Abstract
Jak tyrosine kinases have a unique domain structure containing a kinase domain (JH1) adjacent to a catalytically inactive pseudokinase domain (JH2). JH2 is crucial for inhibition of basal Jak activity, but the mechanism of this regulation has remained elusive. We show that JH2 negatively regulated Jak2 in bacterial cells, indicating that regulation is an intrinsic property of Jak2. JH2 suppressed basal Jak2 activity by lowering the Vmax of Jak2, whereas JH2 did not affect the Km of Jak2 for a peptide substrate. Three inhibitory regions (IR1–3) within JH2 were identified. IR3 (residues 758–807), at the C terminus of JH2, directly inhibited JH1, suggesting an inhibitory interaction between IR3 and JH1. Molecular modeling of JH2 showed that IR3 could form a stable α-helical fold, supporting that IR3 could independently inhibit JH1. IR2 (725–757) in the C-terminal lobe of JH2, and IR1 (619–670), extending from the N-terminal to the C-terminal lobe, enhanced IR3-mediated inhibition of JH1. Disruption of IR3 either by mutations or a small deletion increased basal Jak2 activity, but abolished interferon-γ–inducible signaling. Together, the results provide evidence for autoinhibition of a Jak family kinase and identify JH2 regions important for autoregulation of Jak2.
INTRODUCTION
Protein tyrosine kinases (PTKs) are central mediators of intracellular signaling pathways. Jak2 is a member of the Janus (Jak) family of nonreceptor PTKs (Jak1, Jak2, Jak3, and Tyk2) and is crucial in signaling through multiple cytokine receptors, such as erythropoietin, interleukin-3, and interferon-γ (IFN-γ) receptors (Ihle et al., 1995). Lack of Jak2 causes abrogation of definitive erythropoiesis and embryonic lethality in mice (Neubauer et al., 1998; Parganas et al., 1998).
Jak activation is achieved through cytokine-induced aggregation of receptor chains, leading to reciprocal interaction of receptor-associated Jaks and subsequent phosphorylation of tyrosine residues in the kinase activation loop (A-loop) of Jaks (Gauzzi et al., 1996; Feng et al., 1997; Liu et al., 1997; Zhou et al., 1997). The activated Jak kinases phosphorylate cytokine receptors as well as downstream signaling proteins such as transcription factors called signal transducers and activators of transcription, STATs (Darnell et al., 1994). The activity of Jak2 is strictly controlled by several mechanisms, including protein tyrosine phosphatases, such as SHP-1, PTP1B, and CD45, and suppressors of cytokine signaling proteins that bind Jak2, inhibit its catalytic activity, and promote proteosome-mediated degradation of Jak2 (Klingmuller et al., 1995; Yasukawa et al., 1999; Myers et al., 2001; Irie-Sasaki et al., 2001; Ungureanu et al., 2002). Jak activation induced by cytokine receptors is generally rapid and transient, but constitutive Jak2 activity is observed in a variety of cancers. For example, a chromosomal translocation creating a fusion protein between Jak2 and the dimerization domain of the TEL transcription factor produced a constitutively active Jak2 and lead to acute lymphoblastic leukemia (Lacronique et al., 1997). The critical role of Jaks in cytokine-induced signaling pathways, and their potential role in tumorigenesis, make it important to understand the mechanisms of Jak regulation.
The Jak kinases have a unique domain organization among PTKs in containing two nonidentical kinase domains: adjacent to a C-terminal kinase domain (Jak homology 1 domain, JH1) is a catalytically inactive pseudokinase domain (JH2) (Ziemiecki et al., 1994). The N-terminal Src homology 2 (SH2)-like domain (JH3–4) is followed by a FERM domain (JH4–7) required for cytokine receptor association (Higgins et al., 1996; Al-Lazikani et al., 2001). The JH2 domain shares conserved motifs of protein kinases; however, especially the active site and activation loop regions are modified. The modifications are conserved between Jaks, suggesting an important function for JH2. Indeed, several lines of evidence indicate a crucial role for JH2 in the regulation of Jak activity. In Drosophila Jak homolog Hop, a mutation in the JH2 domain was found to produce a hyperactive kinase and cause hematopoietic neoplasia in the fly, indicative of a negative regulatory role for JH2 (Luo et al., 1997). We also found that JH2 negatively regulated Jak2 and Jak3, and the deletion of JH2 resulted in constitutive Stat signaling (Saharinen et al., 2000; Saharinen and Silvennoinen, 2003). However, in the absence of JH2, the activity of Jak2 and Jak3 deletion mutants could not be induced by cytokines. Thus, the JH2 domain was found to have a dual function: JH2 is required for suppressing basal Jak activity in the absence of cytokine stimulation, and for rendering Jaks competent to respond to cytokine stimulation with increased Jak activity (Saharinen and Silvennoinen, 2003). In line with this, mutations in the Jak3 JH2 domain were found to impair Jak3 signaling, leading to severe combined immunodeficiency, although the Jak3 mutants were constitutively highly phosphorylated (Candotti et al., 1997; Chen et al., 2000). Similarly, mutations in the JH2 domain of Tyk2 were found to result in constitutive Tyk2 phosphorylation, but abrogation of IFN-α signaling (Velazquez et al., 1995; Yeh et al., 2000).
The mechanism by which the JH2 domain regulates Jak kinases has remained poorly understood. The activity of many nonreceptor PTKs is negatively regulated through intramolecular domain–domain interactions (Hubbard et al., 1998). Coexpression experiments have suggested that negative regulation of Jak activity is based on JH1–JH2 interaction, but the involvement of additional regulatory proteins has not been excluded (Chen et al., 2000; Saharinen et al., 2000). Furthermore, no systematic analysis of JH2 regions required for Jak regulation has been undertaken. We present evidence that the JH2-mediated inhibition of basal activity is an intrinsic property of Jak2, and not dependent on additional regulatory proteins. We found that JH2 suppresses the basal activity of Jak2 mainly by lowering the Vmax of Jak2, while leaving the Km for a peptide substrate relatively unchanged. We identify three regions within JH2 that are important for autoinhibition of Jak2, and use molecular modeling of the JH2 domain to gain insight how these regions might regulate Jak2. Finally, we propose a model for the function of JH2 in regulation of Jak2 activity in cytokine receptor complexes.
MATERIALS AND METHODS
Reagents, Cell Culture, and Transfections
293T (American Type Culture Collection, Manassas, VA) and γ2A (Jak2-deficient fibrosarcoma; Watling et al., 1993) cells were grown in DMEM supplemented with 10% fetal bovine serum (Invitrogen, Paisley, United Kingdom) and antibiotics. The cells were stimulated with IFN-γ (R & D Systems, Minneapolis, MN) and transfected using the FuGENE6 transfection reagent (Roche Diagnostics, Indianapolis, IN) according to manufacturer's instructions. Depending on the experiment, 0.5–5 μg of specific cDNAs was used to transfect 60% confluent 10-cm plates of 293T cells, and 100 ng of specific cDNA was used for transfection of a six-well plate well of γ2A cells. The amount of each cDNA transfected was adjusted within a single experiment to obtain similar expression levels of the different cDNA constructs verified by immunoblotting. The cells were harvested 72 h after transfection for immunoprecipitation and after 20 h for luciferase assay. The following antibodies were used: anti-phosphotyrosine (4G10; Upstate Biotechnology, Lake Placid, NY), anti-hemagglutinin (anti-HA, 16B12; Covance, Princeton, NJ), and anti-Stat5 (ST5a-2H2; Zymed Laboratories, South San Francisco, CA).
