Abstract
Excess 17β-estradiol (E2), the most potent of human estrogens, is known to act as a stimulus for the growth of breast tumors. Human estrogenic 17β-hydroxysteroid dehydrogenase type 1 (17β-HSD1), which catalyzes the reduction of inactive estrone (E1) to the active 17β-estradiol in breast tissues, is a key enzyme responsible for elevated levels of E2 in breast tumor tissues. We present here the structure of the ternary complex of 17β-HSD1 with the cofactor NADP+ and 3-hydroxyestra-1,3,5,7-tetraen-17-one (equilin), an equine estrogen used in estrogen replacement therapy. The ternary complex has been crystallized with a homodimer, the active form of the enzyme, in the asymmetric unit. Structural and kinetic data presented here show that the 17β-HSD1-catalyzed reduction of E1 to E2 in vitro is specifically inhibited by equilin. The crystal structure determined at 3.0-Å resolution reveals that the equilin molecule is bound at the active site in a mode similar to the binding of substrate. The orientation of the 17-keto group with respect to the nicotinamide ring of NADP+ and catalytic residues Tyr-155 and Ser-142 is different from that of E2 in the 17β-HSD1–E2 complex. The ligand and substrate-entry loop densities are well defined in one subunit. The substrate-entry loop adopts a closed conformation in this subunit. The result demonstrates that binding of equilin at the active site of 17β-HSD1 is the basis for inhibition of E1-to-E2 reduction by this equine estrogen in vitro. One possible outcome of estrogen replacement therapy in vivo could be reduction of E2 levels in breast tissues and hence the reduced risk of estrogen-dependent breast cancer.
17β-Hydroxysteroid dehydrogenases (17β-HSDs) are a group of enzymes that are involved in interconversion of active and inactive forms of androgens and estrogens (1–7) by NAD(P)(H)-linked oxidoreductive transfer of a hydride to and from the 17-position of steroid molecules. Six distinct 17β-HSD isozymes, numbered 1–6, have been identified and cloned (2–7). These isozymes differ in specificities for substrate and tissue and in the preferred direction of the reaction. In human breast tissues, the most active estrogen, 17β-estradiol (E2), is formed by reduction of the inactive estrogen, estrone (E1), which is catalyzed by 17β-HSD type 1 (17β-HSD1). The estrogenic specificity of 17β-HSD1 as well as its preference for the reduction reaction has been well established (8–10).
17β-HSD1 is expressed in steroidogenic tissues including estrogen target tissues such as normal and malignant endometrium and breast tissues (11–16). Because of its estrogenic specificity and preference for the E1-to-E2 reduction reaction, the enzyme is considered to be primarily responsible for E2 biosynthesis in gonads and in peripheral tissues. This enzyme has been proposed to be involved in maintaining high E2 levels found in breast tumors of postmenopausal women (ref. 17, and references therein). A direct correlation between higher concentrations of E2 and onset of breast cancer, especially in postmenopausal women, is well established (ref. 17, and references therein). There are reports of elevated E2/E1-concentration ratios in breast tumors in comparison with the E2/E1 ratio in circulating blood (18). Furthermore, a number of studies appear to indicate higher levels of 17β-HSD1 activities in the outer quadrant of the breast, where tumors most commonly occur (18–20). 17β-HSD1, therefore, poses an attractive target for structure-based rational drug design for the prevention and control of breast tumor growth.
Detailed knowledge of the biochemistry and molecular biology of 17β-HSD1 has grown rapidly in the last 5 years, culminating in the determination of the three-dimensional structure of the human enzyme (21). This has led to an atomic-level description of the E2-binding pocket of the enzyme and understanding of its mechanism of action and the molecular basis for the estrogen specificity of the enzyme (8–10). In addition, complexes of 17β-HSD1 with E2 and/or NADP+ (22, 23), various mutant complexes (24), and structure-function analysis through site-directed substitutions and enzyme chimeras (9, 10) have been published that further clarify the mechanism of action and provide additional detailed insights into the origin of estrogenic/androgenic specificities of the enzyme. 17β-HSD1 belongs to the short-chain dehydrogenases/reductase family (25), requiring a Tyr-X-X-X-Lys motif and a Tyr-Lys-Ser catalytic triad for activity (21, 26).
We present here the structure of an inhibited ternary complex of 17β-HSD1 with NADP+ and 3-hydroxyestra-1,3,5,7-tetraen-17-one (equilin), which shows that the equilin molecule binds at the catalytic site of 17β-HSD1. Equilin is one of the major components of estrogens used in estrogen replacement therapy, along with estrone and 17α-dihydroequilin. These conjugated estrogens are administered under the commercial name Premarin as salts of their sulfate esters, which are subsequently hydrolyzed to free estrogens. Kinetic data presented also supports in vitro inhibition of the enzyme’s E1-to-E2 reduction activity by equilin.
MATERIALS AND METHODS
For crystallization experiments, human 17β-HSD1 was expressed in Sf9 insect cells as described (9, 10). The expressed enzyme was purified by using red-agarose affinity chromatography in the presence of 0.25 mM NADP+ followed by Mono Q (Pharmacia) anion-exchange chromatography. The equilin inhibition assays were performed by transiently expressing the full-length human 17β-HSD1 cDNA in human embryonic kidney 293 cells under the cytomegalovirus (CMV) promoter by using a pCMV6 vector (27). The reductive activity of 17β-HSD1 was measured in cultured cells with 0.2 μM 3H-labeled substrate, using a 2-hr incubation time. To analyze the inhibitory effects of equilin (Steraloids, Wilton, NH) on 17β-HSD1 activity, the compound was added into the reaction mixture at final concentrations of 1 μM and 10 μM. Amounts of the substrate converted were determined from triplicate measurements of at least three independent experiments. Student’s t-test was used to analyze the significance of differences between the activity results obtained with or without competing equilin. The kinetic data are summarized in Table 1.
Table 1.
Kinetic data for inhibition of 17β-HSD1 by Equilin
| Steroid Values | Concentration, μM | Inhibition, % ± SD | P |
|---|---|---|---|
| Equilin | 1 | 77.2 ± 2.1 | 0.0001 |
| 10 | 97.6 ± 0.4 | 0.0001 |
The enzyme was crystallized by vapor diffusion from 28% polyethylene glycol 4000 in Hepes buffer (pH 7.5) containing 1 mM equilin. The space group was P212121 with a = 44.02 Å, b = 114.16 Å, and c = 114.84 Å, with a complete dimer in the asymmetric unit. The data was collected at an R-AxisIIc image-plate detector under cryogenic conditions. The crystal (≈0.1 mm × 0.1 mm × 0.15 mm) was flash-frozen in vapors of liquid nitrogen by using mother liquor as the cryoprotectant. The data was processed by using denzo (28). A total of 28,469 observations were measured for 11,408 unique reflections. The data was 93.1% complete between 99.0 and 3.00-Å resolution. The completion rate and the F2/σF2 value between 3.23 and 3.00 Å were 92% and 1.7, respectively. The overall Rmerge(F2) was 0.091. The structure was solved by the molecular-replacement technique using the program amore (29). Both the monomer and crystallographic dimer (21) were utilized as search models by using integration radii of 27.7 Å for the monomer and 42.0 Å for the crystallographic dimer (21). A number of trials were conducted for both the monomer and crystallographic dimer by using different resolutions (3.5, 4.0, 4.3, and 4.5 Å), all of which yielded the same solution. The optimal solution (all data between 15 and 3.5 Å) using the crystallographic dimer (21) as the search model yielded a definitive solution (correlation coefficient = 62.7, initial R factor of 38.8%). The refinement of the model was carried out to 3.0-Å resolution by using xplor (30) implemented on a Silicon Graphics Indigo2 workstation. The model building and density fitting were done by using chain (31). Annealed omit maps were generated for refitting the ambiguous regions. Noncrystallographic 2-fold symmetry restraints were used in the refinement. A few well-defined water oxygen atoms that were hydrogen-bonded to protein atoms were included in the final stage of refinement. As in the case of the apoenzyme structure, 284 of 327 amino acid residues could be traced and modeled. The missing 43 C-terminal residues do not have any role in catalysis (10). An additional 18 solvent molecules, 2 NADP+ molecules, and an equilin molecule were added. The cofactor molecules were added midway through the refinement, and the solvent oxygens and equilin were included in the final stage of the refinement. Crystallographic R-factor values (Table 2) are comparable to other structures determined at 3.0-Å resolution (32), where it is not uncommon for R and Rfree to differ by >10%. Table 2 provides a summary of data collection, structure determination, and refinement details.
Table 2.
Refinement statistics of 17β-HSD1–equilin complex
| Protein atoms in the model (2 polypeptide chains) | 4,358 |
| NADP+ (2 molecules) | 96 |
| Ligand (one equilin molecule) atoms | 20 |
| Solvent oxygens | 18 |
| Resolution range | 10.0–3.00 Å |
| Unique data used (F > σF) | 9,322 |
| Crystallographic R | 0.182 |
| Rfree | 0.315 |
| RMS deviations | |
| Bond distance, Å | 0.011 |
| Bond angle, ° | 1.65 |
| Dihedral angle, ° | 24.06 |
| Planarity, ° | 1.61 |
| Ramachandran plot statistics | |
| % residues of 234 nonglycine and nonproline in most favored | 82.1 |
| % in disallowed | 0 |
RESULTS AND DISCUSSION
The 17β-HSD1–equilin complex crystallized in space group P212121 with a dimer in the asymmetric unit. This is in contrast to the apo- and wild-type 17β-HSD1–E2 complexes (21–23) in which the dimer was crystallographically related. A homodimer is known to be the functional unit of the enzyme (33). This holo-form represents a true ternary complex of the wild-type enzyme, with the cofactor and a steroidal ligand.
The kinetic data illustrating the ability of equilin to inhibit the E1-to-E2 conversion by 17β-HSD1 is shown in Table 1. A 77% inhibition of normal 17β-HSD1 activity (E1-to-E2) was achieved with 1 μM equilin, establishing it as a potent inhibitor of E2 synthesis.
Ligand-Binding Interactions.
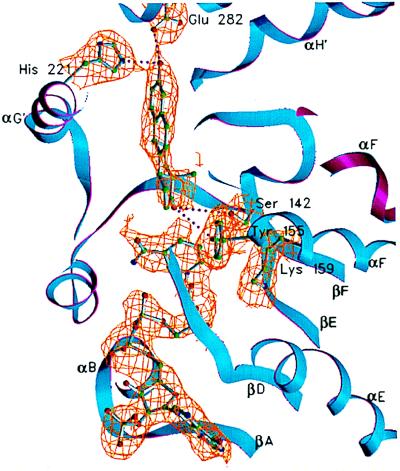
Both equilin and NADP+ have well-defined electron density in the A subunit of the dimeric enzyme. However, the ligand density in the B subunit is poorly defined; therefore, the ligand was not included in the B subunit. The active-site structure of the 17β-HSD1-equilin complex for the A subunit is shown in Fig. 1. The NADP+ and equilin molecules as well as amino acids responsible for binding the ligand are shown in their electron densities. The density for equilin is shown before its inclusion in the protein model. The equilin molecule makes four hydrogen bond contacts with protein atoms. The 17-keto oxygen accepts protons from catalytic residues Tyr-155 and Ser-142 (2.7 and 2.8 Å, respectively) at the catalytic end of the steroid-binding cleft. The 3-hydroxyl group of the ligand makes a bifurcated hydrogen bond to His-221 and Glu-282 (2.9 and 2.9 Å, respectively) at the recognition end of the cleft. With the exception of these two hydrophilic ends, the rest of the steroid-binding cleft almost exclusively is hydrophobic. Residues Val-143, Met-147, Leu-149, Pro-150, Asn-152, and Tyr-218 compose the protein surface in the vicinity of the β-face of the nearly coplanar A-B rings (Fig. 2) of the ligand. Residues Val-225 and Pro-187 are within van der Waals contact distances to the equilin α face, whereas residues Leu-262, Leu-263, and Met-279 are at the floor of the cleft, in the Fig. 1 view. A schematic diagram of the steroid-binding cleft, viewed roughly 90° from Fig. 1 and perpendicular to the β-face of equilin, is shown in Fig. 2. Amino acid side chains involved in hydrogen bond formation with the ligand (as well as those side chains lining the cleft) are depicted. The viewing direction of Fig. 1 is roughly down the putative substrate entry path shown in Fig. 2. In addition to the hydrophobic environment of the cleft described above, Phe-192 and Met-193 from substrate-entry loop (residues 186–201) line the entry path. In the apoenzyme structure, the substrate-entry loop adopts an open conformation providing unrestricted access to the active site cleft (21). Instead, the substrate-entry loop forms a closed conformation in the 17β-HSD1–equilin complex, where the polypeptide chain with residues 186–201 moves toward the catalytic cleft, restricting the access to the active site, as depicted in Fig. 2. In this closed conformation Phe-192 and Met-193 make van der Waals contacts (3.9Å and 4.2 Å, respectively) to the ligand molecule.
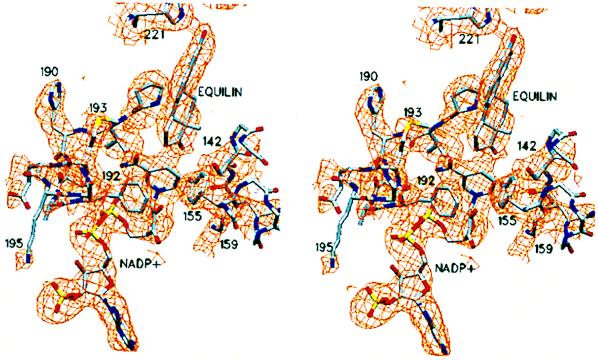
Figure 1.
A view of the active site within the A subunit. NADP+, equilin, and side chains of amino acid residues Ser-142, Tyr-155, and Lys-159 at the catalytic end and His-221 and Glu-282 at the recognition end of the active-site cleft are shown in their 2Fo-Fc electron densities. The equilin molecule was not included in the protein model. The backbone of the A subunit is shown in blue ribbon and that of the B subunit is in magenta. The electron density for the rest of the protein atoms has been excluded by using a 2-Å cutoff distance from the side chain/ligand atoms shown. The visible secondary structure elements are labeled. This figure and Figs. 3–6 were generated by using setor (41).
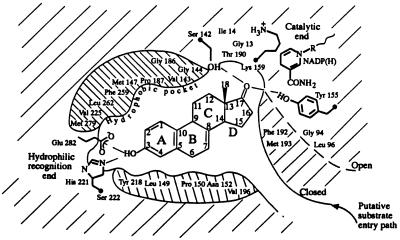
Figure 2.
A schematic diagram of the ligand-binding pocket (viewed roughly perpendicular to the β face of the equilin molecule) derived from the description of the active site provided in the text. The hydrophobic environment depicted above and below the steroid molecule is, in fact, comprised of residues that surround the ligand in the pocket. The open conformation of the substrate-entry path is shown in dashed line. In the ternary complex representing the closed conformation of the entry path, shown in solid line, residues Phe-192 and Met-193 form hydrophobic contacts with the ligand.
In the proposed structure-based hypothesis for the mechanism of hydride transfer (21), an electrophilic attack on the C17-keto oxygen through strong hydrogen-bonding interactions by hydroxyl groups from Tyr-155, Ser-142, or both, as well as correct orientation and proximity of C4 β-hydride of the nicotinamide at the α-face of estrone, are required for the initiation of the transition state of the reaction. Modeling of the transition state with estrone suggested that the hydride transfer should occur through a short distance of about 2 Å between C4 of the nicotinamide ring and C17 of estrone in a direction nearly perpendicular to the planar C17⩵O group of the substrate (21).
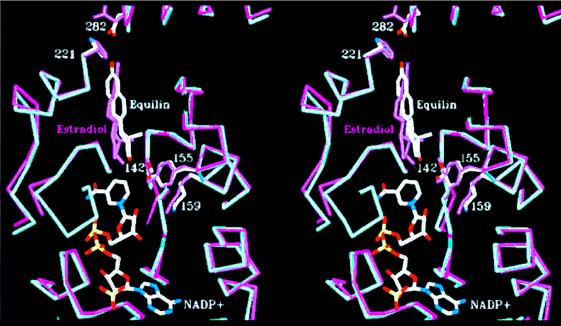
The orientation of C17⩵O of equilin relative to the C4 hydride is more acute (52.7°) than in the above scenario, owing to the differences in puckering of the C-D ring system. The three-dimensional structures of the substrate, estrone, and equilin are strikingly different at the C-D ring systems because of the presence of the C7⩵C8 double bond in equilin. The difference in torsion angle C7-C8-C9-C11 [−179° for estrone (34) and 121° for equilin (35)] caused by the C7⩵C8 double bond results in a 0.9-Å displacement between the C17 carbon atoms. Higher isotropic temperature factors of the atoms of the D-ring of equilin (≈40 Å2) in comparison with those of A-B rings (≈30 Å2) also are suggestive of higher thermal motion at this end. The full effect of these changes on the tertiary structure is illustrated in Fig. 3, in which the structures of both the equilin and estradiol (23) complexes are superimposed by using the main-chain Cα atoms of the catalytic triad (Ser-142, Tyr-155, and Lys-159) and His-221. The distance between the C4 atom of the nicotinamide ring of the cofactor and C17 of equilin is 3.4 Å. However, to evaluate the structural origin of inhibition by equilin, the active site structure of the E1–17β–HSD1 complex should ideally be compared with that of the 17β-HSD1–equilin complex, because E2 is not a substrate, but the product of the catalysis. The origin of the inhibitory property of equilin (from the structural perspective) is therefore a manifestation of its altered C-D-ring structure (and location and orientation of its C17 keto group) with respect to the catalytic machinery at the catalytic end of the active site.
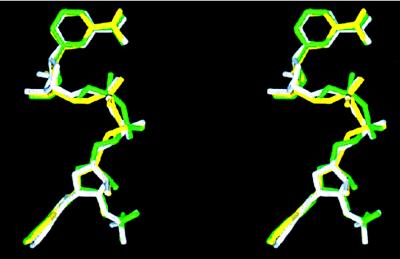
Figure 3.
Least-squares superposition of the structures of ternary complexes with E2 (magenta) and equilin (blue) by using the catalytic triad (Ser-142, Tyr-155, and-Lys 159) and His-221 Cα carbons. E2 and equilin are labeled, and the NADP+ molecule shown is from the equilin structure. The difference in orientation of the two ligand, especially of their B, C, and D rings, is evident.
The Substrate-Entry Loop Structure.
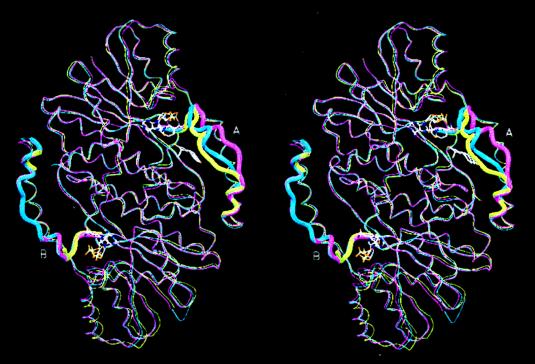
The open and closed conformations of the substrate-entry loop are illustrated in Fig. 4, where the dimeric Cα backbones of the apoenzyme (21), the H221L mutant–NAD complex (24), and the 17β-HSD1–equilin complex are superimposed. The overall tertiary structures are nearly identical except for the substrate-entry loop (shown as thicker cross section of the backbone) between the strand βF and the helix αG′′. The overall rms deviations for 284 Cα atoms of the 17β-HSD1–equilin complex compared with the apo form and the H221L mutant complex are 0.7 Å and 0.8 Å, respectively; the rms deviations for the substrate-entry loop are 3.6 Å (with a maximum of 6.8 Å) at Lys-195 and 1.9 Å (with a maximum of 4.3 Å) at Glu-201, respectively. As shown in Fig. 5, the loop in the A subunit has well defined electron density except for the Lys-195 side chain and packs against both equilin and NADP+ via Phe-192 and Met-193. Both Phe-192 and Met-193 line the substrate-entry path and have van der Waals contacts with the D ring of equilin and the nicotinamide head group of NADP+ (Fig. 5). This closing of the substrate-entry loop effectively traps the ligand in the steroid-binding cleft by occluding the entry path shown in Fig. 2. This combination of an ordered loop and the presence of both the cofactor and ligand is a unique feature of the A subunit of the 17β-HSD1–equilin complex. In contrast, the electron density for the loop in the B subunit is poorly defined after Thr-190. There appear to be multiple densities for His-191, which may be indicative of mixed conformations for the loop structure; therefore, the B loop was modeled by using the apoenzyme structure as a template (21).
Figure 4.
Least-squares superposition of the Cα backbones of the dimers of the 17β-HSD1–equilin complex (blue), the apo-17β-HSD1 (magenta) (21)–E2-complex (yellow) (23) illustrating the differences of the substrate-entry loop structures of the A and B subunits. The substrate-entry loops (residues 186–201) are drawn in wider cross section. The loop in the A subunit of 17β-HSD1–equilin complex is in the closed conformation, whereas that in the B subunit has been modeled after the apo form. NADP+ and equilin of the 17β-HSD1–equilin complex are shown.
Figure 5.
Substrate-entry loop structure of the A subunit with NADP+ and equilin. Residues of the substrate-entry loop are labeled along with the catalytic triad and His-221. The (2Fo-Fc) electron density map contoured at 1σ is shown in orange. The density for all residues before Ser-142 has been removed for clarity.
NADP+-Binding Site and Cofactor Conformation.
The location and overall conformation of the cofactor NADP+ are similar to those in crystal structures of other short-chain dehydrogenases/reductases (36). The bound NADP+ is present in an extended conformation (12.5 Å between rings), with the adenine ring in an anti conformation (χ = −104.6°) and the nicotinamide ring in a syn conformation (χ = 59.1°). This conformation is consistent with the 4-Pro-S hydride transfer from the B face of the nicotinamide ring. Both ribose rings are 2E (C2′-endo) puckered (37). Least-squares superposition of the refined NADP+ of 17β-HSD1–equilin, complex NADP+ from the most recent E2-ternary complex (24), and NAD+ from 3α,20β-HSD (34) (0.65 and 0.67 Å respectively) is shown in Fig. 6. All three cofactors demonstrate similar conformations, consistent with those in other short-chain dehydrogenases/reductase structures (36).
Figure 6.
Least-squares superposition of three cofactor molecules: NADP+ from the E2 ternary structure (green), NADP+ from the 17β-HSD1–equilin structure (white), and NAD+ from the 3α,20β-HSD structure (yellow) illustrating the overall similarity of their conformations.
Specific contacts between the protein and the cofactor NADP+ involve 11 different residues (Ser-11, Ser-12, Ile-14, Arg-37, Leu-64, Val-66, Asn-90, Gly-92, Tyr-155, Thr-190, and Phe-192) as illustrated in Fig. 7. The adenine nucleoside moiety lies in the cleft surrounded by five segments of the polypeptide, βA-to-αB turn (residues 9–12), βB-to-αC turn (residues 36–38), end of βC (residues 64–66), residues 90–93 from βD and Val-113 from αE (Fig. 7). Specific contacts between the protein and adenine moiety include Oγ of both Ser-11 and Ser-12 contacting the O2′-adenine-ribose phosphate group, Leu-64 and Val-66 main-chain atoms interacting with the adenine ring system, and Arg-37 interacting with the O2′-adenine-ribose phosphate allowing for charge neutralization of the 2′ phosphate. One notable interaction that has been observed (24) but is not seen in the present structure is the formation of a salt bridge between Lys-195 and the 2′ phosphate in NADP+.
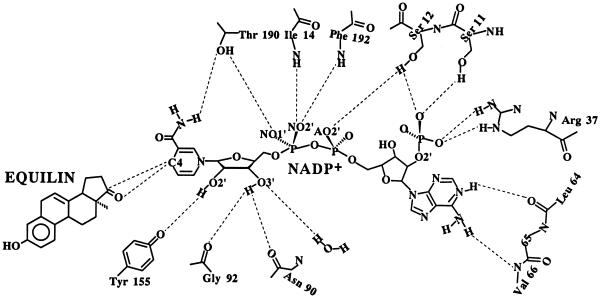
Figure 7.
A schematic diagram showing contacts of NADP+ with protein and equilin atoms. Residues and atoms involved in direct contacts are labeled appropriately.
The pyrophosphate moiety of NADP+ is well defined. Interactions between this region of the cofactor and the protein include Ile-14 and Phe-192 main-chain nitrogens in contact with NO2′, NO1′ with Oγ of Thr-190, and the Oγ of Ser-12 with AO2′ (Fig. 7). Stabilization of the nicotinamide nucleoside moiety is achieved through hydrogen-bonding contacts and hydrophobic interactions. Thr-190 Oγ forms a hydrogen bond with the carbamide group of the nicotinamide ring; Tyr-155 OH interacts with O2′ ribose; Gly-92 and Asn-90 carbonyl groups interact with O3′ ribose; Phe-192 packs against the nicotinamide head group; and C17 of equilin is within van der Waals distance (3.6 Å) of C4 of the nicotinamide ring.
Quaternary Association: The Q Axis Dimer.
The dimer formation is achieved through the interactions of αE and αF, yielding a four-helix bundle nearly perpendicular to the pseudo-2-fold symmetry axis, a highly conserved mode of quaternary association in short-chain dehydrogenases/reductases (36). The rms deviation between the A- and B-subunits’ 284 backbone Cα atoms is 0.4 Å, similar to the estimated random positional error at this resolution. The accessible surface areas of the monomer and dimer of the ternary complex are 12,498 and 20,124 Å2 respectively. Total subunit surfaces involved in the formation of the Q axis dimer amount to 19.4% (4,872 Å2) as opposed to 17.8% of the surface of 3α,20β-HSD (3,415 Å2) (34) used for dimer formation. The discrepancy in initial surface area involved in Q axis dimer formation is caused by the interaction of helices αHs (residues 272–284) of 17β-HSD1 between subunits. Of the total Q axis dimer interface of 17β-HSD1, 55.7% is hydrophobic in nature, which represents a 4% reduction of the hydrophobic surface area on dimerization.
The Q axis dimer formation is dictated primarily by hydrophobic interaction. This conclusion is supported by the fact that 4 leucines and 4 valines from αE (Leu-102, -111, -122, and -126 and Val-107, -110, -115, and -119) and 5 leucines, 2 valines, and 2 phenylalanines from αF (Leu-162, -165, -169, -172, and -173, Val-154 and -178, and Phe-151 and -176) are present at the dimer interface. Of these 17 hydrophobic interactions involved in dimer formation, 9 are conserved between 17β-HSD1 and 3α,20β-HSD. In addition, there are charge and/or polar-group interactions between the two subunits; αE has four interactions between subunits (Glu-100 Oɛ2–Lys-130 NZ, Glu-104 Oɛ1–Gln-123 Nɛ2, Oɛ1, and –Gln-123 Oɛ1, and Glu-104 Oɛ2–Arg-120 NH1) and αF has two (Leu-149 N–Ser-168 Oγ and Asp-153 Oδ1–Leu-169 N).
Several interesting aspects of the dimeric enzyme model are worth mentioning, including differential electron densities between the A and B subunits. Differences in electron densities between the two subunits are isolated to the ligand and extended-loop regions. No notable changes are observed in the remainder of the structure as indicated by an rms deviation between the Cα carbons of the two subunits of 0.4 Å. The difference in ligand density observed between the A and B subunits is consistent with the fact that the substrate-entry loop in the A subunit is well ordered and closes over the cofactor and ligand, whereas in B it remains disordered as in the apo-structure (21). The structural discrepancy between the two subunits may explain why the 17β-HSD1–equilin complex crystallized with a dimer in the asymmetric unit (as opposed to a monomer in the apoenzyme structure). The combined differences in ligand occupancy and loop structure between the A and B subunits may be suggestive of the existence of a negative cooperativity within the dimer, an issue that has not yet been addressed in the existing literature.
Concluding Remark.
17β-HSD1 poses an attractive target for structure-based rational drug design for the prevention and control of breast tumor growth. The tertiary structures of the enzyme and enzyme–substrate complex have yielded information about structural determinants for catalysis and substrate recognition. Kinetic data and the structure of the 17β-HSD1–equilin complex with NADP+ and an equine estrogen, equilin, presented here reveal the molecular basis for inhibition of the enzyme, as opposed to its E1-to-E2 catalysis, by a natural estrogen that closely resembles the substrate. This result clearly demonstrates that in vitro, equilin is a specific inhibitor of the biosynthesis of 17β-estradiol through its binding to human 17β-HSD1, in competition with E1. Although increased levels of 17β-estradiol have been linked to the stimulation of cell proliferation and increased risk of breast cancer, epidemiological studies have failed to correlate unequivocally the use of estrogen replacement therapy with increased incidences of breast cancer or deaths from breast cancer. On the contrary, the recent Cancer Prevention Study II (a large prospective cohort of 422,373 postmenopausal women) by the American Cancer Society revealed a 16% reduction in the relative risk of death from breast cancer in women who had ever used estrogen replacement therapy (38). Other smaller studies in the past have drawn similar conclusions (39, 40). Structural and kinetic data presented here raise the possibility of in vivo inhibition of the estrogen activation process by equilin administered in the form of estrogen replacement therapy and subsequent lowering of the E2 levels.
Acknowledgments
We thank Dr. Walter Pangborn for data collection and Ms. Melda Tugac and Ms. Gloria DelBel for their excellent assistance with illustrations. This work is partially supported by the Roswell Alliance Foundation, by Grant DK26546 from the National Institutes of Health, and by the Research Council for Health of the Academy of Finland and the Technology Development Center of Finland.
ABBREVIATIONS
- HSD
hydroxysteroid dehydrogenase
- 17β-HSD1
17-β HSD type 1
- E1
estrone
- E2
17β-estradiol, equilin, 3-hydroxyestra-1,3,5,7-tetraen-17-one
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 1EQU).
References
- 1.Ryan K J, Engel L L. Endocrinology. 1953;52:287–291. doi: 10.1210/endo-52-3-287. [DOI] [PubMed] [Google Scholar]
- 2.Peltoketo H, Isomaa V, Mäentausta O, Vihko R. FEBS Lett. 1988;239:73–77. doi: 10.1016/0014-5793(88)80548-9. [DOI] [PubMed] [Google Scholar]
- 3.Luu-The V, Lachance Y, Labrie C, Leblanc G, Thomas J L, Strickler R C, Labrie F. Mol Endocrinol. 1989;3:1301–1309. [Google Scholar]
- 4.Wu L, Einstein M, Geissler W M, Chan H K, Elliston K O, Andersson S. J Biol Chem. 1993;268:12964–12969. [PubMed] [Google Scholar]
- 5.Geissler W M, Davis D L, Wu L, Bradshaw K D, Patel S, Mendonca B B, Elliston K O, Wilson J D, Russel D W, Andersson S. Nat Genet. 1994;7:34–39. doi: 10.1038/ng0594-34. [DOI] [PubMed] [Google Scholar]
- 6.Adamski J, Normand T, Leenders F, Monte D, Begue A, Stehelin D, Jungblut P W, Launoit Y. Biochem J. 1995;311:437–443. doi: 10.1042/bj3110437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deyashiki Y, Ohshima K, Nakanishi M, Sato K, Hara A. J Biol Chem. 1995;270:10461–10467. doi: 10.1074/jbc.270.18.10461. [DOI] [PubMed] [Google Scholar]
- 8.Poutanen M, Miettinen M, Vihko R. Endocrinology. 1993;133:2639–2644. doi: 10.1210/endo.133.6.8243287. [DOI] [PubMed] [Google Scholar]
- 9.Puranen T, Poutanen M, Ghosh D, Vihko P, Vihko R. Mol Endocrinol. 1997;11:77–86. doi: 10.1210/mend.11.1.9872. [DOI] [PubMed] [Google Scholar]
- 10.Puranen T, Poutanen M, Ghosh D, Vihko R, Vihko P. Endocrinology. 1997;138:3532–3539. doi: 10.1210/endo.138.8.5303. [DOI] [PubMed] [Google Scholar]
- 11.Fournet-Dulguerov N, MacLusty N J, Leranth C Z, Todd R, Mendelson C R, Simpson E R, Naftolin F. J Clin Endocrinol Metab. 1987;65:757–764. doi: 10.1210/jcem-65-4-757. [DOI] [PubMed] [Google Scholar]
- 12.Mäentausta O, Sormunen R, Isomaa V, Lehto V P, Jouppila P, Vihko R. Lab Invest. 1991;65:582–587. [PubMed] [Google Scholar]
- 13.Dupont E, