Abstract
The mechanism(s) by which microtubule plus-end tracking proteins are targeted is unknown. In the filamentous fungus Aspergillus nidulans, both cytoplasmic dynein and NUDF, the homolog of the LIS1 protein, localize to microtubule plus ends as comet-like structures. Herein, we show that NUDM, the p150 subunit of dynactin, also forms dynamic comet-like structures at microtubule plus ends. By examining proteins tagged with green fluorescent protein in different loss-of-function mutants, we demonstrate that dynactin and cytoplasmic dynein require each other for microtubule plus-end accumulation, and the presence of cytoplasmic dynein is also important for NUDF's plus-end accumulation. Interestingly, deletion of NUDF increases the overall accumulation of dynein and dynactin at plus ends, suggesting that NUDF may facilitate minus-end–directed dynein movement. Finally, we demonstrate that a conventional kinesin, KINA, is required for the microtubule plus-end accumulation of cytoplasmic dynein and dynactin, but not of NUDF.
INTRODUCTION
In eukaryotic cells, the microtubule cytoskeleton is essential for cell cycle progression, the establishment of cell polarity, and cell migration. In most interphase cells, microtubules are polarized in such a way that the minus ends are located at the microtubule organizing center near the nucleus while the plus ends extend to the periphery. Microtubule plus ends are very dynamic, constantly exploring the cytoplasmic space with alternate growing and shrinking phases (reviewed by Desai and Mitchison, 1997; Gundersen, 2002). Kinesin superfamily members and cytoplasmic dynein together play important roles in intracellular trafficking of various proteins, vesicles, and organelles. Because conventional kinesin is a plus-end–directed motor and dynein is a minus-end–directed motor, it is thought that conventional kinesin is responsible for moving materials from the cell body toward cell periphery, whereas cytoplasmic dynein moves materials from the cell periphery back to the cell body (reviewed by Goldstein and Yang, 2000).
We have shown that cytoplasmic dynein is required for the proper distribution of nuclei within the hyphae of the filamentous fungus Aspergillus nidulans (reviewed by Morris et al., 1998b). Cytoplasmic dynein is a complex that consists of heavy chains (HCs), intermediate chains (ICs), light intermediate chains, and light chains (reviewed by King, 2000; Tynan et al., 2000). The HC contains the motor domain (reviewed by Asai and Koonce, 2001), and the other chains may target the motor to various cargoes (Steffen et al., 1997; Tai et al., 1999; Young et al., 2000). The function of cytoplasmic dynein requires dynactin (reviewed by Holleran et al., 1998; Ma et al., 1999; Roghi and Allan, 1999; King and Schroer, 2000; Kumar et al., 2000), a complex that contains multiple subunits, including the 150-kDa dynactin protein and the actin-related proteins Arp1 and Arp11 (reviewed by Holleran et al., 1998; Eckley et al., 1999). Our genetic analyses of nuclear distribution identified multiple nuclear distribution (nud) genes encoding components of the cytoplasmic dynein and dynactin complexes. For example, the nudA, nudI and nudG genes encode the HC, the IC and the 8-kDa light chain of cytoplasmic dynein (Xiang et al., 1995a; Beckwith et al., 1998; Zhang et al., 2002). The nudK gene encodes the actin-related protein Arp1 of the dynactin complex (Xiang et al., 1999). Novel proteins have also been discovered such as NUDC, NUDF, and NUDE (Osmani et al., 1990; Xiang et al., 1995b; Efimov and Morris, 2000). NUDF functions in the cytoplasmic dynein pathway and is homologous to a human protein, LIS1, required for neuronal migration (Xiang et al., 1995b; Willins et al., 1997). Both NUDC and NUDE interact with NUDF/LIS1 (Morris et al., 1998a; reviewed by Morris et al., 1998b; Efimov and Morris, 2000). The human Lis-1 gene was identified as a causal gene for a type 1 lissencephaly, a human disease characterized by brain malformation due to neuronal migration defects (Reiner et al., 1994). Since the discovery that LIS1 homologs participate in dynein function in fungal systems (Xiang et al., 1995b; Geiser et al., 1997), LIS1 and its homologs also have been found to have dynein-related functions in higher eukaryotes (Swan et al., 1999; Faulkner et al., 2000; Liu et al., 2000; Smith et al., 2000; Dawe et al., 2001). More importantly, a direct physical interaction between LIS1/NUDF and the motor domain of cytoplasmic dynein heavy chain has been demonstrated in both mammalian cells and A. nidulans (Sasaki et al., 2000; Hoffmann et al., 2001; Tai et al., 2002), suggesting that LIS1 may regulate dynein motor activity. However, because LIS1 was not isolated as a component of the dynein complex, interaction between LIS1 and dynein may be transient and restricted to special sites in the cell.
Recently, we have shown that GFP-labeled cytoplasmic dynein and its putative regulator NUDF form comet-like structures that accumulate at the dynamic plus ends of microtubules in A. nidulans (Han et al., 2001). This is consistent with discoveries made in mammalian cells that dynactin, and in some cases dynein, localize at the plus ends of microtubules near the cell periphery (Vaughan et al., 1999; Valetti et al., 1999; Habermann et al., 2001; Vaughan et al., 2002). These proteins belong to a growing family of proteins named “plus-end tracking proteins” or +TIPs, which also include the cytoplasmic linker protein CLIP170, the tumor suppressor protein APC, and its binding protein EB1 (Perez et al., 1999; Tirnauer et al., 1999; Brunner and Nurse, 2000; Mimori-Kiyosue et al., 2000a,b; Lin et al., 2001; reviewed by Schuyler and Pellman, 2001; Schroer, 2001). How +TIPs accumulate at microtubule plus ends in vivo is unknown. In this study, we show that in A. nidulans, the p150 subunit of dynactin, which is encoded by the nudM gene, is also localized to microtubule plus ends. We used loss-of-function mutants of cytoplasmic dynein, dynactin, NUDF, and the conventional kinesin KINA to investigate the genetic requirements for dynein, dynactin, and NUDF/LIS1 plus-end localization. Our data support a mutually cooperative role for dynein and dynactin in each other's plus-end localization. Our results also demonstrate that the KINA kinesin is required for cytoplasmic dynein and dynactin localization to microtubule plus ends. However, NUDF most likely uses a KINA-independent mechanism for its plus-end association. Interestingly, the nudF deletion mutant increases the plus-end accumulation of dynein and dynactin, suggesting that NUDF may facilitate dynein's departure from the plus ends of microtubules.
MATERIALS AND METHODS
Aspergillus nidulans Strains, Media, and Techniques
A. nidulans strains used in this study are listed in Table 1. Growth media used were YAG, YAG + UU, and MM + glycerol. Aspergillus growth and transformation were as described previously (Osmani et al., 1987; Waring et al., 1989; Xiang et al., 1995a,b). MM medium for strains with GR5 background was supplemented with 5 μg/ml pyridoxine. Aspergillus protein isolation and Western analysis was done as described previously (Zhang et al., 2002).
