristina Yu, University of California, Santa Cruz, Santa Cruz, CA) and immunoaffinity chromatography followed previously described methods (Kellogg and Alberts, 1992).
SDS-polyacrylamide gels were stained for 12–16 h in 0.1% Coomassie Blue R250 containing 20% methanol and 0.5% acetic acid and then destained in 30% methanol. In-gel digestion with trypsin of excised protein bands, and analysis of the product by matrix-assisted laser desorption/ionization mass spectroscopy (MALDI) were performed according to Williams and Stone (1995) on a Biflex III instrument (Bruker, Bremen, Germany). Spectra were externally calibrated using trypsin-digested horse heart apomyoglobin D. XMASS (Bruker) and/or MoverZ (Proteometrics, Winnipeg, Manitoba, Canada) software was used to analyze spectra, by using trypsin autolytic and keratin peaks as internal calibrants. Monoisotopic peak mass values were submitted to PROWL analysis (Proteometrics) to match peptide masses to protein databases.
Fly Stocks
Lines containing ethylmethanesulfonate-induced lethal mutations on the third chromosome were produced in the laboratory of Dr. Charles Zuker (University of California, San Diego, La Jolla, CA). Brains from homozygous larvae were screened for a variety of mitotic defects. From 1583 mutant lines examined, one (mit-1229) containing a zwilch mutation was recovered. Df(3R)tll-g and Df(3R)tll-e were obtained from the Bloomington Drosophila stock center, whereas Df(3R)A177der22, Df(3R)A177der21, Df(3R)A177.X3, Df(3R)A177der26, Df(3R)3R177.X1, and Df(3R)PGX8 were received from Dr. Erich Frei (University of Zurich, Zurich, Switzerland). Other mutant strains used were zw10S1 (Williams et al., 1992), rodX5 (Karess and Glover, 1989), and a CENP-meta null allele (Yucel et al., 2000); Oregon-R was the wild-type strain. Mutations and deficiencies were balanced over TM6C marked with Stubble (Sb) and Tubby (Tb).
To isolate the lesion causing the mitotic phenotype, mit-1229 was outcrossed to ru h th st cu sr e ca, and the resulting recombinants were rebalanced with TM6C. Homozygotes for one such recombinant, zwilchmit-1229, displayed the mitotic phenotype but was viable.
Nucleic Acids
Deficiency breakpoints were determined as described previously (Williams et al., 1992) by using polymerase chain reaction (PCR)-amplified fragments from appropriate genomic regions as probes. To find the zwilch mutation, DNA was PCR amplified from the zwilchmit-1229 mutant and from a different mutant identified from the same mutagenesis as control. PCR products were cut out of gels, purified (QIAGEN, Valencia, CA), and sequenced using the same primers used in amplification. Differences between mutant and control were rechecked by sequencing duplicate PCR products.
Escherichia coli containing LD17202, a cDNA corresponding to CG18729 (zwilch), was obtained from Research Genetics (Athens, GA). To construct hemagglutinin (HA)-tagged ZW10, a 3X-HA tag was amplified from pGTEP1 (Pasqualone and Huffaker, 1994) and inserted in-frame into the MluI site of the zw10 gene found in pW8-zw10(8B) (Williams et al., 1992). The resulting clone was transformed into flies by P element-mediated germline transformation.
The open reading frame of a human cDNA clone (GenBank AK023175) showing sequence similiarity to Drosophila zwilch was PCR amplified and cloned into pENTR3C (Invitrogen, Carlsbad, CA) to facilitate recombination into a home-made pEGFP vector using the Gateway system. To overexpress His-tagged hZwilch in E. coli, a recombination reaction was performed to transfer the coding sequence from pENTR3C into pDEST17.
Anti-Zwilch Antibodies and Immunoprecipitations
Ssequences encoding the entire 644 amino acids of Zwilch from LD17202 were cloned into the EcoRI-XbaI sites of pMAL-C2 (New England Biolabs, Beverly, MA) in-frame with the gene for maltose binding protein (MBP). The fusion protein was produced in E. coli and purified according to manufacturer's protocols. Both native protein directly eluted from amylose columns and denatured protein eletroeluted from SDS-PAGE gel bands were used to immunize rabbits. Crude serum from each rabbit was affinity purified against MBP-Zwilch fusion protein coupled to CnBr-activated Sepharose beads. Antibodies obtained from both rabbits displayed the same staining in wild-type brains.
His-tagged hZwilch was purified from the inclusion body of an E. coli strain BL21(DE3) Codon Plus (Stratagene, La Jolla, CA) harboring pDEST17-hZwilch under denaturing conditions using a Probond nickel column (Invitrogen). The fusion protein was further purified by SDS-PAGE, and the corresponding gel slices were excised to immunize rats. Some fusion protein was solubilized by dialysis against PBS with a gradually lower concentration of urea. This soluble protein was cross-linked to Affigel-10 (Bio-Rad) to make an affinity column to purify anti-hZwilch antibodies from crude rat antisera. Rabbit anti-hZW10, rat anti-hROD (Chan et al., 2000), rabbit anti-hCENP-I (Liu et al., 2003) and rabbit anti-GFP antibodies (Molecular Probes, Eugene, OR) were also used.
Western blots were performed as described previously (Williams et al., 1992; Chan et al., 2000). Gel exclusion chromatography of Drosophila embryonic extracts was described in Scaerou et al. (2000). Chan et al. (2000) detail the procedures used for immunoprecipitation.
Cytological Preparations
Mitosis in fly larval brains was examined according to Williams et al. (1992) and Williams and Goldberg (1994), by using previously described reagents (Basu et al., 1999; Yucel et al., 2000; Scaerou et al., 2001) and affinity-purified rabbit anti-Zwilch (1/100 dilution in PBS + 0.1% Triton-X). For double-staining experiments, controls demonstrated the absence of bleed-through between channels, and of secondary antibody cross-reaction. Samples were examined using a Pentamax charge-coupled device camera (Roper Scientific, Tucson, AZ) attached to a BX50 microscope (Olympus, Tokyo, Japan) equipped for epifluorescence, and imaged and analyzed with MetaMorph software (Universal Imaging, Downingtown, PA).
Immunofluorescence of paraformaldehyde fixed HeLa cells was done as described previously (Chan et al., 2000). hZwilch was stained with a rat antibody (see above) followed by Alexafluor 488 (Molecular Probes) conjugated goat anti-rat secondary antibody. Centromeres were marked with rabbit antihuman CENP-I stained with Cy5-conjugated goat anti-rabbit secondary antibody.
GFP-hZwilch was transfected into HeLa cells, which were preextracted, fixed, and visualized for GFP fluorescence as described previously (Chan et al., 1999).
RESULTS
Zwilch is Present in a Complex with ZW10 and ROD
Previous studies found that the ZW10 and ROD proteins are closely associated within the context of a large, ∼600- to 800-kDa complex in Drosophila embryos (Scaerou et al., 2001). To identify other potential members of this complex, we performed immunoaffinity chromatography by using a column of immobilized, affinity-purified anti-ZW10 antibody. This antibody is extremely specific, recognizing only a single band on Western blots of extracts from wild-type flies. The band is the correct size for ZW10 (85 kDa) and is not found in extracts from zw10 mutants (Williams et al., 1992; Williams and Goldberg, 1994).
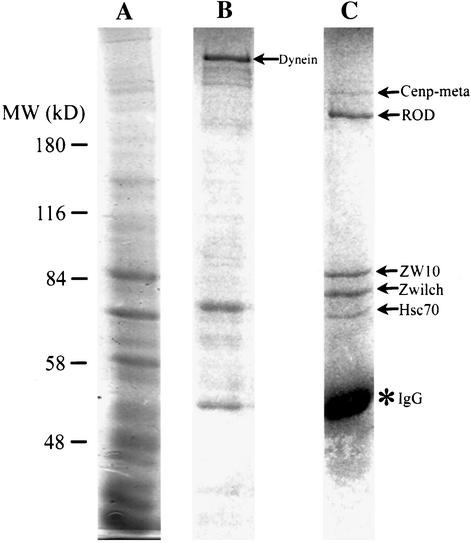
This anti-ZW10 column was used to purify ZW10 and its associated proteins from embryo extracts (Figure 1A). After extensive washing in 100 mM KCl, proteins were successively eluted from the column, first with 1 M KCl and then with 1.5 M MgCl2. Many proteins are seen in the 1 M KCl eluate; most of these seem to bind nonspecifically to IgG or the protein A-Agarose beads, because they also occur in eluates from a preimmune IgG control column (our unpublished data). One prominent component of the anti-ZW10 column 1 M KCl eluate that was not apparent in the control was identified by MALDI mass spectrometry as cytoplasmic dynein (Figure 1B).
Figure 1.
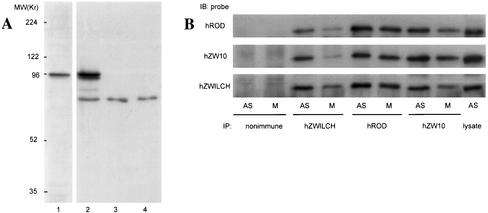
Identification of proteins complexed with ZW10. Embryo extracts were allowed to bind an anti-ZW10 column, which was washed and eluted as described in MATERIALS AND METHODS. Gels were stained with Coomassie Blue and proteins were identified by MALDI. (A) Total protein before column binding. (B) Elution with 1 M KCl. Cytoplasmic dynein elutes in this fraction (arrow); other bands are nonspecific and occur in fractions from control columns (our unpublished data). (C) Elution with 1.5 M MgCl2. Proteins identified by mass spectrometry are indicated (arrows). IgG leaching off the column is also prevalent (*).
In the subsequent 1.5 M MgCl2 elution, five distinct protein bands were observed that were not present in the control. These proteins, migrating at roughly 65, 75, 85, 250 and 300 kDa on SDS-PAGE (Figure 1C), were excised from the gel, digested with trypsin, and identified by MALDI. As expected, the 85-kDa band was ZW10, indicating that the affinity purification with anti-ZW10 was successful. The ∼250-kDa band was positively identified as ROD, confirming that ZW10 and ROD are tightly associated (Scareou et al., 2000). The identification of ZW10 and ROD was verified on Western blots by using anti-ZW10 and anti-ROD (our unpublished data). The 65-kDa band was found to be Hsc70–4, a chaperone that stabilizes protein conformation (Elefant and Palter, 1999). The relationship between Hsc70-4 and the ZW10/ROD complex is currently unclear. A fourth band of ∼300 kDa stains substoichiometrically with respect to the other proteins. MALDI results suggest that this band corresponds to CENP-meta, a kinesin-like protein associated with the kinetochore (Yucel et al., 2000). The possible relationship between CENP-meta and ZW10/ROD is examined later in this article.
Finally, the fifth band at ∼75 kDa was identified as the 644 residue-long product of CG18729, a gene predicted by the Berkeley Drosophila Genome Project. We named this protein Zwilch, because database searches originally failed to identify related proteins in other species (the search came up with “zilch”). Recent database searches have, however, revealed potential Zwilch homologs. The genome of the mosquito Anopholes gambiae contains an obvious homolog (agCP14074), with 28% amino acid identity throughout the coding region. The human and mouse genomes have much more distant relatives (BAB14446 and AAH27435, respectively); the homology between the fly and human proteins (13% amino acid identity overall) is mostly restricted to two short domains (Figure 2). Even though this relationship is remote, we demonstrate below that the fly (dZwilch) and human (hZwilch) proteins are likely to be functionally orthologous. Neither protein shows obvious similarities to known proteins or protein motifs.
Figure 2.
Zwilch orthologs in other species. Alignment of Drosophila, mosquito, and human Zwilch amino acid sequences. Boxes indicate amino acids identical between at least two of the sequences. In the zwilch1229 mutant, a stop codon truncates the protein near the C terminus (arrow).
Isolation of a Mutation in the zwilch Gene
We recently screened >1500 stocks containing ethylmethanesulfonate-induced mutations on the Drosophila third chromosome that cause lethality during late larval or pupal stages for aberrations in larval brain mitoses (see MATERIALS AND METHODS). One stock in which the brains displayed many aneuploid cells was particularly interesting because ZW10 and ROD failed to localize to the kinetochores or kMTs in mitotic figures (see below). This observation suggested that the corresponding gene product might be intimately associated with ZW10 and ROD.
Our attempts to map the mitotic mutation were complicated by the fact that the third chromosome in this stock actually contained two lesions, which we were subsequently able to separate by recombination. One mutation caused the recessive late larval/pupal lethality, but the brains of dying third instar larvae homozygous for this mutation did not contain aneuploid cells. The second mutation (mit-1229) caused sterility of homozygous females; more importantly, the brains of homozygous mutant larvae of either sex displayed many aneuploid cells. We mapped mit-1229 to the 100B region by crossing the stock to deletions that covered most of the third chromosome, using female sterility to indicate a lack of complementation. The mit-1229 mutation was delimited to the area between the distal breakpoint of Df(3R)A177der22 and the distal breakpoints of Df(3R)A177.X3, Df(3R)der26, and Df(3R)PGX8 (Zilian et al., 1999). We determined the position of the deletion breakpoints at the molecular level by examining Southern blots of whole genomic DNA from deletion heterozygotes. This analysis located mit-1229 to a 150-kb region of the third chromosome containing 11 predicted genes (FlyBase, 1999; http://flybase.bio.indiana.edu/).