DNA Constructs
The amino acids encoded by the Jak2 constructs are shown in Figures 4A, 5A, 6A, and 7A. The numbering refers to mouse Jak2 (GenBank accession L16956) sequence. Expression vectors for Jak2, JH2Δ-Jak2, AflIIΔ-Jak2, JH1–2-Jak2, and JH1-Jak2 have been described previously and they contain an HA tag in their C terminus (Saharinen et al., 2000). JH1-Jak2 and JH1–2-Jak2 were further cloned into pGEX-4T-1 (Amersham Biosciences AB, Uppsala, Sweden), creating HA-tagged glutathione S-transferase (Gst) fusion proteins of the tyrosine kinase and the double kinase domains of Jak2. JH1-HA was also cloned in pEF-BOS expression vector for use in this study (Mizushima and Nagata, 1990). BglIIΔ-Jak2 has been described previously (Kohlhuber et al., 1997), and in this study an HA tag was added to the C terminus of BglIIΔ-Jak2. 761Δ-Jak2 was cloned using recombinant polymerase chain reaction (PCR) into pCIneo (Promega, Madison, WI), creating a deletion construct lacking amino acids 762–774 of the full-length Jak2. New translation initiation codons were introduced by PCR into HA-tagged Jak2 to create 758-Jak2 (pEF-BOS), 725-Jak2 (pEF-BOS), 671-Jak2 (pEF-BOS), 619-Jak2 (pCIneo), and 584-Jak2 (pCIneo), where the first amino acid is numbered according to its location in the full-length Jak2. Alanine mutations were created into the 758-Jak2 construct by using PCR. The protein domains were localized with the Smart program (Schultz et al., 1998). All PCR products were confirmed by sequencing (Applied Biosystems, Foster City, CA). Expression vector for Stat5A was a kind gift from Dr. Tim Wood (Karolinska Institute, Stockholm, Sweden). Luciferase reporter construct containing the Stat1 binding site from the promoter of the IRF-1 gene was a kind gift from Dr. Richard Pine (Pine et al., 1994).
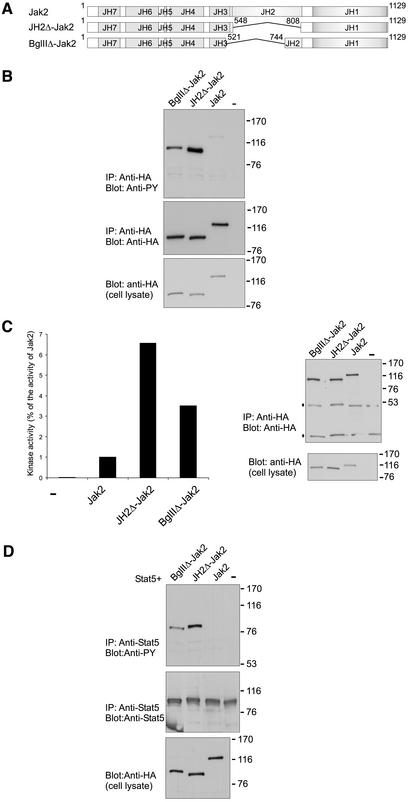
Figure 4.
Comparison of the effect of two different JH2 deletions on the activity of Jak2. (A) Schematic presentation of Jak2 constructs. (B) Expression plasmids for Jak2-HA, JH2Δ-HA, and BglIIΔ-HA were transfected into 293T cells and cell lysates were immunoprecipitated using anti-HA antibody. Aliquots of the immunoprecipitates were separated in 7.5% SDS-PAGE and analyzed in anti-phosphotyrosine (top) and anti-HA immunoblots (middle). Aliquots of cell lysates were separated in 7.5% SDS-PAGE and analyzed in anti-HA immunoblot (bottom). (C) Expression plasmids for Jak2-HA, JH2Δ-HA, and BglIIΔ-HA were transfected into 293T cells, and cell lysates were immunoprecipitated using anti-HA antibody. Aliquots of the immunoprecipitates were subjected to in vitro kinase assay by using [γ-32P]ATP and Stat5-derived peptide as a substrate. The peptides were separated in 20% SDS-PAGE followed by quantification using PhosphorImager. Aliquots of the immunoprecipitates (top) and cell lysates (bottom) were also separated in 7.5% SDS-PAGE and analyzed in anti-HA immunoblot. (D) Stat5 expression plasmid was transfected either alone or together with expression plasmids for Jak2-HA, JH2Δ-HA, and BglIIΔ-HA into 293T cells, and cell lysates were immunoprecipitated using anti-Stat5 antibody. Aliquots of the immunoprecipitates were separated in 7.5% SDS-PAGE and analyzed in anti-phosphotyrosine (top) and anti-Stat5 (middle) immunoblots. Cell lysates were separated in 7.5% SDS-PAGE and analyzed by immunoblotting with anti-HA antibody (bottom). The mobilities of the molecular mass markers (in kilodaltons) are shown on the right.
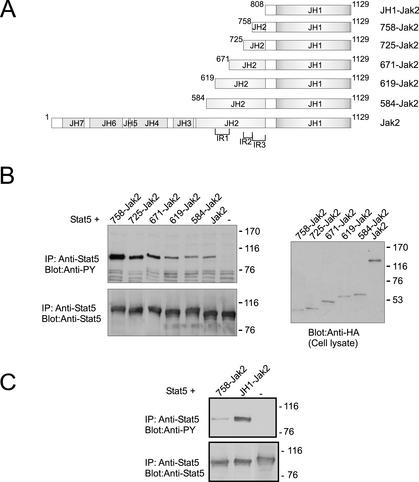
Figure 5.
Mapping of the inhibitory region in JH2. (A) Schematic presentation of Jak2 constructs. (B) Stat5 expression plasmid was transfected into 293T cells either alone or together with HA-tagged Jak2, 584-Jak2, 619-Jak2, 671-Jak2, 725-Jak2, or 758-Jak2, as indicated. Cell lysates were immunoprecipitated using anti-Stat5 antibody, and immunoprecipitates were separated in 7.5% SDS-PAGE followed by immunoblotting with anti-phosphotyrosine (top) and anti-Stat5 antibodies (bottom). Cell lysates were separated in 4–15% SDS-PAGE and analyzed by immunoblotting with anti-HA antibody (right). (C) Stat5 expression plasmid was transfected into 293T cells either alone or together with HA-tagged JH1-Jak2 or 758-Jak2. Cell lysates were immunoprecipitated using anti-Stat5 antibody, and immunoprecipitates were separated in 7.5% SDS-PAGE followed by immunoblotting with anti-phosphotyrosine (top) and anti-Stat5 (bottom) antibodies. The mobilities of the molecular mass markers (in kilodaltons) are shown on the right.
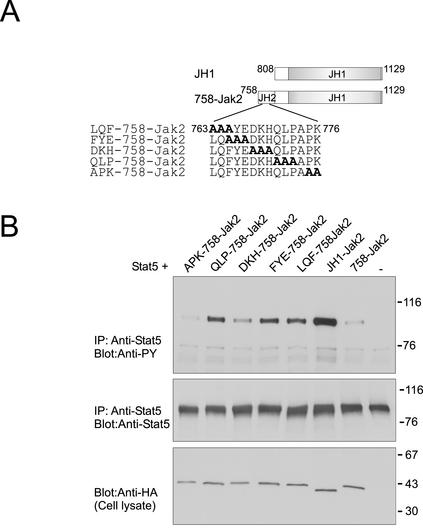
Figure 6.
Mutations in IR3 increase the activity of Jak2. (A) Schematic presentation of Jak2 constructs. (B) Stat5 expression plasmid was transfected into 293T cells either alone or together with HA-tagged JH1-Jak2, 758-Jak2, LQF-758-Jak2, FYE-758-Jak2, DKH-758-Jak2, QLP-758-Jak2, or APK-758-Jak2, as indicated. Cell lysates were immunoprecipitated using anti-Stat5 antibody, and immunoprecipitates were separated in 7.5% SDS-PAGE followed by immunoblotting with anti-phosphotyrosine (top) and anti-Stat5 antibodies (middle). Cell lysates were separated in 4–15% SDS-PAGE and analyzed by immunoblotting with anti-HA antibody (bottom). The mobilities of the molecular mass markers (in kilodaltons) are shown on the right.
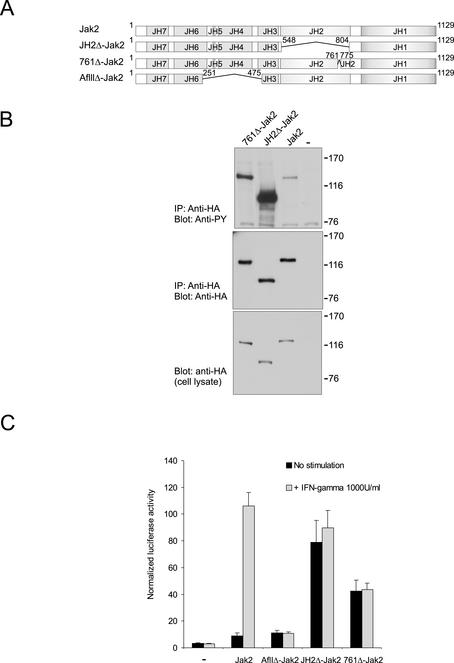
Figure 7.
IR3-deletion results in increased basal Jak2 activity and lack of cytokine-inducible signaling. (A) Schematic presentation of Jak2 constructs. (B) Expression plasmids for Jak2-HA, JH2Δ-HA, and 761Δ-HA were transfected into 293T cells, and cell lysates were immunoprecipitated using anti-HA antibody. Aliquots of the immunoprecipitates were separated in 7.5% SDS-PAGE and analyzed in anti-phosphotyrosine (top) and anti-HA immunoblots (middle). Aliquots of cell lysates were separated in 7.5% SDS-PAGE and analyzed in anti-HA immunoblot (bottom). (C) γ2A cells were transfected with Stat1-dependent luciferase reporter vector, pRLTK control vector, and Jak2, JH2Δ-Jak2, AflIIΔ-Jak2, or 761Δ-Jak2 constructs or with empty vector as a control. Five hours after transfection, the cells were changed into serum-free medium and starved for 15 h. The cells were stimulated with IFN-γ (1000 U/ml) for 5 h or left unstimulated. Luciferase activity was measured as described in MATERIALS AND METHODS. Shown is the mean from three independent experiments and the SEs of the mean.
Cell Lysis, Immunoprecipitation, and Immunoblotting
Cells were lysed in kinase lysis buffer (10 mM Tris-HCl, pH 7.5, 1% Triton X-100, 20% glycerol, 5 mM EDTA, 50 mM NaCl, 50 mM NaF, 1 mM Na3VO4) supplemented with protease inhibitors. Equal amounts of protein from cell lysates were always used for immunoprecipitations and Western blotting of cell lysates. Protein concentrations were determined using the protein assay system from Bio-Rad (Hercules, CA). The immunoprecipitation protocol has been described previously (Saharinen et al., 1997). The immunoprecipitates were subjected to Western blotting or used for kinase assay. The immunoprecipitates and cell lysates were separated in SDS-PAGE (Ready Gels; Bio-Rad) and transferred to nitrocellulose membrane. Immunodetection was performed using specific primary antibodies, biotinylated anti-mouse or anti-rabbit secondary antibodies (DAKO A/S, Glostrup, Denmark) and streptavidin-biotin horseradish peroxidase-conjugate (Amersham Biosciences AB) followed by enhanced chemiluminescence.
Kinase and Luciferase Assays
For kinase assay, the immunoprecipitates were washed four times with kinase lysis buffer and twice with kinase assay buffer (10 mM HEPES, pH 7.4, 50 mM NaCl, 5 mM MgCl2, 5 mM MnCl2, 50 mM NaF, 0.1 mM Na3VO4). The immunoprecipitates were suspended in kinase assay buffer containing dithiothreitol (1 mM). The Stat5 (AKAADGYVKPQIKQVV) derived peptide (1 mg/ml) was used as substrate. [γ-32P]ATP (10 μCi; Amersham Biosciences AB) was added to the reactions followed by 10-min incubation at room temperature and boiling in reducing Laemmli sample buffer. For kinetic analysis of catalytic activity, the kinase assays were carried out in the kinase assay buffer with dithiothreitol (1 mM), cold ATP (250 μM) (Cell Signaling Technology, Beverly, MA), 6 μCi of [γ-33P]ATP (Amersham Biosciences AB), and Jak2-derived peptide (VLPQDKEYYKVKEPGES) corresponding to the double tyrosine motif in the kinase activation loop of Jak2. The JH1 and JH1–2 proteins extracted from 293T cells were used at 25–100 nM final concentrations. The peptide concentrations used were between 3 and 2000 μM. The reactions were started by adding the ATP mixture containing both the unlabeled and [γ-33P]ATP and were allowed to proceed for different times at 26°C, and stopped by boiling in reducing Laemmli sample buffer. All kinase reactions were separated in 20% SDS-PAGE followed by quantification of radioactivity by using PhosphorImager (FujiFilm, Dusseldorf, Germany).
Luciferase activity was determined using dual-luciferase reporter assay system (Promega) according to manufacturer's instructions. The Stat-dependent luciferase activity was normalized to the activity of the cotransfected plasmid constitutively expressing Renilla luciferase.
Expression of Gst Fusion Proteins
The Escherichia coli strain XL1-Blue (Stratagene, La Jolla, CA) was transformed with expression plasmids for JH1-Gst and JH1–2-Gst. Overnight cultures were diluted 1:200 in Luria media containing ampicillin and incubated at 37°C for 3 h. A sample from JH1–2-Gst culture was taken after 15-min induction of Gst fusion protein expression with 1 mM isopropyl β-d-thiogalactoside (final concentration) (Sigma-Aldrich, St. Louis, MO). To obtain equal expression level of JH1-Gst protein, a sample was taken after 15-min incubation in the absence of isopropyl β-d-thiogalactoside. After centrifugation of the samples, the cell pellet was directly lysed by boiling in reducing Laemmli sample buffer.
Molecular Modeling
The Jak2 JH2 domain was modeled based on the structure of the activated insulin receptor tyrosine kinase (Irk) at 1.9-Å resolution (Hubbard, 1997; Protein Data Bank [Abola et al., 1997] entry 1ir3). The sequence alignment was performed with Clustal W (Thompson et al., 1994) and MULTICOMP (Vihinen et al., 1992) program packages. The final alignment of the Jak2 JH2 and Irk kinase domains was obtained by manual combination of information from multiple sequence analysis of protein kinases and secondary structural information from the three-dimensional structures of several tyrosine kinases. The model was built with the program InsightII (Accelrys, San Diego, CA). A side chain rotamer library was used to model amino acid substitutions and for the modeling of insertions and deletions was used a database of loops in an unbiased selection of PDB. Fragments obtained were evaluated on three criteria, including the root mean square deviation from the anchor points, sequence similarity, and interference with the protein core region. The models were refined by energy minimization with the program Discover in stepwise manner by using Amber force field. First, hydrogen atoms were relaxed, then the side chains of the newly built loops were relaxed and the rest of the molecule was fixed. Then the borders of insertions and deletions and the Cα atoms of the conserved regions, and finally only the Cα atoms of the conserved regions were harmonically constrained.
RESULTS
Regulation of Jak2 by JH2 Domain in E. coli
We have previously shown that the JH2 domain is critical for the function of Jak2 by negatively regulating basal Jak2 activity and that deletion of JH2 results in constitutive Jak2 activity (Saharinen et al., 2000). Due to the lack of catalytic activity, JH2 is expected to regulate Jak2 through interactions with other JH domains, other proteins, or possibly with nonprotein ligands. A number of other nonreceptor PTKs are known to be negatively regulated by intramolecular domain–domain interactions, which maintain the inactive conformation of the kinase domain (Hubbard et al., 1998). Therefore, one possible mechanism for the inhibition of basal Jak2 activity is that JH2 directly interacts with JH1, modulating its activity. Alternatively, inhibition of Jak2 might involve additional regulatory proteins interacting with JH2.
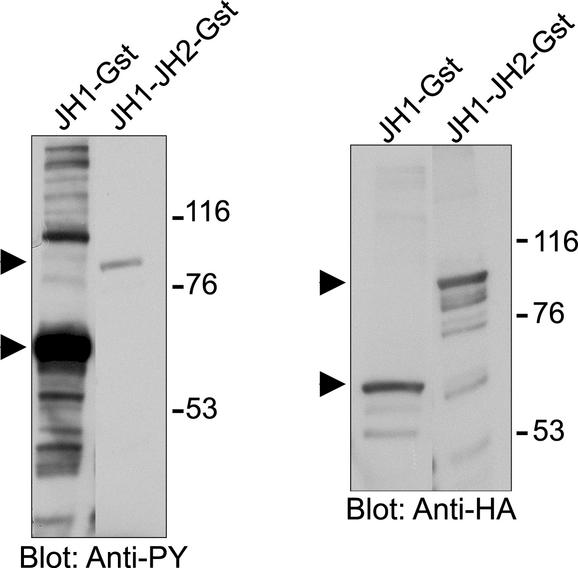
To distinguish between these possibilities, we analyzed the effect of JH2 on the activity of Jak2 in bacterial cells. Bacteria do not have PTKs and therefore are not expected to have regulatory proteins targeted against tyrosine kinases. We have previously found that, when expressed in mammalian cells, the isolated JH1 domain has significantly increased activity compared with full-length Jak2, whereas JH1–2-Jak2, containing the JH1 and JH2 domains, has similar activity to Jak2 (Saharinen et al., 2000). Therefore, we expressed JH1 and JH1–2 domains as Gst fusion proteins in E. coli and analyzed their activities by determining their phosphotyrosine levels in an anti-phosphotyrosine immunoblot (Figure 1). Both JH1-Gst and JH1–2-Gst were tyrosine phosphorylated, indicating that the expressed proteins were active in bacterial cells. However, phosphorylation of JH1-Gst was significantly higher than phosphorylation of JH1–2-Gst, although the two proteins were expressed at similar levels, as shown by anti-HA immunoblot of cell lysates. The phosphorylation of several bacterial proteins was detected only in cells expressing JH1-Gst. These results indicated that the activity of JH1-Gst was higher than that of JH1–2-Gst and that JH2 inhibited the activity of JH1 also in prokaryotic cells. Thus, the JH2-mediated regulation most likely is an intrinsic property of Jak2 and does not require additional regulatory proteins, strongly supporting for an autoinhibitory mechanism in regulation of Jak2 by JH2.
Figure 1.
Autoinhibition of Jak2 in E. coli. Gst-fusion proteins for JH1 (JH1-Gst) and JH1–2 (JH1–2-Gst) domains of Jak2 were expressed in bacterial cells as described in MATERIALS AND METHODS. The cells were lysed by boiling in reducing Laemmli sample buffer. Lysates were separated in 7.5% SDS-PAGE, and analyzed in anti-HA (right) and anti-phosphotyrosine (left) immunoblots. Arrows indicate the migration of JH1-Gst and JH1–2-Gst. The mobilities of the molecular mass markers (in kilodaltons) are shown on the right.
JH2 Domain Suppresses Basal Activity by Lowering Vmax of Jak2
To analyze the regulatory mechanism used by JH2 in suppressing Jak2 activity, we performed kinetic analysis on the catalytic activity of JH1 and JH1–2 proteins. JH1 and JH1–2 were expressed in 293T cells, the Jak2 proteins were immunoprecipitated using anti-HA antibody, and subjected to anti-HA immunoblotting (Figure 2A) as well as to in vitro kinase assay by using a peptide substrate corresponding to the double tyrosine motif in the kinase activation loop of Jak2. First, we analyzed the time course of peptide phosphorylation by JH1 and JH1–2, by varying the reaction time and keeping the kinase, peptide, and ATP concentrations constant. Figure 2B shows that JH1 exhibited a linear relationship between time and peptide phosphorylation during the 60-min assay period. JH1 had also much higher activity than JH1–2, at all time points analyzed.
Figure 2.
Kinetic analysis of the catalytic activity of Jak2. (A) Expression plasmids for JH1-HA and JH1–2-HA were transfected into 293T cells, and cell lysates were immunoprecipitated using anti-HA antibody. Aliquots of the immunoprecipitates (left) and cell lysates (right) were separated in 4–15% SDS-PAGE and analyzed in an anti-HA immunoblot. The mobilities of the molecular mass markers (in kilodaltons) are shown on the right. (B) Immunoprecipitated JH1-HA and JH1–2-HA were subjected to in vitro kinase assay by using Jak2-derived peptide (1.2 mM) as a substrate. The reactions were stopped at various time points and the peptides were separated in 20% SDS-PAGE followed by quantification using PhosphorImager. (C) Immunoprecipitated JH1-HA and JH1–2-HA were subjected to in vitro kinase assay by using as substrate a range of different Jak2 peptide concentrations, as indicated. The reaction time was 30 min. The peptides were separated in 20% SDS-PAGE followed by quantification with PhosphorImager. (D) Data from C are shown as percentages of maximal JH1 or JH1–2 activities (the values for JH1 are divided by the highest JH1 value, and the values for JH1–2 are divided by the highest JH1–2 value). In B and C, [γ-33P]ATP was used, and the total concentration of ATP was 250 μM.
We next wanted to gain insight into the kinetic parameters of peptide phosphorylation by JH1 and JH1–2. The immunoprecipitated JH1 and JH1–2 proteins were incubated in varying peptide concentrations, while keeping the reaction time and the ATP concentration constant. Figure 2C shows that JH1 had much higher activity compared with JH1–2, over the range of substrate concentrations used. The activity of JH1 and JH1–2 did not further increase, even when the peptide concentration was increased from 600 μM to 2 mM (our unpublished data). From Figure 2C it can be estimated that the maximal velocity (Vmax) is higher for JH1 than for JH1–2. When the activities from Figure 2C are plotted as percentages of the highest activities observed for JH1 and JH1–2 (the JH1 values are divided by the highest activity obtained for JH1 and the JH1–2 values are divided by the highest activity obtained for JH1–2) (Figure 2D), it can be seen that JH1 and JH1–2 showed almost identical hyperbolic activity curves in response to different peptide concentrations. Because JH1 and JH1–2 achieved their half-maximal activities in similar substrate concentrations, the Km values for JH1 and JH1–2 are expected to be very similar. Thus, we conclude that JH2 suppresses the catalytic activity mainly by decreasing the Vmax of Jak2, without affecting the Km of the kinase.
Molecular Modeling of JH2 Domain of Jak2
The three-dimensional structures of Jak2 and the isolated domains of Jak2 are currently unresolved, and thus the nature of possible inhibitory domain–domain interactions in Jak2 remains unknown. To be able to understand the molecular mechanism of JH2-mediated regulation of Jak2, we constructed a model of JH2 by using homology-based molecular modeling (Figure 3).
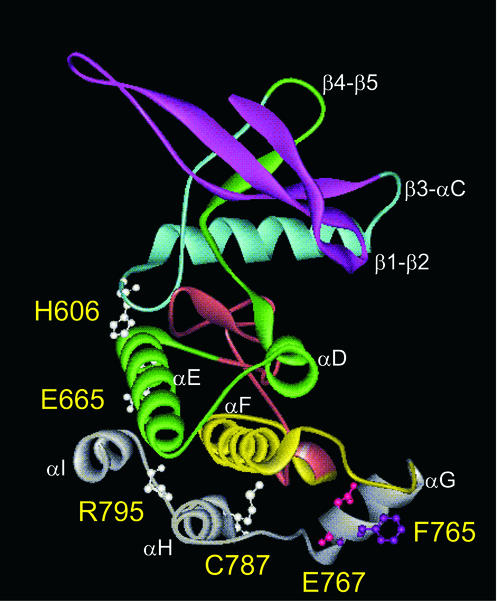
Figure 3.
Molecular model of the three-dimensional structure of the JH2 domain of human Jak2. Different colors represent sequential deletions of 584-Jak2 (magenta), 619-Jak2 (blue), 671-Jak2 (green), 725-Jak2 (red), and 758-Jak2 (yellow) constructs. Ball and stick representation of indicated amino acids. IR1 (green), IR2 (yellow), and IR3 (gray).
The sequence alignment between the Jak2 JH2 domain and the Irk kinase domain was refined based on multiple sequence analysis of several tyrosine kinase domains, and locations of hallmark residues and conserved secondary structures in kinases. According to the sequence alignment (our unpublished data) there were five deletions of one, two, two, six, and nine residues, and two insertions of one and eight residues in the JH2 domain of Jak2 compared with the Irk kinase domain. The structure in Irk is not complete for the activation loop region, and thus the deletion in that region in JH2 could not be modeled. Another part that was not modeled in JH2 was the insertion of eight residues in the catalytic loop between β6 and β7. These functionally important regions are highly variable with respect to length and amino acid sequence between kinase domains. The sequence identity of the Jak2 JH2 domain with the Irk is 23%.
The JH2 domain model has five-stranded antiparallel β-sheet in the N-terminal (N) lobe and mostly α-helical C-terminal (C) lobe. The upper domain in active kinase domains is responsible for ATP binding. The ATP-binding residues and the glycine-rich loop are not conserved in JH2. In addition, several other key residues are not conserved in the JH2 domain in the catalytic site and substrate binding regions. The Jak2 JH2 model bears significant similarity to the previously modeled Jak3 JH2 domain (Vihinen et al., 2000).
Localization of Inhibitory Regions in JH2
To determine inhibitory regions in the JH2 domain, the activities of two different JH2 deletion mutants of Jak2 were compared. The entire JH2 domain is deleted in JH2Δ-Jak2, whereas in BglIIΔ-Jak2 the 60 C-terminal amino acids of JH2 are present (Kohlhuber et al., 1997) (Figure 4A). JH2Δ-Jak2 and BglIIΔ-Jak2 were transiently expressed in 293T cells, and the Jak2 proteins were immunoprecipitated using anti-HA antibody. The immunoprecipitates were subjected to anti-phosphotyrosine and anti-HA immunoblotting (Figure 4B) as well as to in vitro kinase assay by using a peptide substrate corresponding to the tyrosine phosphorylation site in Stat5 (Y694) (Figure 4C) (Gouilleux et al., 1994). Tyrosine phosphorylation as well as kinase activities of JH2Δ-Jak2 and BglIIΔ-Jak2 were greatly increased compared with Jak2. However, JH2Δ-Jak2 showed even higher phosphorylation and kinase activity than BglIIΔ-Jak2. To analyze downstream signaling mediated by the Jak2 deletion mutants, JH2Δ-Jak2 and BglIIΔ-Jak2 were coexpressed with Stat5A. JH2Δ-Jak2 as well as BglIIΔ-Jak2 exhibited increased activity in tyrosine phosphorylation of Stat5 compared with Jak2, but phosphorylation of Stat5 was higher by JH2Δ-Jak2 than by BglIIΔ-Jak2 (Figure 4D). These results indicated that the region deleted in BglIIΔ-Jak2 possessed inhibitory functions, because BglIIΔ-Jak2 was more active than Jak2. However, BglIIΔ-Jak2 was less active than JH2Δ-Jak2, suggesting that the C-terminal 60 residues of JH2 contained an additional inhibitory region. The molecular model suggested that the C-terminal end of JH2 might have a stable fold in the absence of other JH2 regions, thus supporting the finding that this region alone could inhibit JH1.
To analyze the inhibitory regions in JH2 in more detail, we sequentially deleted regions from the N terminus of the JH1–2-Jak2 construct. The 584-Jak2, 619-Jak2, 671-Jak2, 725-Jak2, and 758-Jak2 deletion constructs are shown in Figure 5A and illustrated in colors in the model structure of JH2 in Figure 3. The molecular model of JH2 shows that much of the central β-sheet structure in the N lobe of JH2 (β1-β3) is deleted in 584-Jak2. In 619-Jak2, the only α-helix (αC) located in the N lobe is further deleted. Thus, almost the entire N lobe is deleted in 619-Jak2. 671-Jak2 deletes the first two α-helices (αD and αE) of the C lobe in addition to the entire N lobe, and 725-Jak2 further deletes the two small β-strands (β7-β8) and the sequence forming much of the activation loop. 758-Jak2 contains only the last 50 amino acids of JH2. These 50 residues form three α−helices (αG, αH, and αI) in the model structure of JH2.
The above-described Jak2 constructs were transiently coexpressed with Stat5, and their ability to activate Stat5 was compared with that of Jak2 (Figure 5B). 584-Jak2 and 619-Jak2 showed equal activity in phosphorylating Stat5 compared with Jak2, indicating that the regions deleted in these constructs (amino acids 536–617 in full-length Jak2) did not take part in inhibition. 671-Jak2 and 725-Jak2 exhibited similar activity in phosphorylation of Stat5, but their activity was increased compared with Jak2, 584-Jak2, or 619-Jak2. Thus, in 671-Jak2, the deletion of one β-strand in the N lobe and αD and αE in the C lobe, comprising residues 619–670, resulted in increased Jak2 activity. However, in 725-Jak2 construct the additional deletion of β-strands β7 and β8 and the sequence corresponding to the activation loop in kinase domains did not further increase the activity of 671-Jak2. 758-Jak2 showed the highest activity in phosphorylation of Stat5. 758-Jak2 differs from 725-Jak2 by the lack of residues 725–757, including the helix F. To conclude, these results identified two inhibitory regions in JH2 that were termed inhibitory region 1 (IR1) containing amino acids 619–670 and IR2 containing residues 725–757.
We next compared the activity of 758-Jak2 to that of JH1-Jak2, containing only the kinase domain and the preceding linker region, by coexpressing with Stat5 (Figure 5C). JH1-Jak2 showed significantly increased activity in tyrosine phosphorylation of Stat5 compared with 758-Jak2. Thus, the C-terminal 50 amino acids of JH2 forming helices G, H, and I in 758-Jak2 inhibited JH1, and therefore residues 758–807 were termed as IR3. This result is in accordance with the result obtained in comparing JH2Δ-Jak2 to BglIIΔ-Jak2, where the C-terminal 60 amino acids of JH2 inhibited Jak2 activity (Figure 4).
To confirm the role of the IR3 region in regulation of Jak2, we performed alanine substitution mutagenesis within IR3 in the context of the 758-Jak2 construct, shown in Figure 6A. The effects of the mutations in the 758-Jak2 construct were analyzed by coexpressing with Stat5 (Figure 6B). Compared with 758-Jak2, increased Stat5 tyrosine phosphorylation was detected upon transfection with LQF-758-Jak2, FYE-758-Jak2, and QLP-758-Jak2. Stat5 phosphorylation in DKH-758-Jak2– and APK-758-Jak2–transfected cells was comparable with that in 758-Jak2–transfected cells. Thus, substitution of amino acids 763–767 (LQFYE) and 771–773 (QLP) with alanines in IR3 increased the activity of 758-Jak2. The molecular model of JH2 indicated that residues L763, Q764, F765, E767, Q771 and P773 are on the surface of the domain, thereby enabling interactions between these residues and JH1. The molecular model also showed that residues 763–767 are located in αG, and the helical structure might be an important structural feature and locally distorted by alanine mutations, thus causing deregulation of inhibition.
We next analyzed the role of IR3 in the context of full-length Jak2. We deleted the 13-amino acid region in IR3, where we had introduced the alanine mutations, creating the 761Δ-Jak2 construct (Figure 7A), and compared the activity of 761Δ-Jak2 with either Jak2 or JH2Δ-Jak2. Tyrosine phosphorylation of 761Δ-Jak2 was increased compared with Jak2, although not to the extent of JH2Δ-Jak2 (Figure 7B).
To analyze the function of 761Δ-Jak2 in cytokine signaling, where activation of Jak2 is dependent on cytokine-induced receptor dimerization, we transiently expressed Jak2, JH2Δ-Jak2, or 761Δ-Jak2 together with a Stat1-dependent luciferase reporter construct in a Jak2-negative cell line γ2A (Watling et al., 1993). We have previously found that expression of JH2Δ-Jak2 in γ2A cells results in constitutive, cytokine-independent activation of Stat1 (Saharinen et al., 2000). As a control, we expressed AflIIΔ-Jak2, which lacks the JH4–5 domains and small fragments of domains 3 and 6. AflIIΔ-Jak2 cannot signal through the IFN-γ receptor due to its inability to associate with IFNγRII (Kohlhuber et al., 1997).
Transfection of Jak2 or AflIIΔ-Jak2 did not activate the reporter gene, and stimulation with IFN-γ induced significant Stat1 activity in Jak2-transfected cells, but not in AflIIΔ-Jak2–transfected cells (Figure 7C). Transfection of JH2Δ-Jak2 resulted in ligand-independent Stat1 activation, which was not further induced upon stimulation with IFN-γ. Also, transfection of 761Δ-Jak2 resulted in increased basal Stat1 activation, although not to the extent observed by transfection of JH2Δ-Jak2. Furthermore, Stat activity was not induced by IFN-γ in 761Δ-Jak2–transfected cells. Thus, the deletion in IR3 increased basal activity of Jak2 and abolished IFN-γ-inducible Jak2-Stat1 signaling. These results indicate that IR3 is involved in inhibition of basal Jak2 activity, but in accordance with results in Figures 4 and 5, this region is not solely responsible for inhibition. Rather, inhibition of Jak2 is dependent on multiple regions in JH2.
DISCUSSION
In this study, we present evidence for autoregulation of a Jak family member, Jak2. The finding that Jak2 is inhibited by its pseudokinase domain also in bacterial cells indicates that regulation by the pseudokinase domain is an intrinsic property of the Jak2 molecule and not dependent on additional regulatory proteins.
The participation of PTKs in diverse cellular functions by linking receptor activation to downstream signaling events makes regulation of tyrosine kinase activity extremely important. The activities of several nonreceptor PTKs belonging to distinct kinase families are modulated by N-terminal protein domains. The crystal structures of the inactive forms of Src family members revealed regulatory intramolecular domain–domain interactions and confirmed the models of Src regulation that were based on previously obtained biochemical data. In c-Src and Hck, the SH2 domain interacts with a C-terminal tyrosine residue and the SH3 domain binds to the linker between the SH2 and kinase domains, resulting in inactive conformation of the activation loop and blockage of the substrate-binding site (Sicheri et al., 1997; Xu et al., 1997, 1999). In Tec family kinases Itk and Btk, the SH3 domain interacts with the adjacent proline-rich region (Andreotti et al., 1997; Hansson et al., 2001; Okoh and Vihinen, 2002). In c-Abl, the N-terminal region is responsible for the inhibition of the kinase domain through intramolecular interaction (Pluk et al., 2002). The significance of the above-described intramolecular regulation is emphasized by mutations that abrogate the domain–domain interactions and, consequently, result in ligand-independent activation of the kinases. In v-Src, mutation of the C-terminal regulatory tyrosine causes cell transformation. Thus, regulation of kinase activity by intramolecular interactions seems to be a general mechanism for many nonreceptor PTKs. Based on this study, Jak2 can be added to the list of tyrosine kinases with autoregulatory properties.
Due to the lack of complete three-dimensional structure for any of the Jak kinases, there is no mechanistic explanation for the inhibitory function of the JH2 domain in Jak2 thus far. The comparison of the kinetics of the catalytic activities of JH1 and JH1–2 indicated that the presence of JH2 reduced the activity (Vmax) of Jak2 by severalfold. JH2 might inhibit JH1 by interacting with the active site, thereby blocking the access of ATP and/or substrates to the catalytic site. If JH2 acted by competing with the substrate in access to the active site, it would be expected to increase the Km value of Jak2. Our results show that the Km values of JH1 and JH1–2 for the Jak2 peptide, as approximated from Figure 2D, are very similar. Exact determination of the kinetic parameters would require the use of purified proteins, and we cannot totally rule out that JH2 would affect the Km value of Jak2. Nevertheless, the results suggest that JH2 does not merely compete with the substrate but may inhibit the activity of JH1 by inducing a conformational change in JH1, resulting in distortion of the structures essential for catalysis. Helix C in the N lobe of kinase domains is the target for regulation in many tyrosine kinases. Intramolecular interactions affect the orientation and position of helix C, which in turn may affect the structure and position of the activation loop, and also regulate the accessibility to the substrate and ATP binding sites.
In the EphB2 receptor tyrosine kinase, autoregulation is dependent on the interaction between the kinase domain and a helical juxtamembrane domain on the N-terminal side of the kinase domain (Wybenga-Groot et al., 2001). In EphB2, interaction of two juxtamembrane helices with the kinase domain results in distortion of the N lobe and prevents the A-loop from attaining its active conformation (Wybenga-Groot et al., 2001). Thus, the regulatory interactions in EphB2 are mediated via conformational change alone and do not involve conventional SH2/SH3 domain-mediated interactions. The JH2-based regulation of Jak2 may rely on interactions alike to those found in the EphB2 receptor. Two juxtamembrane tyrosines of EphB2, in their unphosphorylated states, also interact with the kinase domain (Dodelet and Pasquale, 2000). Ligand-induced change in the configuration of the receptor enables phosphorylation of these tyrosines, and this contributes to activation of EphB2, probably by destabilizing the structure of the juxtamembrane domain (Wybenga-Groot et al., 2001).
The inactive conformation of many kinases can be relieved by binding of a substrate, which disrupts the intramolecular contacts. For example, Src family kinases can be activated by substrates containing ligands for SH2/SH3 domains that bind to the regulatory elements in the kinase (Moarefi et al., 1997). The requirement of the Stat SH2 domain for activation by Jak2 (Gupta et al., 1996), but not by the JH2 deletion mutant (Saharinen et al., 2000), suggests that the SH2 domain may be essential for relieving the inhibited state of Jak2. Interestingly, an SH2-containing protein, SH2-Bβ, can bind to and activate Jak2 (Rui and Carter-Su, 1999). The mechanism for SH2-Bβ–mediated activation of Jak2 is not known but may involve the modulation of JH2 function (O'Brien et al., 2001).
We used deletion analysis to identify regions within JH2 that might be required for autoinhibition. Three distinct regions were identified that when deleted, resulted in increased activity of Jak2. Those refer to amino acids 619–670 (IR1), 725–757 (IR2), and 758–807 (IR3). IR2 and IR3 are located in the bigger lobe of JH2, whereas IR1 extends from the N lobe to the C lobe. The finding that IR3 was able to inhibit the kinase domain alone suggests that this region may directly interact with the kinase domain. The model of the JH2 structure also suggests that IR3 may fold as an independent unit. IR1 and IR2 were found to increase IR3-mediated inhibition. IR1 and IR2 may make additional inhibitory contacts with JH1, or IR1 and IR2 may be crucial for the structural context of IR3 and thereby enhance the inhibitory function of IR3.
Previously, a number of mutations have been characterized in the JH2 domains of Jak2, Jak3, and Tyk2 causing aberrant kinase function. Substitution of E695K in JH2 results in hyperactivation of Drosophila Jak, and the corresponding mutation (E665K) has a similar, but less pronounced effect also in Jak2 (Luo et al., 1997). E665 is located in helix D in the model of Jak2 JH2, and the entire helix localizes to IR1. Four mutations in the JH2 domain of Tyk2 have been characterized, resulting in abrogation of IFN-α signaling (Yeh et al., 2000). Interestingly, two of these Tyk2 mutants were also constitutively tyrosine phosphorylated (Yeh et al., 2000). H669 (mutated to P) in Tyk2 corresponds to residue H606 in Jak2, which is located close to, but outside of the N-terminal border of IR1. R856 (mutated to G) in Tyk2 corresponds to R795 in Jak2 located within the minimal 50 amino acid inhibitory region, IR3. Similarly, C759R mutation in Jak3 JH2 derived from a severe combined immunodeficiency patient resulted in nonfunctional but constitutively highly phosphorylated Jak3 (Chen et al., 2000). In Jak2, C759 corresponds to C787 in IR3.
The alanine mutations introduced into IR3 indicated an inhibitory role for residues 763–767 (LQFYE) and 771–773 (QLP). Residues FYE are well conserved between the Jak JH2 domains, whereas the other inhibitory residues show less conservation. Molecular modeling suggested that residues 763L, 764Q, 765F, E767, 771Q, and 773P in IR3 might be exposed on the surface of the JH2 domain, thus being able to interact with JH1 and other molecules. Y766, on the other hand, projects inward in the model and is not likely to interact with JH1, and consequently, may not be phosphorylated. One explanation for the activating effects of alanine substitutions in IR3 is that the mutations distort the α-helical structure (αG), which might affect, directly or indirectly, the conformation of JH1.
In addition to inhibition of Jak2 activity, the JH2 domain is required for induction of IFN-γ– and interleukin-2–dependent signaling in Jak2 and Jak3, respectively (Saharinen and Silvennoinen, 2003). In these studies, the deletion of JH2 from Jak2 and Jak3 resulted in constitutive Stat signaling in the absence of cytokine but lack of cytokine-dependent induction of Jak activity. Interestingly, the deletion of only 13 residues in IR3 similarly increased basal Jak2 activity, but it abolished IFN-γ–dependent induction of signaling. Furthermore, we have found that deletions in the N lobe of JH2 abrogate IFN-γ–inducible activation of Stat1, with an increase in basal Stat activity (Saharinen, unpublished data). Thus, for obtaining proper regulation of Jak2 activity in response to cytokine stimulation, the integrity of the entire JH2 domain seems to be required. These conclusions are in accordance with the previously characterized mutations in Jak3 and Tyk2 that result in constitutive tyrosine phosphorylation of the mutants but abrogation of signal transduction from cytokine receptors (Candotti et al., 1997; Chen et al., 2000; Yeh et al., 2000).
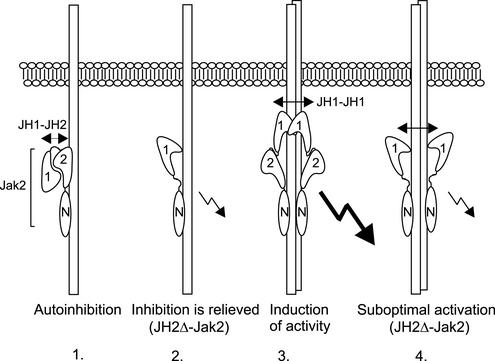
Herein, we provide evidence that JH2 regulates basal activity of Jak2 through an autoinhibitory mechanism, without the need for additional regulatory proteins, and identify three regions in JH2 important for JH1 inhibition. Previously, we have found that JH2 interacts with JH1, but this interaction is weaker than a homotypic interaction between two JH1 domains (Saharinen et al., 2000). Thus, during ligand-induced juxta-positioning of Jaks, a JH1–JH1 interaction might be sufficient to displace the inhibitory JH2–JH1 interaction, resulting in increased Jak2 activity. The subsequent induction of maximal Jak activity requires the JH2 domain. The mechanism by which JH2 mediates inducible Jak activation remains to be resolved, but JH2 may stabilize the activated state of Jak2, or be required for formation of an active Jak-receptor complex. These results are suggestive of a model for JH2 function in activation of Jak2, where 1) in the absence of ligand Jak2 is autoinhibited through an intramolecular JH2–JH1 interaction; 2) upon cytokine induced receptor aggregation, the inhibitory JH2–JH1 interaction is displaced, possibly through formation of a JH1–JH1 interaction, resulting in increase in Jak activity; and 3) induction of maximal Jak2 activation requires a functional JH2 domain, via a still unknown mechanism (Figure 8).
Figure 8.
A model for the function of the JH2 domain in activation of Jak2 by cytokine receptors. In the absence of cytokine, Jak2 is autoinhibited through a possibly intramolecular JH2–JH1 interaction (1). Cytokine binding results in receptor aggregation and displacement of the inhibitory JH1–JH1 interaction (2), possibly through engagement of JH1 in a homotypic JH1–JH1 interaction, leading to increased Jak activity. Induction of maximal activity of Jak2 requires a functional JH2 domain, via a still unknown mechanism (3). In the absence of JH2, maximal Jak activity is not achieved by cytokine stimulation (4).
ACKNOWLEDGMENTS
We thank Drs. Ian Kerr, Richard Pine, and Tim Wood for kindly providing the reagents specified in MATERIALS AND METHODS. This study was supported by the Academy of Finland, the Biomedicum Foundation, the Ella and Georg Ehrnrooth Foundation, the Emil Aaltonen Foundation, the Research and Science Foundation of Farmos, the Finnish Cancer Organization, the Ida Montin Foundation, the Instrumentarium Scientific Fund, the Maud Kuistila Foundation, the Sigrid Juselius Foundation, and the Medical Research Fund of Tampere University Hospital.
Abbreviations used:
- IFN
interferon
- Irk
insulin receptor tyrosine kinase
- JH
Jak homology
- PCR
polymerase chain reaction
- PTK
protein tyrosine kinase
- SH
Src homology
- Stat
signal transducer and activator of transcription
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0342. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0342.
REFERENCES
- Abola EE, Sussman JL, Prilusky J, Manning NO. Protein Data Bank archives of three-dimensional macromolecular structures. Methods Enzymol. 1997;277:556–571. doi: 10.1016/s0076-6879(97)77031-9. [DOI] [PubMed] [Google Scholar]
- Al-Lazikani B, Sheinerman FB, Honig B. Combining multiple structure and sequence alignments to improve sequence detection and alignment: application to the SH2 domains of Janus kinases. Proc Natl Acad Sci USA. 2001;98:14796–14801. doi: 10.1073/pnas.011577898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti AH, Bunnell SC, Feng S, Berg LJ, Schreiber SL. Regulatory intramolecular association in a tyrosine kinase of the Tec family. Nature. 1997;385:93–97. doi: 10.1038/385093a0. [DOI] [PubMed] [Google Scholar]
- Candotti F, et al. Structural and functional basis for JAK3-deficient severe combined immunodeficiency. Blood. 1997;90:3996–4003. [PubMed] [Google Scholar]
- Chen M, Cheng A, Candotti F, Zhou YJ, Hymel A, Fasth A, Notarangelo LD, O'Shea JJ. Complex effects of naturally occurring mutations in the JAK3 pseudokinase domain: evidence for interactions between the kinase and pseudokinase domains. Mol Cell Biol. 2000;20:947–956. doi: 10.1128/mcb.20.3.947-956.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Dodelet VC, Pasquale EB. Eph receptors and ephrin ligands: embryogenesis to tumorigenesis. Oncogene. 2000;19:5614–5619. doi: 10.1038/sj.onc.1203856. [DOI] [PubMed] [Google Scholar]
- Feng J, Witthuhn BA, Matsuda T, Kohlhuber F, Kerr IM, Ihle JN. Activation of Jak2 catalytic activity requires phosphorylation of Y1007 in the kinase activation loop. Mol Cell Biol. 1997;17:2497–2501. doi: 10.1128/mcb.17.5.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauzzi MC, Velazquez L, McKendry R, Mogensen KE, Fellous M, Pellegrini S. Interferon-α-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J Biol Chem. 1996;271:20494–20500. doi: 10.1074/jbc.271.34.20494. [DOI] [PubMed] [Google Scholar]
- Gouilleux F, Wakao H, Mundt M, Groner B. Prolactin induces phosphorylation of Tyr694 of Stat5 (MGF), a prerequisite for DNA binding and induction of transcription. EMBO J. 1994;13:4361–4369. doi: 10.1002/j.1460-2075.1994.tb06756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Yan H, Wong LH, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-α signals. EMBO J. 1996;15:1075–1084. [PMC free article] [PubMed] [Google Scholar]
- Hansson H, Okoh MP, Smith CI, Vihinen M, Hard T. Intermolecular interactions between the SH3 domain and the proline-rich TH region of Bruton's tyrosine kinase. FEBS Lett. 2001;489:67–70. doi: 10.1016/s0014-5793(00)02438-8. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Mohammadi M, Schlessinger J. Autoregulatory mechanisms in protein-tyrosine kinases. J Biol Chem. 1998;273:11987–11990. doi: 10.1074/jbc.273.20.11987. [DOI] [PubMed] [Google Scholar]
- Ihle JN, Witthuhn BA, Quelle FW, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- Irie-Sasaki J, et al. CD45 is a JAK phosphatase and negatively regulates cytokine receptor signaling. Nature. 2001;409:349–354. doi: 10.1038/35053086. [DOI] [PubMed] [Google Scholar]
- Klingmuller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- Kohlhuber F, et al. A JAK1/JAK2 chimera can sustain alpha and gamma interferon responses. Mol Cell Biol. 1997;17:695–706. doi: 10.1128/mcb.17.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacronique V, Boureux A, Valle VD, Poirel H, Quang CT, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard OA. A TEL-JAK2 fusion protein with constitutive kinase activity in human leukemia. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- Liu KD, Gaffen SL, Goldsmith MA, Greene WC. Janus kinases in interleukin-2-mediated signaling: JAK1 and JAK3 are differentially regulated by tyrosine phosphorylation. Curr Biol. 1997;7:817–826. doi: 10.1016/s0960-9822(06)00369-1. [DOI] [PubMed] [Google Scholar]
- Luo H, Rose P, Barber D, Hanratty WP, Lee S, Roberts TM, D'Andrea AD, Dearolf CR. Mutation in the Jak kinase JH2 domain hyperactivates Drosophila and mammalian Jak-Stat pathways. Mol Cell Biol. 1997;17:1562–1571. doi: 10.1128/mcb.17.3.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, Miller WT. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- Myers MP, Andersen JN, Cheng A, Tremblay ML, Horvath CM, Parisien JP, Salmeen A, Barford D, Tonks NK. TYK2 and JAK2 are substrates of protein-tyrosine phosphatase 1B. J Biol Chem. 2001;276:47771–47774. doi: 10.1074/jbc.C100583200. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- O'Brien KB, O'Shea JJ, Carter-Su C. SH2-B family members differentially regulate JAK family tyrosine kinases. J Biol Chem. 2001;277:8673–8681. doi: 10.1074/jbc.M109165200. [DOI] [PubMed] [Google Scholar]
- Okoh MP, Vihinen M. Interaction between Btk TH and SH3 domain. Biopolymers. 2002;63:325–334. doi: 10.1002/bip.10049. [DOI] [PubMed] [Google Scholar]
- Parganas E, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Pine R, Canova A, Schindler C. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma, and is likely to autoregulate the p91 gene. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, Dorey K, Superti-Furga G. Autoinhibition of c-Abl. Cell. 2002;108:247–259. doi: 10.1016/s0092-8674(02)00623-2. [DOI] [PubMed] [Google Scholar]
- Rui L, Carter-Su C. Identification of SH2-bbeta as a potent cytoplasmic activator of the tyrosine kinase Janus kinase 2. Proc Natl Acad Sci USA. 1999;96:7172–7177. doi: 10.1073/pnas.96.13.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saharinen P, Ekman N, Sarvas K, Parker P, Alitalo K, Silvennoinen O. The Bmx tyrosine kinase induces activation of the Stat signaling pathway, which is specifically inhibited by protein kinase Cδ. Blood. 1997;90:4341–4353. [PubMed] [Google Scholar]
- Saharinen P, Silvennoinen O. The pseudokinase domain is required for suppression of basal activity of Jak2 and Jak3 tyrosine kinases and for cytokine-inducible activation of signal transduction. J Biol Chem. 2002;277:47954–47963. doi: 10.1074/jbc.M205156200. [DOI] [PubMed] [Google Scholar]
- Saharinen P, Takaluoma K, Silvennoinen O. Regulation of the Jak2 tyrosine kinase by its pseudokinase domain. Mol Cell Biol. 2000;20:3387–3395. doi: 10.1128/mcb.20.10.3387-3395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungureanu D, Saharinen P, Junttila I, Hilton DJ, Silvennoinen O. Regulation of Jak2 through the ubiquitin-proteasome pathway involves phosphorylation of Jak2 on Y1007 and interaction with SOCS-1. Mol Cell Biol. 2002;22:3316–3326. doi: 10.1128/MCB.22.10.3316-3326.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watling D, et al. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-γ signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- Velazquez L, Mogensen KE, Barbieri G, Fellous M, Uze G, Pellegrini S. Distinct domains of the protein tyrosine kinase tyk2 required for binding of interferon-α/β and for signal transduction. J Biol Chem. 1995;270:3327–3334. doi: 10.1074/jbc.270.7.3327. [DOI] [PubMed] [Google Scholar]
- Vihinen M, Euranto A, Luostarinen P, Nevalainen O. MULTICOMP: a program package for multiple sequence comparison. Comput Appl Biosci. 1992;8:35–38. doi: 10.1093/bioinformatics/8.1.35. [DOI] [PubMed] [Google Scholar]
- Vihinen M, Villa A, Mella P, Schumacher RF, Savoldi G, O'Shea JJ, Candotti F, Notarangelo LD. Molecular modeling of the Jak3 kinase domains and structural basis for severe combined immunodeficiency. Clin Immunol. 2000;96:108–118. doi: 10.1006/clim.2000.4880. [DOI] [PubMed] [Google Scholar]
- Wybenga-Groot LE, Baskin B, Ong SH, Tong J, Pawson T, Sicheri F. Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region. Cell. 2001;106:745–757. doi: 10.1016/s0092-8674(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–638. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- Yasukawa H, et al. The JAK-binding protein JAB inhibits Janus tyrosine kinase activity through binding in the activation loop. EMBO J. 1999;18:1309–1320. doi: 10.1093/emboj/18.5.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh TC, Dondi E, Uze G, Pellegrini S. A dual role for the kinase-like domain of the tyrosine kinase Tyk2 in interferon-α signaling. Proc Natl Acad Sci USA. 2000;97:8991–8996. doi: 10.1073/pnas.160130297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou YJ, Hanson EP, Chen YQ, Magnuson K, Chen M, Swann PG, Wange RL, Changelian PS, O'Shea JJ. Distinct tyrosine phosphorylation sites in JAK3 kinase domain positively and negatively regulate its enzymatic activity. Proc Natl Acad Sci USA. 1997;94:13850–13855. doi: 10.1073/pnas.94.25.13850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemiecki A, Harpur A, Wilks A. JAK protein tyrosine kinases: their role in cytokine signaling. Trends Cell Biol. 1994;4:207–212. doi: 10.1016/0962-8924(94)90143-0. [DOI] [PubMed] [Google Scholar]