Table 1.
A. nidulans strains used in this work (all the strains have the veA1 marker)
| Strain | Genotype* | Source |
|---|---|---|
| GR5 | pyrG89; wA3; pyroA4 | G. S. May |
| R153 | wA3; pyroA4 | C. F. Roberts |
| WX116 | nudM116; pyrG89; yA1 | Xiang et al. (1999) |
| GFP-nudA | GFP-nudA-pyr4; pyrG89; wA3; pyroA4 | Xiang et al. (2000) |
| GFP-nudF | GFP-nudF-pyr4; pyrG89; wA3; pyroA4 | Han et al. (2001) |
| JZ10 or J1 | GFP-nudI-pyr4; pyrG89; wA3; pyroA4 | Zhang et al. (2002) |
| JZ3 | GFP-nudM-pyr4; pyrG89; wA2; pyroA4 | This work |
| SL5 | GFP-nudA-pyr4; nudM116; pyrG89; possibly yA1 and/or wA3 | This work |
| SL6 | GFP-nudF-pyr4; nudM116; pyrG89; possibly yA1 and/or wA3 | This work |
| ΔF/GFP-nudA | ΔnudF-pyr4; GFP-nudA-pyr4; pyrG89; wA3; pyroA4 | Zhang et al. (2002) |
| ΔF/GFP-nudI | ΔnudF-pyr4; GFP-nudI-pyr4; pyrG89; wA3 | This work |
| ΔF/GFP-nudM | ΔnudF-pyr4; GFP-nudM-pyr4; pyrG89; wA3 | This work |
| JZ105 | GFP-nudF-pyr4; nudK317; pyrG89; wA3 | This work |
| JZ108 | GFP-nudF-pyr4; ΔnudA-pyrG; pyrG89; possibly wA3 | This work |
| ΔA/GFP-tubA | GFP-tubA-pyr4; ΔnudA-pyrG; pyrG89; possibly wA3 and pyroA4 | This work |
| XX97 | GFP-nudF-pyr4; nudA2; pyrG89; wA3 | This work |
| XX108 | GFP-nudF-pyr4; pyrG89; nudI416; wA3 | This work |
| JZ4 | GFP-nudA-pyr4; ΔkinA-pyr4; pyrG89; wA3 | This work |
| SL1 | GFP-nudF-pyr4; ΔkinA-pyr4; pyrG89; yA1 | This work |
| SL7 | GFP-nudM-pyr4; ΔkinA-pyr4; pyrG89; wA3 | This work |
| SL9 | GFP-nudI-pyr4; ΔkinA-pyr4; pyrG89; wA3 | This work |
| SNR7a | ΔkinA-pyr4; yA2; pyroA4 | R. Fischer |
Cloning of the nudM p150 Dynactin Gene from A. nidulans
We first searched the A. nidulans genome database (http://microbial.cereon.com/) for sequence homologs of the RO3 protein, the p150 dynactin of Neurospora crassa (Tinsley et al., 1996). Several sequences were identified. We then designed two oligos: AAATGGCCGAGCTTACCATC (underlined sequence indicates the first codon ATG) and TTCGAGACCCACAACCTC for amplifying the genomic DNA encoding the p150 dynactin. By using the GR5 (wild-type strain) genomic DNA as a template, we amplified a region of 4 kb and cloned it into the PCR2.1 vector (Invitrogen, Carlsbad, CA). Sequencing of this clone confirmed that it encodes the p150 dynactin from A. nidulans because it shows high sequence homology with dynactin genes in other organisms. Strong protein sequence homology between the A. nidulans dynactin and the Neurospora crassa Ro-3 dynactin also helped us in identifying the only intron at the 5′-coding region of the gene. For further sequence analysis, the intron sequence was removed based on the presence of the conserved splicing sites. For sequencing the whole p150 dynactin coding region, we also designed two oligos, CTCGATGCACCTCTTACG and AGATGATTGACATAGTGG, and used them for polymerase chain reaction on genomic DNA template. This allowed us to obtain a clone of the C terminus of dynactin that overlaps with the 4-kb clone.
We transformed the 4-kb fragment of p150 dynactin into various new nud mutants (Xiang et al., 1999). The autoreplicating plasmid pAid was cotransformed for selection. We found that the 4-kb p150 dynactin fragment complemented the nudM116 mutation. Our cloned p150 dynactin gene most likely represents the nudM gene rather than an extragenic suppressor of the nudM116 mutation, based on the following evidence. The 4-kb fragment used to transform the nudM116 mutant is not a full-length gene containing promoter and termination sequences. It starts from ATG and ends in the middle of the C terminus. Thus, it is most likely that this fragment repaired the mutation by homologous recombination rather than integrated into another site as an extracopy number suppressor. Furthermore, we analyzed a cross between a ts+ transformant and a wild-type strain R153. (To facilitate such a cross, we first streaked the ts+ transformant on YUU plates to allow the spontaneous loss of the cotransformed extrachromosomal vector pAid carrying the pyrG gene). This cross yielded 100% ts+ progeny (n = 420), consistent with the notion that the original nudM116 mutation was repaired by the p150 dynactin genomic DNA that underwent homologous recombination.
Construction of GFP-nudM (p150 Dynactin) Strain
To construct the GFP-nudM strain in which the endogenous nudM gene is replaced by a functional GFP-nudM fusion gene, we used a strategy similar to what has been described previously for the construction of the GFP-nudI strain (Zhang et al., 2002). For construction of the GFP-nudM plasmid, a 1.4-kb DNA sequence of the N-terminal region of nudM was amplified with oligos D-NotI AAGCGGCCGCTGGCCGAGCTTACCATC and D-SmaI AACCCGGGTCTTCTCCATCTGCAAC as primers. The polymerase chain reaction product was digested with NotI/SmaI and ligated to the NotI and SmaI sites of pLB01 (Liu and Morris, 2000). The resulting plasmid contains sequence of a codon-modified GFP (Fernandez-Abalos et al., 1998), fused to the N terminus of NUDM and the expression of the fusion protein is under the control of the alcA promoter (Waring et al., 1989). On transformation, homologous integration of the nudM sequence on the plasmid into the genomic nudM sequence generates a truncated nudM gene with its own promoter and a GFP-nudM fusion gene under the control of the alcA promoter. The ability to repress the alcA promoter by using glucose allows us to select for transformants in which homologous integration occurred based on their nud-like colony morphology on glucose (similarly described by Zhang et al., 2002). One such transformant for which the single integration event was confirmed by Southern analysis was designated the GFP-nudM strain and used for further observation. GFP-NUDM is functional in vivo because the GFP-nudM cells grow as well as the wild-type cells and exhibit normal nuclear distribution on glycerol medium that is nonrepressing to the alcA promoter (our unpublished data). A similar strategy was also used to create a GFP-nudK (Arp1 of the dynactin complex) strain. However, adding GFP to the N terminus of NUDK Arp1 causes Arp1 to lose its function as the GFP-nudK strain grows like a nud mutant on a glycerol plate. Therefore, we focused on the GFP-nudM (p150 dynactin) strain to study dynactin localization in A. nidulans.