One of these genes is CG18729, which encodes the Zwilch protein. On sequencing CG18729 from the mit-1229 chromosome, we found a nonsense mutation that would shorten the 644 residue wild-type Zwilch protein to 603 amino acids (Figure 2). Truncation of this protein was later verified using anti-Zwilch antibodies in Western blots of mit-1229 larvae (see below). Additional results presented below strengthen the argument that the mutant phenotype indeed results from the nonsense mutation in CG18729. We hereafter call this mutation zwilch1229, though we acknowledge it remains a formal possibility that the mutant phenotype is caused by a lesion in one of the other 10 predicted genes in the region.
The zwilch Mutant Phenotype
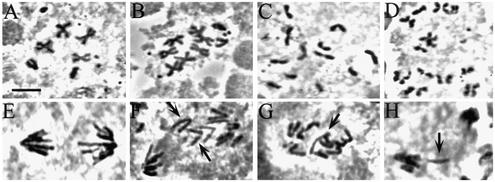
Animals homozygous for zwilch1229 show mitotic and meiotic defects very similar to those previously seen in zw10 and rod mutants. Many larval brain cells are aneuploid (21%; n = 564), and 20% of anaphases (n = 87) show aberrations such as chromosomes that lag at the metaphase plate or an unequal distribution of chromosomes to the two spindle poles (Figure 3). These defects occur even though chromosome congression to the metaphase plate, as well as spindle and chromosome morphology, seem normal (our unpublished data). Thus, just as in zw10 and rod mutants (Williams and Goldberg, 1994; Scaerou et al., 1999), the mitotic problems that produce aneuploidy in zwilch1229 mutants most likely begin at anaphase onset. Chromosome behavior aberrations are not restricted to mitotic divisions: mutant testes show high levels of chromosome missegregation during both meiotic divisions (our unpublished data).
Figure 3.
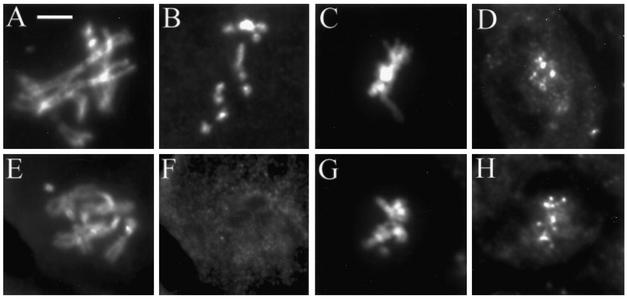
Mitotic defects in the zwilch mutant. Wild-type (A and E) and zwilch mutant (B, C, D, F, G, and H) brains were treated with colchicine and hypotonic solutions (A–D) or left untreated (E and F) and squashed in aceto-orcein. (A) Wild-type metaphase. Note the attachment of sister chromatids at the centromeres. (B) Aneuploid cell with seven autosomes. (C and D) PSCS in the zwilch mutant. (E) Wild-type anaphase. (F–H) Mutant anaphases with lagging chromatids. Bar, 5 μm.
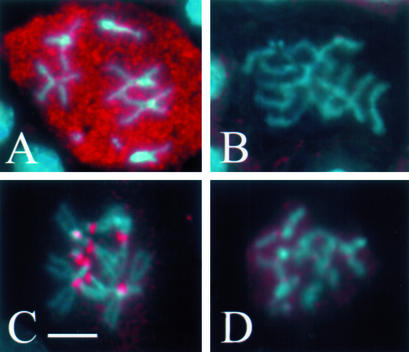
Several criteria indicate that the spindle checkpoint is defective in zwilch1229 mutant mitoses, as is true for zw10 and rod mutants (Smith et al., 1985; Williams et al., 1992; Basto et al., 2000; Chan et al., 2000; Savoian et al., 2000). In wild-type cells with a functioning checkpoint, treatment with microtubule inhibitors such as colchicine produces a metaphase-like arrest in which sister chromatids remain attached and the levels of cyclin B remain elevated (Whitfield et al., 1990; González et al., 1991). But in zwilch1229 colchicine-treated brains, 40% of the mitotic cells (n = 564) display precocious sister chromatid separation (PSCS; Figure 3). Furthermore, cyclin B levels are drastically reduced in cells exhibiting PSCS compared with cells with attached sister chromatids in the same mutant brain (Figure 4). Additional evidence for a checkpoint bypass in zwilch1229 mutants was provided by staining with BUB1, a component of the checkpoint apparatus. In wild type, BUB1 localizes to the kinetochores at prometaphase and metaphase, but exhibits markedly reduced staining on kinetochores during anaphase (Basu et al., 1999). In zwilch1229 mutant cells, BUB1 localizes properly to the kinetochores of chromosomes with attached sister chromatids, yet mutant cells with PSCS have drastically reduced accumulations of BUB1 at the kinetochores, like anaphase in normal, untreated cells (Figure 4). These findings all demonstrate a failure of the spindle checkpoint in zwilch1229 mutants: instead of the expected metaphase arrest, mutant cells treated with colchicine enter anaphase and exit mitosis.
Figure 4.
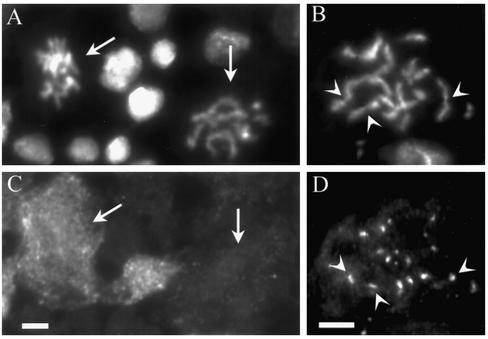
Bypass of the spindle checkpoint in zwilch mutants. Zwilch mutant brains were treated with colchicine (to activate the spindle checkpoint) and stained to visualize cyclin B (A and B) or Bub1 (C and D), both pseudocolored red. Chromosomes were stained with Hoechst (blue). In cells exhibiting PSCS (B and D), levels of cyclin B (B) and Bub1 (D) are drastically reduced. Bar, 5 μm.
It is important to note that classical genetic tests suggest that the zwilch1229 mutation is hypomorphic rather than completely null, because the phenotype of zwilch1229/Df hemizygotes is more severe than that of zwilch1229 homozygotes. The brains of hemizygotes display higher levels of aneuploidy (25%), defective anaphases (25%), and PSCS (52%) than those described above for homozygotes.
Immunochemical Characterization of Zwilch
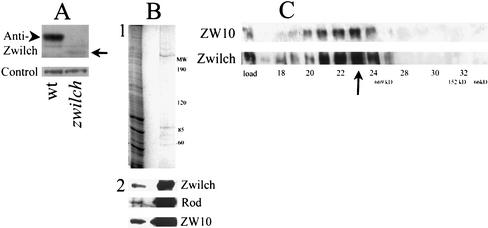
Polyclonal antibodies raised against Zwilch epitopes recognize a band of 75 kDa on Western blots of extracts from wild-type Drosophila larvae (Figure 5A). This protein is prominent in larval brains but is missing in the larval brains from zwilch1229 mutants, where it is replaced by greatly reduced amounts of a 60-kDa band. The size of this truncated protein is exactly that predicted from the position of the nonsense mutation within the mutant gene. Furthermore, using this antibody, we have verified that the Zwilch protein in the final affinity column eluate is indeed the product of CG18729 (Figure 5B).
Figure 5.
Zwilch protein. (A) Anti-Zwilch antibody identifies Zwilch on a Western blot of third instar larvae from wild-type and from zwilch mutants (top). A 70-kDa band in wild-type (arrowhead) is replaced by much lower levels by a 60-kDa product in the mutant (arrow). The amount of protein in each lane was assayed with an unrelated antibody (bottom). (B) Zwilch is purified on the anti-ZW10 column. (1) Coomassie Blue staining of total embryonic extract (left lane) and a low pH elution from the column (right lane). (2) Western blot of the same gel probed with anti-Zwilch, anti-ROD, and anti-ZW10. (C) Zwilch and ZW10 cofractionate on a Superose 6 sizing column. Equal volumes of each fraction were analyzed on a Western blot probed with anti-Zwilch (bottom). The blot was then reprobed with anti-ZW10. A sample of the total extract is indicated in the load lane. The arrow indicates the peak fraction for both Zwilch and ZW10. The void volume was at fraction 13, and the salt front in fraction 42. Standards: 669 kDa, thyroglobulin; 66 kDa, bovine serum albumin; 158 kDa, aldolase.
We next constructed affinity columns using anti-Zwilch to isolate other proteins with which Zwilch interacts. The results obtained were nearly identical to those from the anti-ZW10 column: the final 1.5 M MgCl2 eluate contained Zwilch, ZW10, ROD, and Hsc70-4 in roughly equal stoichiometries. The CENP-meta protein could not be seen by Coomassie Blue staining, but very small amounts were detected on Western blots with anti-CENP-meta (our unpublished data).
We demonstrated previously that ZW10 and ROD cofractionate in a 700- to 900-kDa peak from total embryo extracts analyzed by gel exclusion chromatography (Scaerou et al., 2000). Herein, we show that Zwilch also elutes from the sizing column in exactly the same fractions, consistent with the idea that Zwilch is contained within the same large protein complex as ZW10 and ROD (Figure 5C).
Zwilch Colocalizes with ZW10 on Kinetochores and KMTs
Affinity-purified anti-Zwilch was used in immunofluorescence experiments to examine Zwilch's intracellular localization during mitosis in wild-type brains. Like ZW10 and ROD, Zwilch is found on kinetochores during prometaphase and metaphase, on kMTs during metaphase, and again on kinetochores during anaphase (Figure 6, A–E). We did not detect immunofluorescence signals on kinetochores or kMTs in zwilch1229 mutant brains (Figure 6F). Anti-Zwilch also lightly stains the centrosomes, but because the centrosomal signal is also visible in zwilch1229 mutants, this probably represents a cross-reaction with an unrelated protein.
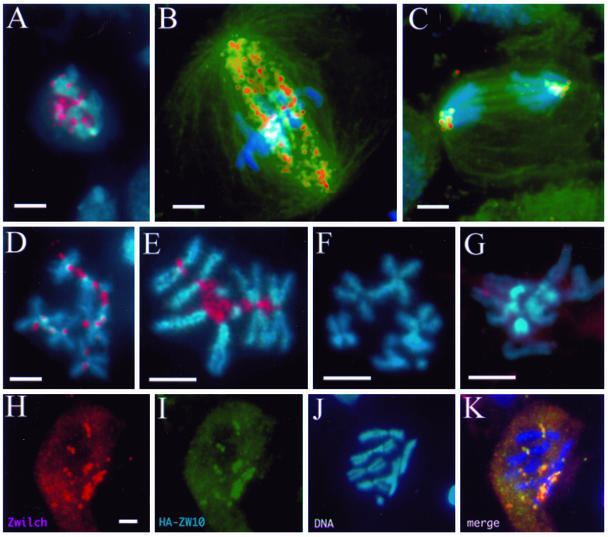
Figure 6.
Zwilch localizes dynamically to kinetochores and kinetochore microtubules. Larval brain neuroblasts were stained for Zwilch (red), chromosomes (blue), and α-tubulin (green; in B and C only). (A–C) Wild-type neuroblasts. At prometaphase (A), Zwilch occupies the kinetochore. At metaphase (B), Zwilch is also present on kinetochore microtubules. Zwilch is seen only at kinetochores during anaphase (C). Zwilch also localizes strongly to kinetochores in cells arrested with colchicine (D and E). In zwilch (F) and rod (G) mutant cells treated with colchicine, Zwilch is not detected at the kinetochore, and is instead diffuse in the cytoplasm. (H–K) Zwilch (H; red) and Zw10 (I; green) colocalize at the kinetochores of the chromosomes (J). The merged image (K) is yellow where Zwilch and ZW10 colocalize. Bars, 5 μm; H–K are at the same magnification.
We next examined whether Zwilch colocalizes with ZW10, as might be predicted from their biochemical association. Because anti-ZW10 and anti-Zwilch were generated from the same species (rabbit), we used rat anti-HA to detect ZW10 in cells expressing an HA-tagged ZW10 protein. This tagged protein rescues the lethality of zw10 mutants, and the pattern of anti-HA staining in larval cells expressing the fusion protein is identical to that seen with antibodies against endogenous ZW10. HA-ZW10 colocalized exactly with Zwilch at the kinetochores (Figure 6, H–K).
Recruitment of Zwilch, ZW10, and ROD to Kinetochores Is Mutually Interdependent
We have previously shown that the targeting of ZW10 to kinetochores requires ROD and that the association of ROD with kinetochores requires ZW10 (Williams and Goldberg, 1994). We now find that Zwilch is similarly indispensable for the recruitment of ZW10 and ROD to kinetochores: we could not detect ZW10 or ROD at the kinetochores in zwilch1229 mutants (Figure 7; our unpublished data). Conversely, ZW10 and ROD are both needed for the kinetochore and kMT localization of Zwilch (Figure 6G; our unpublished data).