Placing the GFP-nudA, GFP-nudI, GFP-nudM, and GFP-nudF Fusion Alleles into Various Mutants
Standard genetic techniques in A. nidulans were used for genetic crosses. To place a GFP fusion in the background of a specific nud mutation, a nud mutant carrying the pyrG89 mutation was crossed to the strain harboring the GFP fusion. The GR5 parental strain carrying the GFP fusions has the pyrG89 marker, and therefore requires uridine and uracil for growth. The GFP fusions are genetically marked by the Neurospora pyr4 gene that was integrated into the genome along with GFP. This pyr4 gene allows the cells to grow without uridine and uracil. Thus, if any pyrG89 strain is crossed to the GFP strains, only those progeny that contain the GFP-fusion can grow without uridine and uracial. Progeny carrying both the GFP fusion gene and the nud mutation were identified by their ability to grow without uridine and uracil supplementation and by their temperature-sensitive nud phenotype at 42°C. To identify progeny carrying a GFP fusion in the background of a particular nud deletion strain in which the deletion allele is also marked by pyr4 or pyrG, we rely on phenotypic analyses. Specifically, for the cross between the ΔnudF or ΔnudA strain and the GFP-nudA, GFP-nudI, GFP-nudM or GFP-nudF strains, the progeny that carry the GFP fusion proteins in the ΔnudF or ΔnudA background were selected by the presence of the nud phenotype on a glycerol plate at 32°C and the presence of green fluorescence under the microscope using ΔnudF or ΔnudA as negative control, and confirmed by Western analysis demonstrating the presence of the GFP-fusion proteins. For the crosses between the ΔkinA strain and the GFP-nudA, GFP-nudI, GFP-nudM, or GFP-nudF strains, the progeny that carry the GFP fusion proteins in the ΔkinA background were selected by the presence of the nud phenotype of the GFP strains on YUU plates (repressive medium for the alcA promoter) at 32°C and the smaller colony phenotype of the ΔkinA mutant on a glycerol plate. In addition, Southern blot analyses were used to confirm the presence of the ΔkinA allele in strains with various GFP fusion proteins.
Image Acquisition and Analyses, Immunostaining, and Western Blotting
Cells were grown in ΔTC3 culture dishes (Bioptechs, Butler, PA) in 1.5 ml of MM + glycerol + pyridoxine medium at 32°C overnight. In contrast to ethanol or threonine that causes overexpression of the alcA-driven genes, glycerol is a noninducing but derepressing carbon source (Waring et al., 1989), which does not cause the alcA-driven dynein or NUDF to be overproduced compared with the endogenous protein in a wild-type strain (our unpublished data). Images were captured using an IX70 inverted fluorescence microscope (Olympus, Tokyo, Japan) (with a 63× objective) linked to a 5-MHz MicroMax cooled charge-coupled device camera (Princeton Scientific Instruments, Monmouth Junction, NJ) as described previously (Xiang et al., 2000). For measuring the intensity of GFP-dynein or dynactin comets in wild-type and ΔnudF cells, individual GFP comets were segmented and the intensity sum of each segment was measured using the Measuring Segment tool in the IPLab software. Because the GFP-labeled dynein and dynactin comets that are the closest to the hyphal tip are usually the brightest and have similar signal intensity among wild-type cells, we only used values obtained from these comets in each sequence. Data were processed using SigmaPlot and Microsoft Excel.
For visualizing microtubules in cells expressing GFP-NUDM, we used the DM1A anti-α-tubulin antibody (Sigma-Aldrich, St. Louis, MO) for immunostaining, with a procedure similar to what was described previously (Willins et al., 1995; Han et al., 2001). For digestion of the cell wall, β-d-glucanase (16 mg/ml) and driselase (10 mg/ml) (Interspex Products, Foster City, CA) were used (Jung et al., 2000). Although the GFP comets (dynein, dynactin, and NUDF) are present in nearly 100% of the living cells, after fixation, cell wall digestion, and staining, only a very low percentage of the cells show GFP comets. Because this procedure significantly lowered the GFP-comet signal intensity, we used a polyclonal anti-GFP antibody (Chemicon International, Temecula, CA) to stain GFP after microtubule staining. The anti-GFP antibody was used at 1/60. The anti-tubulin antibody was used at 1/200. Cy3-anti-mouse (1/1000; Sigma-Aldrich) and fluorescein isothiocyanate-anti-rabbit secondary antibodies (1/1000; Sigma-Aldrich) were used to visualize microtubules and GFP-NUDM in the same cell. Using this modified procedure, we were able to see GFP-NUDM signals at microtubule ends near the hyphal tip in ∼20% of fixed cells. 4,6-Diamidino-2-phenylindole staining and Western blotting analyses were done as described previously (Zhang et al., 2002).
RESULTS
Cytoplasmic Dynein and Dynactin Require Each Other for Microtubule Plus-End Localization
Our genetic analysis indicated that the nudM gene encodes the A. nidulans p150 dynactin protein (GenBank accession no. AY158343). NUDM shows 43.9% overall sequence identity to the N. crassa dynactin protein RO3 (Tinsley et al., 1996) and contains the CAP-Gly microtubule-binding motif at its N terminus (the GCG program). We have constructed a GFP-nudM strain in which the endogenous nudM gene was replaced by a functional GFP-nudM fusion gene. The GFP-NUDM fusion protein forms dynamic comet-like structures that locate on the distal plus ends of microtubules near the hyphal tip (Figure 1, A and B), a behavior similar to that exhibited by GFP-NUDA (cytoplasmic dynein heavy chain), GFP-NUDI (cytoplasmic dynein IC), and GFP-NUDF (the LIS1-like protein) (Xiang et al., 2000; Han et al., 2001; Zhang et al., 2002).
Figure 1.
(A) Dynamics of GFP-NUDM dynactin comets in vivo. Bar, ∼5 μm. (B) Immunostaining of GFP-NUDM and microtubules. This image resulted from a merge of a pseudocolored microtubule image (red) and a GFP-NUDM image (green).
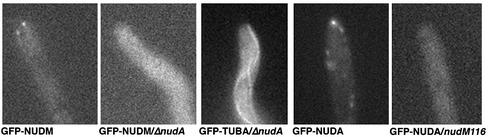
To test whether cytoplasmic dynein plays a role in the localization of dynactin, we crossed the GFP-nudM strain to the nudA deletion mutant, ΔnudA. In the ΔnudA mutant, the region covering the first four ATP-binding sites of the heavy chain is replaced by the pyrG-selective marker gene and the heavy chain protein is not detectable (Xiang et al., 1995a). Interestingly, in most of the ΔnudA cells, the GFP-NUDM dynactin comets were no longer observed near the hyphal tip (Figure 2). Because microtubules in the ΔnudA mutant reach the hyphal tip (Figure 2), our result suggests that cytoplasmic dynein is required for dynactin's plus-end accumulation. Similar to our previous observation that an Arp1 mutation abolishes GFP-NUDA localization (Xiang et al., 2000), herein we found that the nudM116 mutation of p150 dynactin also abolishes GFP-NUDA localization (Figure 2), further supporting dynactin's role in dynein localization. Together, these results indicate that dynein and dynactin are dependent upon each other for their accumulation at microtubule plus ends.