Figure 7.
CENP-meta, but not ZW10, is targeted to kinetochores in zwilch mutants. Wild-type (A–D) and zwilch mutant (E–H) brains treated with colchicine and stained to visualize DNA (A, C, E, and G), ZW10 (B and F) or CENP-meta (D and H). Bar, 5 μm.
The failure of ZW10 and ROD to bind kinetochores in zwilch1229 mutants is not due to gross defects in kinetochore structure. The localization of other centromere/kinetochore proteins, such as CID (Blower and Karpen, 2001) and MEI-S332 (Moore et al., 1998), are unaffected in zwilch1229 brains (our unpublished data). Furthermore, even though the spindle checkpoint is defective in mutant cells, checkpoint components such as BUB3 (Basu et al., 1998) and BUB1 (Basu et al., 1999) are still able to associate with kinetochores in zwilch1229 mutants (Figure 4). Besides ZW10 and ROD, cytoplasmic dynein is the only other kinetochore component examined that fails to bind kinetochores in zwilch1229 cells. This latter result is anticipated as ZW10 and ROD are themselves required for the kinetochore recruitment of dynein, probably through an interaction between ZW10 and the p50 subunit of the dynein-associated dynactin complex (Starr et al., 1998).
CENP-meta is Required for the Spindle Checkpoint, but It Is Targeted to Kinetochores Independently of ZW10/ROD/Zwilch
The data presented in Figure 1C suggest that CENP-meta, one of two Drosophila proteins related to mammalian CENP-E, might interact with ZW10, ROD, and Zwilch. This possibility is intriguing because CENP-meta is a known constituent of the kinetochore during mitosis (Yucel et al., 2000). Another argument for such an interaction emerged when we examined mitotic figures in the colchicine-treated larval brains of animals homozygous for a CENP-meta null mutation. We found that, just as in zw10, rod, and zwilch mutants, these larval brains displayed high levels of aneuploidy (∼50% of cells) and PSCS (∼75% of mitotic cells). In the cells with PSCS, cyclin B levels are considerably diminished with respect to cells with attached sister chromatids (Figure 8, A and C). CENP-meta is thus needed for the spindle checkpoint to block sister chromatid separation and mitotic exit when the spindle is disrupted.
Figure 8.
CENP-meta mutant. CENP-meta mutant larval brains were treated with colchicine and stained to visualize chromosomes (A and B), cyclin B (C), or Zwilch (D). In A and C, the left arrow points to a cell whose sister chromatids are still attached, and with high cyclin B levels. A cell in the same field whose sister chromatids are separated (right arrow) has degraded cyclin B. (B and D) In PSCS cells in the CENP-meta mutant, Zwilch localizes to the kinetochores of the separated chromatids (arrowheads). Bar, 5 μm.
Despite these similarities in the behavior of CENP-meta and ZW10/ROD/Zwilch, we subsequently found that the recruitment of CENP-meta to the kinetochore is independent of the other three proteins, and vice versa. For example, Zwilch is able to localize properly to the kinetochore in CENP-meta mutants, even in those cells exhibiting PSCS (Figure 8, B and D). The same is true of CENP-meta in zw10 or zwilch mutants (Figure 7; our unpublished data).
Characterization of Human Zwilch
As shown in Figure 2, the human protein tentatively called hZwilch shares only limited amino acid sequence similarity with Drosophila Zwilch. This tenuous homology brings into question whether they are functionally orthologous. To address this issue, antibodies were raised against hZwilch epitopes. These antibodies identified an ∼70-kDa protein that is approximately the calculated size of the 67-kDa open reading frame (Figure 9A).
Figure 9.
hROD, hZW10, and hZwilch form a complex in vivo. (A) Specificity of anti-hZwilch. Cell lysates made from GFP-hZwilch–transfected HeLa cells (lanes 1 and 2) and untransfected asynchronous (lane 3) or mitotic (lane 4) HeLa cell lysates were probed with anti-GFP (lane 1) and anti-hZwilch (lanes 2–4). GFP-hZwilch occurs as a 96-kDa protein (arrowhead), whereas hZwilch migrates at ∼75 kDa (arrow). (B) Asynchronous (AS) or mitotic (M) HeLa cell lysates were immunoprecipitated with nonimmune IgG, anti-hZwilch, anti-hROD, or anti-hZW10, and then probed with these antibodies on Western blots as indicated.
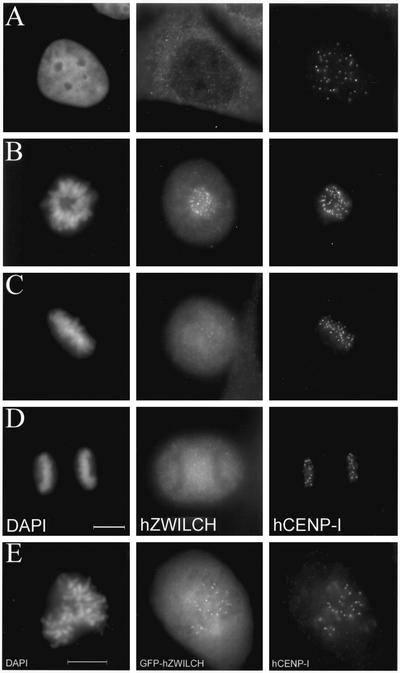
Two observations strongly indicate that the human and fly proteins play closely related, though perhaps not identical, roles. First, hZwilch is targeted to the kinetochores during mitosis in HeLa cells (Figure 10). Immunofluorescence staining with anti-hZwilch reveals that the protein is cytoplasmic in interphase cells, but it is seen only at kinetochores from prophase through metaphase. In contrast with dZwilch, we have not detected any hZwilch signal on kMTs during metaphase nor on kinetochores during anaphase. The localization pattern of hZwilch was independently confirmed by visualizing the distribution of a GFP-hZwilch fusion protein expressed in HeLa cells (Figure 10). We further found that hZwilch forms a biochemical complex with hROD and hZW10: immunoprecipitation with antibodies against any one of these three proteins coimmunoprecipitates the other two proteins (Figure 9B). The combined data strongly suggest that we have identified the human ortholog of dZwilch.
Figure 10.
Localization of hZwilch. (A–D) hZwilch transiently associates with kinetochores in mitosis. Cells were stained simultaneously with 4,6-diamidino-2-phenylindole (column 1), anti-hZwilch (column 2), and anti-hCENP-I (column 3). (A) Interphase. (B) Prometaphase. (C) Metaphase. (D) Anaphase. (E) GFP-hZwilch also transiently localizes at kinetochores in mitosis. At prometaphase, GFP-hZwilch (middle) colocalizes with hCENP-I (right) at the kinetochore. Bars, 10 μm.
DISCUSSION
A Complex Containing ZW10, ROD, and Zwilch Proteins
We describe herein the use of immunoaffinity chromatography to identify proteins tightly associated with Drosophila ZW10. Our results demonstrate the existence of a stable complex including not only ZW10 and ROD but also the newly identified component Zwilch. These three proteins are retained on anti-ZW10 affinity columns through extensive washes and are eluted only under stringent conditions in near equimolar stoichiometries. All three proteins are found in the same high-molecular-weight fractions (∼700–900 kDa) from a sizing column. The three proteins display the same cell cycle-dependent pattern of localization to kinetochores and kMTs. Indeed, ZW10, ROD, and Zwilch are colocalized in all Drosophila cells we have examined, and their kinetochore targeting is completely interdependent. Finally, mutations in the genes encoding these three proteins cause the same syndrome of phenotypes, including high frequencies of aneuploidy, aberrations during anaphase such as lagging chromatids, and disruption of the spindle checkpoint.
This multisubunit complex and its functions are conserved in humans and perhaps all metazoan organisms. Injection of antibodies against hZW10 and hROD into HeLa cells causes both mitotic abnormalities and a bypass of a nocodazole-induced mitotic block (Chan et al., 2000), the same phenotypes associated with mutations in fly zw10, rod, and zwilch. We have previously demonstrated a biochemical association between hZW10 and hROD (Chan et al., 2000; Scaerou et al., 2001) and now show that hZwilch is part of the same complex (Figure 9). The three human proteins are recruited to the kinetochores early in prophase, where they colocalize through metaphase (Figure 10; Scaerou et al., 2001). However, the fate of these three proteins may then diverge subsequent to anaphase onset. We have been unable to visualize hZwilch on the kMTs during metaphase, in contrast with hZW10 or hROD. Similarly, we have not observed hZwilch on kinetochores during anaphase, though hZW10 and hROD clearly bind kinetochores at least early in anaphase. Our previous studies offer a precedent for a divergence in the behaviors of these proteins late in mitosis: hROD accumulates at the spindle poles in late anaphase and telophase, when hZW10 is primarily at the spindle midzone. These differences might be explained by problems due to epitope masking or differences in signal-to-noise ratios, but the possibility that the three proteins play dissimilar roles late in mitosis remains intriguing. These considerations also raise the possibility that the complex we have characterized exists in different forms at different stages of the cell cycle.
The Zwilch Protein and zwilch Gene
The Zwilch amino acid sequence has been less conserved during evolution than that of ZW10 or ROD. The homologies between dZwilch and hZwilch are modest and are mostly restricted to two small domains. Even the two dipteran proteins, fly and mosquito Zwilch, show considerable divergence. This lack of conservation makes it difficult to detect Zwilch homologs even in organisms with completely sequenced genomes. For example, multiple reiterations of PSI-BLAST (National Center for Biotechnology Information) are required to reveal a putative C. elegans Zwilch, the hypothetical protein Y39G10AR.2. Computerized searches have not uncovered any candidate Zwilch homolog in Arabidopsis thaliana, although we suspect such a protein must exist. In any event, it will be of particular future interest to identify proteins that might associate with the two short Zwilch regions that are most conserved.
We did not anticipate that animals homozygous for the zwilch1229 mutation would be viable. This was surprising because all known zw10 and rod alleles cause substantial lethality; indeed, the genetic screen that uncovered zwilch1229 was searched for recessive mutations causing late larval or pupal lethality. (The only reason we could identify zwilch1229 was the fortuitous occurrence of a recessive lethal mutation elsewhere on the same mutagenized chromosome.) The simplest explanation is that the zwilch1229 allele is not null, but instead retains residual activity that allows development into the adult stage. In support of this idea, zwilch1229 behaves genetically as a hypomorph rather than a null, and reduced levels of truncated protein are seen in zwilch1229 homozygotes. Also consistent with this hypothesis, we sporadically see anti-ZW10 or anti-ROD staining slightly above background levels at kinetochores in zwilch1229 mutant cells. The viability of zwilch1229 animals is foreshadowed by previous reports that balanced stocks carrying null zw10 or rod alleles produce rare homozygous mutant adult escapers that are sterile and die soon after eclosion (Karess and Glover, 1989; Williams et al., 1992). We presume that the proportion of aneuploid cells in these escapers permits adult viability but not fertility; the more robust viability of zwilch1229 homozygotes would be explained by a slightly lower incidence of aneuploidy.
Does the Complex Contain Other Components?
The 700- to 900-kDa ZW10/ROD/Zwilch complex is considerably larger than the sum of the three components' masses (85 + 240 + 72 = 397 kDa). Perhaps the complex contains two molecules of each protein, consistent with their apparent equimolar stoichiometry, but it might instead contain other proteins. Some putative constituents might bind less tightly to the complex; these could have eluted from the affinity column with 1 M KCl and have been lost in the nonspecific background in these fractions (Figure 1B). We could nonetheless identify one protein, cytoplasmic dynein, in the 1 M KCl eluate. Considerable biochemical and genetic evidence indicates that cytoplasmic dynein is unlikely to be a core component of the ZW10/ROD/Zwilch complex, but this result was not unexpected. ZW10 and ROD are needed to target cytoplasmic dynein to the kinetochore (Starr et al., 1998); this requirement is indirect and is probably mediated through an interaction between ZW10 and the p50 subunit of dynactin complex. We did not see p50 or other dynactin components in the 1 M KCl fractions. We presume these proteins were present in insufficient quantities to form discrete bands against the background; unfortunately, no antibodies to these Drosophila proteins are available to probe the samples on Western blots.
More stringent elutions (1.5 M MgCl2) suggest two other possible components of the ZW10/ROD/Zwilch complex. The chaperone Hsc70-4 might conceivably stabilize the complex. However, because Hsc70-4 is so abundant in the original embryo extract, and because we and others (Papoulas et al., 1998) have detected Hsc70-4 in corresponding eluates from affinity columns with antibodies to several other Drosophila proteins, we are not convinced that Hsc70-4 is an integral part of the complex. The second candidate in these fractions is CENP-meta, a kinesin-like protein found at kinetochores that is closely related to mammalian CENP-E. Although there are similarities in the behavior of CENP-meta and ZW10/ROD/Zwilch, strong arguments weigh against CENP-meta's participation in the complex. First, much less CENP-meta is found in the 1 M MgCl2 eluate than the other three proteins. Second, the mitotic phenotype of mutations in the CENP-meta gene is distinguishable from those for the other three genes. CENP-meta mutations result in incomplete chromosome congression to the metaphase plate (Yucel et al., 2000), a phenotype not seen in zwilch, zw10, or rod mutants. Finally, and most importantly, CENP-meta associates with kinetochores in the absence of ZW10, ROD, or Zwilch proteins and vice versa.
In summary, we see no compelling arguments in favor of subunits of the complex beyond ZW10, ROD, and Zwilch, but our data do not allow us to exclude this possibility. Much work remains to characterize the complex and to establish which functions of the complex each protein performs.
The Spindle Checkpoint and the Kinetochore Assembly Pathway
Our data point to at least three relatively independent branches for the assembly of the Drosophila kinetochore: one branch includes ZW10, ROD, and Zwilch; the second BUBR1 and BUB3; and the third CENP-meta. It remains possible, however, that CENP-meta's recruitment to the kinetochore depends on the BUB/MAD proteins, as is the case for CENP-E (the vertebrate homolog of CENP-meta), whose kinetochore targeting requires BubR1 (Chen, 2002).
It is striking that cells mutant for zw10, rod, zwilch, and CENP-meta all display defects in the spindle checkpoint normally activated by the addition of microtubule poisons. This phenotype is similar to that seen for mutants in canonical checkpoint genes such as bub1 and bub3. Although the exact nature of the checkpoint signal emitted by improperly attached kinetochores is still unknown, it is generally accepted that the canonical BUB and MAD proteins are intimately involved in the elaboration of this signal. The roles of the other kinetochore proteins in the checkpoint are much less clear, as illustrated by the fact that Xenopus extracts depleted for CENP-E (the homolog of CENP-meta) are defective in the checkpoint (Abrieu et al., 2000), whereas suppression of CENP-E synthesis in mammalian cells paradoxically activates the checkpoint (Yao et al. 2000). The ZW10/ROD/Zwilch complex and CENP-meta could provide independent inputs that inhibit the anaphase promoting complex (APC). Alternatively, these proteins might act through the BUB and MAD proteins. If true, this influence must be at some level separate from how these proteins assemble onto the kinetochore, because BUBR1 and BUB3 can associate with kinetochores in cells lacking the other proteins.
It has been proposed that the ZW10/ROD complex inactivates the checkpoint through dynein, which is known to carry Mad2 away from the kinetochore and thus silence the “wait anaphase” signal (Howell et al., 2001; Wojcik et al., 2001). If this model were correct then one would predict that the loss of dynein from kinetochores lacking ZW10, ROD, and Zwilch would prevent Mad2 from dissociating from kinetochores, so the checkpoint could not be turned off. However, in both Drosophila and human cells, depletion of ZW10 and ROD has the opposite effect: the checkpoint is actually inactivated. Furthermore, kinetochores in HeLa cells depleted of hZW10, hROD, and dynein retained other checkpoint proteins that could respond normally to microtubule interactions. Even in the absence of dynein, kinetochores that established bipolar attachments were selectively depleted of Mad1, Mad2, and other checkpoint proteins (Chan et al., 2000). We therefore believe that ZW10/ROD/Zwilch is likely to play a more direct role in the checkpoint independent from its function in targeting dynein to the kinetochores. Further characterization of the ZW10/ROD/Zwilch complex will be needed to clarify whether it acts in parallel or in conjunction with the canonical checkpoint proteins.
ACKNOWLEDGMENTS
We thank William Sullivan, Kristina Yu, and Doug Kellogg (University of California, Santa Cruz) for providing embryos and technical advice for the affinity chromatography; and Jim Kerwin (Cornell University, Ithaca, NY) for assisting with MALDI analysis. We also thank the following for generously supplying antibodies for use in these studies: Claudio Sunkel (Universidade do Porto, Porto, Portugal), Christian Lehner (University of Bayreuth, Bayreuth, Germany), and Don Cleveland (University of California, San Diego, La Jolla, CA). Charles Zuker and Edmund Koundakjian (University of California, San Diego) were very generous in sharing collection of mutagenized Drosophila stocks. Drosophila deficiency stocks were received from Erich Frei (University of Zurich). We thank J. Hittle, B. Conner, and the Lab Animal Facility at FCCC for excellent technical support. This work was supported by National Institutes of Health grant GM-48430 (to M.L.G.); work in the laboratory of T.J.Y. was supported by National Institutes of health grant GM-44762, Core Grant 06927, and an Appropriation from the Commonwealth of Pennsylvania. S.T.L. was supported by the Greenwald Fellowship.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–09–0624. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–09–0624.
REFERENCES
- Abrieu A, Kahana JA, Wood KW, Cleveland DW. CENP-E as an essential component of the mitotic checkpoint in vitro. Cell. 2000;102:817–826. doi: 10.1016/s0092-8674(00)00070-2. [DOI] [PubMed] [Google Scholar]
- Amon A. The spindle checkpoint. Curr Opin Genet Dev. 1999;9:69–75. doi: 10.1016/s0959-437x(99)80010-0. [DOI] [PubMed] [Google Scholar]
- Basto R, Gomes R, Karess RE. Rough Deal and ZW10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol. 2000;2:939–943. doi: 10.1038/35046592. [DOI] [PubMed] [Google Scholar]
- Basu J, Bousbaa H, Logarinho E, Li Z, Williams BC, Lopes C, Sunkel CE, Goldberg ML. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu J, Logarinho E, Herrmann S, Bousbaa H, Li Z, Chan GK, Yen TJ, Sunkel CE, Goldberg ML. Localization of the Drosophila checkpoint control protein Bub3 to the kinetochore requires Bub1 but not ZW10 or Rod. Chromosoma. 1998;107:376–385. doi: 10.1007/s004120050321. [DOI] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. Role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan GKT, Jablonski SA, Starr DA, Goldberg ML, Yen TJ. Human ZW10 and Rod kinetochore proteins are novel components of the mitotic checkpoint. Nat Cell Biol. 2000;2:944–947. doi: 10.1038/35046598. [DOI] [PubMed] [Google Scholar]
- Chan GK, Jablonski SA, Sudakin V, Hittle JC, Yen TJ. Human BUBR1 is a mitotic checkpoint kinase that monitors CENP-E functions at kinetochores and binds the cyclosome/APC. J Cell Biol. 1999;146:941–954. doi: 10.1083/jcb.146.5.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J Cell Biol. 2002;158:487–496. doi: 10.1083/jcb.200204048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefant F, Palter KB. Tissue-specific expression of dominant negative mutant Drosophila HSC70 causes developmental defects and lethality. Mol Biol Cell. 1999;10:2101–2117. doi: 10.1091/mbc.10.7.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FlyBase. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/27.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González C, Casal-Jimenez J, Ripoll P, Sunkel CE. The spindle is required for the process of sister chromatid separation in Drosophila neuroblasts [published erratum appears in Exp. Cell Res. (1991) 193, 438] Exp Cell Res. 1991;192:10–15. doi: 10.1016/0014-4827(91)90150-s. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, Hoffman DB, Farrar EM, Rieder CL, Salmon ED. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Jablonski SA, Chan GK, Cooke CA, Earnshaw WC, Yen TJ. The hBUB1 and hBUBR1 kinases sequentially assemble onto kinetochores during prophase with hBUBR1 concentrating at the kinetochore plates in mitosis. Chromosoma. 1998;107:386–396. doi: 10.1007/s004120050322. [DOI] [PubMed] [Google Scholar]
- Karess RE, Glover DM. rough deal: a gene required for proper mitotic segregation in Drosophila. J Cell Biol. 1989;109:2951–2961. doi: 10.1083/jcb.109.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg DR, Alberts BM. Purification of a multiprotein complex containing centrosomal proteins from the Drosophila embryo by chromatography with low-affinity polyclonal antibodies. Mol Biol Cell. 1992;3:1–11. doi: 10.1091/mbc.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. Science. 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Liu, S.T., Hittle, J.C., Jablonski, S.A., Campbell, M.S., Yoda, K., and Yen, T.J. (2003). Human CENP-I specifies localization of CENP-F, MAD1, and MAD2 to kinetochores and is essential for accurate mitosis. Nature Cell Biol. (in press). [DOI] [PubMed]
- Martinez-Exposito MJ, Kaplan KB, Copeland J, Sorger PK. Retention of the BUB3 checkpoint protein on lagging chromosomes. Proc Natl Acad Sci USA. 1999;96:8493–8498. doi: 10.1073/pnas.96.15.8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP, Page AW, Tang TT, Kerrebrock AW, Orr-Weaver TL. The cohesion protein MEI-S332 localizes to condensed meiotic and mitotic centromeres until sister chromatids separate. J Cell Biol. 1998;140:1003–1012. doi: 10.1083/jcb.140.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Beek SJ, Moseley SL, McCallum CM, Sarte M, Shearn A, Tamkun JW. The Drosophila trithorax group proteins BRM, ASH1 and ASH2 are subunits of distinct protein complexes. Development. 1998;125:3955–3966. doi: 10.1242/dev.125.20.3955. [DOI] [PubMed] [Google Scholar]
- Pasqualone D, Huffaker TC. STU1: a suppressor of alpha-tubulin mutation, encodes a novel and essential component of the yeast mitotic spindle. J Cell Biol. 1994;127:1973–1984. doi: 10.1083/jcb.127.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savoian MS, Goldberg ML, Riederm CL. The rate of poleward chromosome movement is attentuated in Drosophila zw10 and rod mutants. Nat Cell Biol. 2000;2:948–952. doi: 10.1038/35046605. [DOI] [PubMed] [Google Scholar]
- Scaerou F, Aguilera I, Saunders R, Kane N, Blottiere L, Karess R. The rough deal protein is a new kinetochore component required for accurate chromosome segregation in Drosophila. J Cell Sci. 1999;112:3757–3768. doi: 10.1242/jcs.112.21.3757. [DOI] [PubMed] [Google Scholar]
- Scaerou F, Starr DA, Piano F, Papoulas O, Karess RE, Goldberg ML. The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J Cell Sci. 2001;114:3103–3114. doi: 10.1242/jcs.114.17.3103. [DOI] [PubMed] [Google Scholar]
- Smith DA, Baker BS, Gatti M. Mutations in genes controlling essential mitotic functions in Drosophila melanogaster. Genetics. 1985;110:647–670. doi: 10.1093/genetics/110.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Hays TS, Goldberg ML. ZW10 helps recruit dynactin and dynein to the kinetochore. J Cell Biol. 1998;142:763–774. doi: 10.1083/jcb.142.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Williams BC, Li Z, Etemad-Moghadam B, Dawe RK, Goldberg ML. Conservation of the centromere/kinetochore protein ZW10. J Cell Biol. 1997;138:1289–1301. doi: 10.1083/jcb.138.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Ha E, McKeon F. The human homologue of Bub3 is required for kinetochore localization of Bub1 and a Mad3/Bub1-related protein kinase. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters JC, Chen RH, Murray AW, Salmon ED. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield WGF, González C, Maldonado-Codina G, Glover DM. The A and B type cyclins are accumulated and destroyed in temporally distinct events that define separable phases of the G2/M transition. EMBO J. 1990;9:2563–2572. doi: 10.1002/j.1460-2075.1990.tb07437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Gatti M, Goldberg ML. Bipolar spindle attachments affect redistributions of ZW10, a Drosophila centromere/kinetochore component required for accurate chromosome segregation. J Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Goldberg ML. Determinants of Drosophila zw10 protein localization and function. J Cell Sci. 1994;107:785–798. doi: 10.1242/jcs.107.4.785. [DOI] [PubMed] [Google Scholar]
- Williams BC, Karr TL, Montgomery JM, Goldberg ML. The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, K.R., and Stone, K.L. (1995). In gel digestion of SDS-PAGE-separated proteins: observations from internal sequencing of 25 proteins. In: Techniques in Protein Chemistry VI, ed. Crabb, J.W. Orlando, FL: Academic Press Inc., 143–152.
- Wojcik E, Basto R, Serr M, Scaerou F, Karess R, Hays RT. Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol. 2001;3:1001–1007. doi: 10.1038/ncb1101-1001. [DOI] [PubMed] [Google Scholar]
- Yao X, Abrieu A, Zheng Y, Sullivan K, Cleveland DW. CENP-E forms a link between attachment of spindle microtubules to kinetochores and the mitotic checkpoint. Nat Cell Biol. 2000;2:484–491. doi: 10.1038/35019518. [DOI] [PubMed] [Google Scholar]
- Yucel J, Marszalek J, McIntosh J, Goldstein L, Cleveland D, Philp A. CENP-meta, an essential kinetochore kinesin required for the maintenance of metaphase chromosome alignment in Drosophila. J Cell Biol. 2000;150:1–11. doi: 10.1083/jcb.150.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]
- Zilian O, Frei E, Burke R, Brentrup D, Gutjahr T, Bryant PJ, Noll M. double-time is identical to discs overgrown, which is required for cell survival, proliferation and growth arrest in Drosophila imaginal disc. Development. 1999;126:5409–5420. doi: 10.1242/dev.126.23.5409. [DOI] [PubMed] [Google Scholar]