Figure 2.
Images of GFP-NUDM dynactin in wild-type (GFP-NUDM) and the ΔnudA mutant (GFP-NUDM/ΔnudA) cells, GFP-TUBA (α tubulin) in the ΔnudA mutant (GFP-TUBA/ΔnudA), and GFP-NUDA dynein heavy chain in wild-type (GFP-NUDA) and the nudM116 mutant (GFP-NUDA/nudM116) cells.
Presence of Cytoplasmic Dynein Is Important for Localization of NUDF
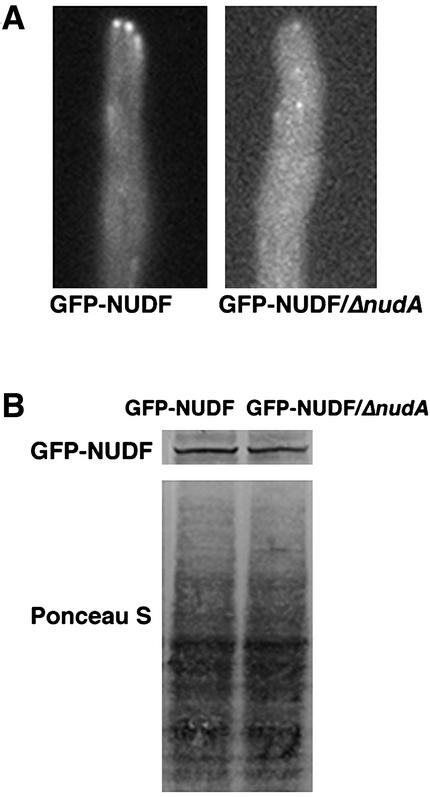
We have noticed that the signal intensities of GFP-NUDF comets are relatively lower in various loss-of-function mutants of cytoplasmic dynein (nudA2 in the heavy chain, nudI416 in the IC) and dynactin (nudK317 in Arp1, nudM116 in the p150 dynactin). The decrease in intensity is most obvious in the dynein heavy chain deletion mutant ΔnudA (Figure 3). Because NUDF/LIS1 interacts directly with the dynein heavy chain (Sasaki et al., 2000; Hoffmann et al., 2001; Tai et al., 2002), we tested whether the NUDF protein level is decreased in the absence of the dynein heavy chain. Our Western blot and densitometry analyses showed that the GFP-NUDF protein level was not significantly decreased in ΔnudA (Figure 3; our unpublished data). Together, these results suggest that the presence of cytoplasmic dynein may enhance NUDF's association with microtubule plus ends. This is consistent with previous observations in mammalian cells that the signal intensity of the kinetochore-localized LIS1 decreases in dynein-defective mitotic cells (Coquelle et al., 2002; Tai et al., 2002).
Figure 3.
(A) Images of GFP-NUDF (LIS1-like protein) in wild-type and the ΔnudA mutant cells. (B) Western blot analysis on the level of GFP-NUDF in protein extract from the wild-type and the ΔnudA mutant cells. A monoclonal anti-GFP antibody (BD Biosciences Clontech) was used at 1/500. The level of protein loading was shown by Ponceau S staining of the membrane.
nudF Deletion Mutant Increases Signal Intensity of GFP-Cytoplasmic Dynein and Dynactin Comet-like Structures
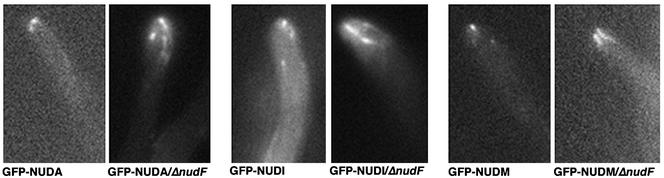
NUDF is apparently not required for cytoplasmic dynein heavy chain targeting to the microtubule plus ends because bright GFP-NUDA comets are obvious at the hyphal tip in the background of a nudF deletion mutant (Zhang et al., 2002; Figure 4). Herein, we show that the GFP-NUDI and GFP-NUDM comets are also bright in the nudF deletion mutant, suggesting that NUDF is also not required for the plus end localization of dynein IC as well as the p150 dynactin (Figure 4). During our observation of many cells, we have had the impression that the GFP-NUDA, GFP-NUDI, and GFP-NUDM comets are brighter in the nudF deletion mutant background compared with that in the wild-type background. To show the differences in intensity quantitatively, we measured the fluorescence intensity of individual comets close to the hyphal tip of each cell. Our measurements showed that the sum of fluorescence of individual GFP-NUDA, GFP-NUDI and GFP-NUDM comets in the nudF deletion cells is significantly higher than that in wild-type cells (Figure 5). These results suggest that although NUDF does not play a role in dynein/dynactin targeting to microtubule plus ends, it could be required for the dynamic departure of dynein/dynactin from the microtubule plus ends. In some ΔnudF cells, a cloud of background fluorescence around the hyphal tip can be observed (Figure 4). Although we do not have a good interpretation of this result, it is possible that overaccumulation of the proteins causes them to fall off the microtubule ends.
Figure 4.
Images of GFP-NUDA (dynein HC), GFP-NUDI (dynein IC), and GFP-NUDM (p150 dynactin) in wild-type and the ΔnudF mutant cells.
Figure 5.
Graphic presentation of the fluorescence intensities of GFP comets near the hyphal tip. Values of mean and SD were generated based on 12 individual comets of each strain.
Deletion of kinA Kinesin Gene Diminishes Plus-End Comets of Both Cytoplasmic Dynein and Dynactin, but Not NUDF
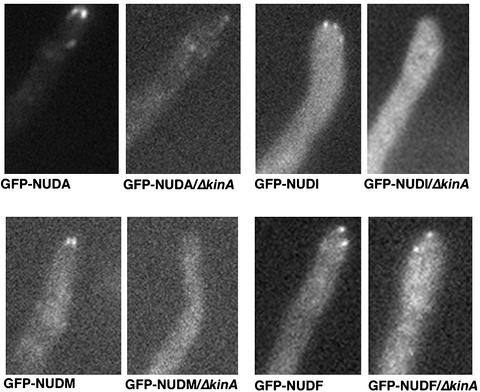
Because cytoplasmic dynein is a minus-end–directed microtubule motor, its in vivo localization to the plus ends of microtubules may depend on the transport function of a plus-end motor such as kinesin. So far, at least seven kinesins have been found in the A. nidulans genome (Fischer, unpublished data; Liu, personal communication), and KINA is the only identified conventional kinesin. Although the kinA deletion mutant grows better than the dynein mutants, it does exhibit a partial defect in nuclear positioning (Requena et al., 2001). In addition, although microtubules look normal in the kinA deletion mutant, genetic evidence suggests that they are more stable than in wild-type cells, which is also similar to what has been described for the cytoplasmic dynein mutants (Willins et al., 1995; Han et al., 2001; Requena et al., 2001). To test whether KINA is involved in cytoplasmic dynein localization to the plus ends in vivo, we crossed a kinA deletion strain (ΔkinA) to the GFP-nudA (cytoplasmic dynein HC) strain and the GFP-nudI strain (cytoplasmic dynein IC), and observed GFP-NUDA as well as GFP-NUDI in the ΔkinA background. Interestingly, cytoplasmic dynein comet-like structures near the hyphal tip are much less obvious in the ΔkinA mutant (Figure 6). These results demonstrated that KINA is important for cytoplasmic dynein accumulation to the plus ends of microtubules.
Figure 6.
Images of GFP-NUDA (dynein HC), GFP-NUDI (dynein IC), GFP-NUDM (p150 dynactin), and GFP-NUDF (LIS1-like protein) in wild-type and the ΔkinA mutant cells.
Because dynactin and NUDF/LIS1 also accumulate at the plus ends of microtubules, it was of interest to test whether their localization is also KINA dependent. To do this, we crossed ΔkinA to the GFP-nudM (p150 dynactin) strain and to the GFP-nudF strain and observed GFP-NUDM as well as GFP-NUDF in the ΔkinA background. Our results show that similar to GFP-NUDA, GFP-NUDM comets structures near the hyphal tip are hardly visible in the ΔkinA mutant (Figure 6), indicating that KINA is also important for the plus-end accumulation of dynactin. However, GFP-NUDF comets were easily observed near the hyphal tip (Figure 6), suggesting that the plus-end localization of NUDF does not require the KINA kinesin. However, consistent with the notion that KINA is required for dynein/dynactin localization and that the presence of dynein/dynactin may enhance NUDF's association with microtubule plus ends, we found that the signal intensity of many GFP-NUDF comets was lower.
To address whether KINA functions only in the cytoplasmic dynein genetic pathway, a previous study has analyzed the double mutant containing a temperature-sensitive nudA allele, nudA1, and the ΔkinA allele (Requena et al., 2001). Although the nuclear migration phenotype of the double mutant resembles that of the nudA1 single mutant, the double mutant grows more poorly than the nudA1 single mutant (Requena et al., 2001). In this study, we compared an alcA-promoter–based nudA conditional null mutant (GFP-nudA) with the corresponding double null mutant (GFP-nudA/ΔkinA). On a glucose-containing medium that shuts off the alcA promoter, the double null mutant exhibited a more severe growth defect than that of the nudA or the kinA single null mutant (Figure 7A). However, the nuclear migration phenotype of the double null mutant is similar to that exhibited by the nudA single null mutant when germlings of similar lengths are compared (Figure 7B). These results suggest that, in addition to being involved in dynein-related functions such as nuclear migration, KINA also participates in pathway(s) that regulate other cellular processes.
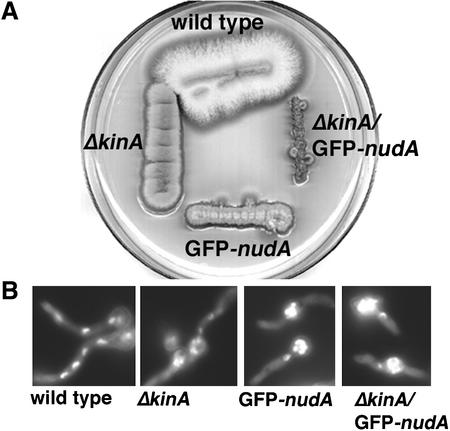
Figure 7.
NudA/kinA double mutant analyses. (A) Growth of the wild-type, ΔkinA, GFP-nudA, and ΔkinA/GFP-nudA (JZ4) strains on YUU that represses the expression of the alcA-driven GFP-nudA fusion gene, the only intact version of nudA in the genome. The plate was incubated at 32°C for 2 d. (B) 4,6-Diamidino-2-phenylindole staining of the wild-type, ΔkinA, GFP-nudA, and ΔkinA/GFP-nudA (JZ4) cells that were grown in YUU at 32°C for 7.5 h.
DISCUSSION
In this study, we have provided the genetic evidence that KINA, a conventional kinesin, is involved in the localization of both cytoplasmic dynein and dynactin to the plus ends of microtubules in A. nidulans. In the deletion mutant of kinA, the microtubule plus-end accumulation of both cytoplasmic dynein (the NUDA heavy chain and the NUDI IC) and dynactin (the NUDM p150 dynactin) are significantly diminished. Although it is possible that KINA may transport cytoplasmic dynein and dynactin toward the microtubule plus end, other possibilities also exist. For example, KINA itself may be targeted to the plus ends, and its presence there is required for dynein/dynactin to associate to the plus ends. However, a previous study has shown that KINA itself is not a plus-end–associated protein. Although in many cells the GFP-KINA fusion protein does not show specific localization, it does light up microtubule-like structures in some cells (Requena et al., 2001). Similarly, a homologous conventional kinesin in Ustilago maydis is also a cytoplasmic protein, which exhibits localization to microtubule-like structures only when cells are treated to deplete ATP (Lehmler et al., 1997). At this point, we have not detected any physical interaction between KINA and cytoplasmic dynein. Therefore, it is also possible that the effect of kinA deletion on dynein and dynactin localization is indirect. For example, the deletion of kinA may change the microtubule ends in such a way that dynein and dynactin can no longer be associated. In fact, genetic evidence has shown that the kinA deletion mutant slows down microtubule dynamics (Requena et al., 2001). However, because dynein and dynactin are clearly accumulated at the plus ends of microtubules in the nudF mutant, in which microtubules are also less dynamic (Han et al., 2001), it is unlikely that dynein/dynactin's failure to accumulate at the microtubule plus ends in the kinA mutant is caused by microtubules being too stable.
Interestingly, the KINA-dependent mechanism may not be the only mechanism for dynein/dynactin's accumulation to the plus ends of microtubules. In some kinA deletion cells, faint dynein and dynactin comets moving toward the hyphal tip can still be observed. This suggests the participation of a redundant player. The fact that the kinA deletion mutant exhibits a nuclear distribution defect, which is less severe than that of a dynein heavy chain null mutant, is also consistent with KINA being a player in the dynein pathway whose function is partially redundant with other protein(s) yet to be identified.
It would be interesting to know whether dynein/dynactin's localization to the microtubule plus ends in higher eukaryotic cells also depends on conventional kinesin. Several studies have suggested that cytoplasmic dynein may be present on cargoes that are transported by kinesin (Hirokawa et al., 1990; reviewed by Goldstein and Yang, 2000; Ma and Chisholm, 2002; Gross et al., 2002). A very recent study in Drosophila has also demonstrated the role of kinesin in dynein localization (Brendza et al., 2002). Whether other +TIPs use kinesins to get to the plus ends is also an interesting question. APC, a tumor suppressor protein that associates with another plus-end protein EB1, physically interacts with KAP3, a KIF3A-KIF3B kinesin superfamily-associated protein, and this interaction is important for APC's accumulation to the membrane protrusions (Jimbo et al., 2002). Our results show that NUDF's localization is not dependent upon the conventional kinesin KINA, suggesting that +TIPs may use different mechanisms for their microtubule plus-end targeting. The CLIP170 protein that physically interacts with LIS1 may be targeted to the microtubule plus end by copolymerization with tubulins (Diamantopoulos et al., 1999; Perez et al., 1999; Coquelle et al., 2002). However, its localization to the plus end rather than along the length of a microtubule would require its dissociation from the early-polymerized segments, and how that may be achieved is not clear. The growing end distinguishes itself from the rest of the microtubule in both its structure and the presence of a GTP cap (reviewed by Desai and Mitchison, 1997). Although the size of the GTP cap is estimated to be very small (Panda et al., 2002), such a cap combined with the structure feature of a growing end may be sufficient to prevent the proteins from falling off the end.
In this study, we have also found that cytoplasmic dynein is required for the plus-end localization of dynactin. This result seems contradictory to the observation made in mammalian cells that only dynactin but not dynein accumulates at the microtubule plus end under physiological conditions (Vaughan et al., 1999; Habermann et al., 2001). However, it is possible that cytoplasmic dynein is required for the initial targeting but not for maintaining the association of dynactin to the microtubule plus end. In contrast to A. nidulans in which the plus-end localization of cytoplasmic dynein has been observed under physiological temperatures (Xiang et al., 2000; Han et al., 2001), the plus-end localization of cytoplasmic dynein in mammalian cells has only been observed upon a shift to a lower temperature (Vaughan et al., 1999). This may indicate that mammalian cytoplasmic dynein but not dynactin leaves the microtubule plus end frequently under physiological conditions.
Interestingly, NUDF/LIS1 may be required for the departure of dynein and dynactin from the microtubule plus end. The fluorescence intensities of the dynein and dynactin comets near the hyphal tip are significantly higher in the absence of NUDF. Because cytoplasmic dynein is involved in retrograde vesicle transport in filamentous fungi (Seiler et al., 1999), the plus-end dynein/dynactin in filamentous fungi may also represent a cargo-loading site as has been suggested and recently demonstrated in mammalian cells (Vaughan et al., 1999, 2002; Valetti et al., 1999; Habermann et al., 2001). Because LIS1 physically interacts with both the cargo-binding region and the first AAA (ATPases associated with cellular activities) repeat of the dynein heavy chain, it was proposed that LIS1 may be involved in the coordination between motor activity and cargo binding in vivo (Tai et al., 2002). Thus, it is possible that in the absence of NUDF, dynein and dynactin fail to depart from the microtubule plus ends toward the minus end, which results in an overaccumulation of dynein and dynactin at the plus end (Figure 8).
Figure 8.
Model providing an explanation for the observation that accumulation of dynein/dynactin at the microtubule plus ends is increased in the nudF deletion mutant. NUDF/LIS1 may facilitate dynein-mediated cargo departure from the plus end toward the minus end of the microtubule. Failure of such departure is expected to cause a more intense plus-end accumulation of dynein/dynactin, which is what has been found in absence of NUDF.
The function of the plus-end cytoplasmic dynein may be multifold. Recent work has shown that in the budding yeast Saccharomyces cerevisiae, cytoplasmic dynein IC is also localized to the microtubule plus end (Beach and Bloom, personal communication). The microtubules and their motors in S. cerevisiae are used only for spindle function and nuclear migration (Cottingham et al., 1999), and thus the plus-end dynein may be involved in regulating microtubule–cortex interaction and microtubule dynamics, which are important for nuclear migration and spindle orientation in fungi (Oakley and Morris, 1981; Willins et al., 1995; Carminati and Stearns, 1997; Shaw et al., 1997; Cottingham et al., 1999; Adames and Cooper, 2000; Han et al., 2001; Requena et al., 2001; Tran et al., 2001). Our previous observation that microtubules are less dynamic in both the nudF and dynein mutant cells indicates that NUDF may also regulate microtubule dynamics (Han et al., 2001). In some ΔnudF cells, disorientated dynein/dynactin comets can be observed, which is also consistent with some microtubules curling around and growing back from the hyphal tip in the nudF and dynein mutant cells (see movie GFP-NUDI/ΔnudF in Supplementary Materials; Han et al., 2001).
Note added in proof.
Since the acceptance of this article, the articles listed below, covering two related topics, have been published.
Topic A: A nidulans and S. cerevisiae dynein localization:
Efimov, V.P. (2003). Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol. Biol. Cell. 14, 871–888.
Lee, W.L., Oberle, J.R., and Cooper, J.A. (2003). The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J. Cell Biol. 160, 355–364.
Liu, B., Xiang, X., Lee, Y.R. (2003). The requirement of the LC8 dynein light chain for nuclear migration and septum positioning is temperature dependent in Aspergillus nidulans. Mol. Microbiol. 47, 291–301.
Sheeman, B., Carvalho, P., Sagot, I., Geiser, J., Kho, D., Hoyt, M.A., and Pellman, D. (2003). Determinants of S. cerevisiae dynein localization and activation. Implications for the mechanism of spindle positioning. Curr Biol. 13, 364–372.
Xiang, X. (2003). L1S1 at the microtubule plus end and its role in dynein-mediated nuclear migration. J. Cell Biol. 160, 289–290.
Topic B: The kinesin-dynein relationship in Drosophila
Duncan, J.E., Warrior, R. (2002). The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12, 1982–1991.
Januschke, J., Gervais, L., Dass, S., Kaltschmidt, J.A., Lopez-Schier, H., Johnston, D.S., Brand, A.H., Roth, S., and Guichet, A. (2002). Polar transport in the Drosophila oocyte requires dynein and kinesin I cooperation. Curr Biol. 12, 1971–1981.
Palacios, I.M., and Johnston, D.S. (2002). Kinesin light chain-independent function of the kinesin heavy chain in cytoplasmic streaming and posterior localisation in the Drosophila oocyte. Development. 129, 5473–5485.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. Teresa Dunn, John. A. Hammer, Vladimir P. Efimov, and Tian Jin for helpful reading of the manuscript. We thank Dr. Bo Liu for communicating information about A. nidulans kinesins. We also thank Drs. Kerry Bloom and Dale Beach for communicating the budding yeast results on dynein IC localization. The p150 dynactin gene was initially identified in Cereon Microbe Sequence database (http://microbial.cereon.com). We thank the Monsanto Company for allowing us to use this database and we also thank the stuff members for help. This work is supported by a National Science foundation grant and a Uniformed Services University of the Health Sciences intramural grant (to X.X.).
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0516. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0516.
REFERENCES
- Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai DJ, Koonce MP. The dynein heavy chain: structure, mechanics and evolution. Trends Cell Biol. 2001;11:196–202. doi: 10.1016/s0962-8924(01)01970-5. [DOI] [PubMed] [Google Scholar]
- Beckwith SM, Roghi CH, Liu B, Morris NR. The “8-kD” cytoplasmic dynein light chain is required for nuclear migration and for dynein heavy chain localization in Aspergillus nidulans. J Cell Biol. 1998;143:1239–1247. doi: 10.1083/jcb.143.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza R, Serbus L, Saxton W, Duffy J. Posterior localization of dynein and dorsal-ventral axis formation depend on kinesin in Drosophila oocytes. Curr Biol. 2002;12:1541–1545. doi: 10.1016/s0960-9822(02)01108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Carminati JL, Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coquelle FM, et al. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Gheber L, Miller DL, Hoyt MA. Novel roles for Saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawe AL, Caldwell KA, Harris PM, Morris NR, Caldwell GA. Evolutionarily conserved nuclear migration genes required for early embryonic development in Caenorhabditis elegans. Dev Genes Evol. 2001;211:434–441. doi: 10.1007/s004270100176. [DOI] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Diamantopoulos GS, Perez F, Goodson HV, Batelier G, Melki R, Kreis TE, Rickard JE. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J Cell Biol. 1999;144:99–112. doi: 10.1083/jcb.144.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Gill SR, Melkonian KA, Bingham JB, Goodson HV, Heuser JE, Schroer TA. Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov VP, Morris NR. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner NE, Dujardin DL, Tai CY, Vaughan KT, O'Connell CB, Wang Y, Vallee RB. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- Fernandez-Abalos JM, Fox H, Pitt C, Wells B, Doonan JH. Plant-adapted green fluorescent protein is a versatile vital reporter for gene expression, protein localization and mitosis in the filamentous fungus, Aspergillus nidulans. Mol Microbiol. 1998;27:121–130. doi: 10.1046/j.1365-2958.1998.00664.x. [DOI] [PubMed] [Google Scholar]
- Geiser JR, Schott EJ, Kingsbury TJ, Cole NB, Totis LJ, Bhattacharyya G, He L, Hoyt MA. Saccharomyces cerevisiae genes required in the absence of the CIN8-encoded spindle motor act in functionally diverse mitotic pathways. Mol Biol Cell. 1997;8:1035–1050. doi: 10.1091/mbc.8.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein LS, Yang Z. Microtubule-based transport systems in neurons: the roles of kinesins and dyneins. Annu Rev Neurosci. 2000;23:39–71. doi: 10.1146/annurev.neuro.23.1.39. [DOI] [PubMed] [Google Scholar]
- Gundersen GG. Evolutionary conservation of microtubule-capture mechanisms. Nat Rev Mol Cell Biol. 2002;3:296–304. doi: 10.1038/nrm777. [DOI] [PubMed] [Google Scholar]
- Gross SP, Welte MA, Block SM, Wieschaus EF. Coordination of opposite-polarity microtubule motors. J Cell Biol. 2002;156:715–724. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann A, Schroer TA, Griffiths G, Burkhardt JK. Immunolocalization of cytoplasmic dynein and dynactin subunits in cultured macrophages: enrichment on early endocytic organelles. J Cell Sci. 2001;114:229–240. doi: 10.1242/jcs.114.1.229. [DOI] [PubMed] [Google Scholar]
- Han G, Liu B, Zhang J, Zuo W, Morris NR, Xiang X. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr Biol. 2001;11:719–724. doi: 10.1016/s0960-9822(01)00200-7. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Sato-Yoshitake R, Yoshida T, Kawashima T. Brain dynein (MAP1C) localizes on both anterogradely and retrogradely transported membranous organelles in vivo. J Cell Biol. 1990;111:1027–1037. doi: 10.1083/jcb.111.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann B, Zuo W, Liu A, Morris NR. The LIS1-related protein NUDF of Aspergillus nidulans and its interaction partner NUDE bind directly to specific subunits of dynein and dynactin. J Biol Chem. 2001;276:38877–38884. doi: 10.1074/jbc.M106610200. [DOI] [PubMed] [Google Scholar]
- Holleran EA, Karki S, Holzbaur EL. The role of the dynactin complex in intracellular motility. Int Rev Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, Akiyama T. Identification of a link between the tumor suppressor APC and the kinesin superfamily. Nat Cell Biol. 2002;4:323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- Jung MK, Ovechkina Y, Prigozhina N, Oakley CE, Oakley BR. The use of β-d-glucanase as a substitute for Novozyme 234 in immunofluorescence, and protoplasting. Fungal Genet Newsl. 2000;47:65–66. [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- King SM. The dynein microtubule motor. Biochim Biophys Acta. 2000;1496:60–75. doi: 10.1016/s0167-4889(00)00009-4. [DOI] [PubMed] [Google Scholar]
- Kumar S, Lee IH, Plamann M. Cytoplasmic dynein ATPase activity is regulated by dynactin-dependent phosphorylation. J Biol Chem. 2000;275:31798–31804. doi: 10.1074/jbc.M000449200. [DOI] [PubMed] [Google Scholar]
- Lehmler C, Steinberg G, Snetselaar KM, Schliwa M, Kahmann R, Bolker M. Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J. 1997;16:3464–3473. doi: 10.1093/emboj/16.12.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, de Carvalho P, Kho D, Tai CY, Pierre P, Fink GR, Pellman D. Polyploids require Bik1 for kinetochore-microtubule attachment. J Cell Biol. 2001;155:1173–1184. doi: 10.1083/jcb.200108119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Morris NR. A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol Gen Genet. 2000;263:375–387. doi: 10.1007/s004380051181. [DOI] [PubMed] [Google Scholar]
- Liu Z, Steward R, Luo L. Drosophila Lis1 is required for neuroblast proliferation, dendritic elaboration and axonal transport. Nat Cell Biol. 2000;2:776–783. doi: 10.1038/35041011. [DOI] [PubMed] [Google Scholar]
- Ma S, Chisholm RL. Cytoplasmic dynein-associated structures move bidirectionally in vivo. J Cell Sci. 2002;115:1453–1460. doi: 10.1242/jcs.115.7.1453. [DOI] [PubMed] [Google Scholar]
- Ma S, Trivinos-Lagos L, Graf R, Chisholm RL. Dynein intermediate chain mediated dynein-dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium. J Cell Biol. 1999;147:1261–1274. doi: 10.1083/jcb.147.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000a;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J Cell Biol. 2000b;148:505–518. doi: 10.1083/jcb.148.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM, Albrecht U, Reiner O, Eichele G, Yu-Lee LY. The lissencephaly gene product Lis1, a protein involved in neuronal migration, interacts with a nuclear movement protein, NudC. Curr Biol. 1998a;8:603–606. doi: 10.1016/s0960-9822(98)70232-5. [DOI] [PubMed] [Google Scholar]
- Morris NR, Efimov VP, Xiang X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998b;8:467–470. doi: 10.1016/s0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- Oakley BR, Morris NR. A β-tubulin mutation in Aspergillus nidulans that blocks microtubule function without blocking assembly. Cell. 1981;24:837–845. doi: 10.1016/0092-8674(81)90109-4. [DOI] [PubMed] [Google Scholar]
- Osmani SA, May GS, Morris NR. Regulation of the mRNA levels of nimA, a gene required for the G2-M transition in Aspergillus nidulans. J Cell Biol. 1987;104:1495–1504. doi: 10.1083/jcb.104.6.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani AH, Osmani SA, Morris NR. The molecular cloning and identification of a gene product specifically required for nuclear movement in Aspergillus nidulans. J Cell Biol. 1990;111:543–551. doi: 10.1083/jcb.111.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda D, Miller HP, Wilson L. Determination of the size and chemical nature of the stabilizing “cap” at microtubule ends using modulators of polymerization dynamics. Biochemistry. 2002;41:1609–1617. doi: 10.1021/bi011767m. [DOI] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, Dobyns WB, Caskey CT, Ledbetter DH. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- Requena N, Alberti-Segui C, Winzenburg E, Horn C, Schliwa M, Philippsen P, Liese R, Fischer R. Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol Microbiol. 2001;42:121–132. doi: 10.1046/j.1365-2958.2001.02609.x. [DOI] [PubMed] [Google Scholar]
- Roghi C, Allan VJ. Dynamic association of cytoplasmic dynein heavy chain 1a with the Golgi apparatus and intermediate compartment. J Cell Sci. 1999;112:4673–4685. doi: 10.1242/jcs.112.24.4673. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Shionoya A, Ishida M, Gambello MJ, Yingling J, Wynshaw-Boris A, Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- Schroer TA. Microtubules don and doff their caps: dynamic attachments at plus and minus ends. Curr Opin Cell Biol. 2001;13:92–96. doi: 10.1016/s0955-0674(00)00179-4. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D. Microtubule “plus-end-tracking proteins”: the end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- Seiler S, Plamann M, Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr Biol. 1999;9:779–785. doi: 10.1016/s0960-9822(99)80360-1. [DOI] [PubMed] [Google Scholar]
- Shaw SL, Yeh E, Maddox P, Salmon ED, Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behavior and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Steffen W, Karki S, Vaughan KT, Vallee RB, Holzbaur EL, Weiss DG, Kuznetsov SA. The involvement of the intermediate chain of cytoplasmic dynein in binding the motor complex to membranous organelles of Xenopus oocytes. Mol Biol Cell. 1997;8:2077–2088. doi: 10.1091/mbc.8.10.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A, Nguyen T, Suter B. Drosophila lissencephaly-1 functions with Bic-D and dynein in oocyte determination and nuclear positioning. Nat Cell Biol. 1999;1:444–449. doi: 10.1038/15680. [DOI] [PubMed] [Google Scholar]
- Tai AW, Chuang JZ, Bode C, Wolfrum U, Sung CH. Rhodopsin's carboxy-terminal cytoplasmic tail acts as a membrane receptor for cytoplasmic dynein by binding to the dynein light chain Tctex-1. Cell. 1999;97:877–887. doi: 10.1016/s0092-8674(00)80800-4. [DOI] [PubMed] [Google Scholar]
- Tai CY, Dujardin DL, Faulkner NE, Vallee RB. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley JH, Minke PF, Bruno KS, Plamann M. p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol Biol Cell. 1996;7:731–742. doi: 10.1091/mbc.7.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, O'Toole E, Berrueta L, Bierer BE, Pellman D. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J Cell Biol. 1999;145:993–1007. doi: 10.1083/jcb.145.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoue S, Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;153:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan SH, Purohit A, Doxsey SJ, Vallee RB. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J Biol Chem. 2000;275:32763–32768. doi: 10.1074/jbc.M001536200. [DOI] [PubMed] [Google Scholar]
- Valetti C, Wetzel DM, Schrader M, Hasbani MJ, Gill SR, Kreis TE, Schroer TA. Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol Biol Cell. 1999;10:4107–4120. doi: 10.1091/mbc.10.12.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150Glued to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT, Tynan SH, Faulkner NE, Echeverri CJ, Vallee RB. Colocalization of cytoplasmic dynein with dynactin and CLIP-170 at microtubule distal ends. J Cell Sci. 1999;112:1437–1447. doi: 10.1242/jcs.112.10.1437. [DOI] [PubMed] [Google Scholar]
- Waring RB, May GS, Morris NR. Characterization of an inducible expression system in Aspergillus nidulans using alcA and tubulin-coding genes. Gene. 1989;79:119–130. doi: 10.1016/0378-1119(89)90097-8. [DOI] [PubMed] [Google Scholar]
- Willins DA, Xiang X, Morris NR. An α tubulin mutation suppresses nuclear migration mutations in Aspergillus nidulans. Genetics. 1995;141:1287–1298. doi: 10.1093/genetics/141.4.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willins DA, Liu B, Xiang X, Morris NR. Mutations in the heavy chain of cytoplasmic dynein suppress the nudF nuclear migration mutation of Aspergillus nidulans. Mol Gen Genet. 1997;255:194–200. doi: 10.1007/s004380050489. [DOI] [PubMed] [Google Scholar]
- Xiang X, Han G, Winkelmann DA, Zuo W, Morris NR. Dynamics of cytoplasmic dynein in living cells and the effect of a mutation in the dynactin complex actin-related protein arp1. Curr Biol. 2000;10:603–606. doi: 10.1016/s0960-9822(00)00488-7. [DOI] [PubMed] [Google Scholar]
- Xiang X, Osmani AH, Osmani SA, Xin M, Morris NR. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol Biol Cell. 1995b;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Roghi C, Morris NR. Characterization and localization of the cytoplasmic dynein heavy chain in Aspergillus nidulans. Proc Natl Acad Sci USA. 1995a;92:9890–9894. doi: 10.1073/pnas.92.21.9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang X, Zuo W, Efimov VP, Morris NR. Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr Genet. 1999;35:626–630. doi: 10.1007/s002940050461. [DOI] [PubMed] [Google Scholar]
- Young A, Dictenberg JB, Purohit A, Tuft R, Doxsey SJ. Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol Biol Cell. 2000;11:2047–2056. doi: 10.1091/mbc.11.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Han G, Xiang X. Cytoplasmic dynein intermediate chain and heavy chain are dependent upon each other for microtubule end localization in Aspergillus nidulans. Mol Microbiol. 2002;44:381–392. doi: 10.1046/j.1365-2958.2002.02900.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.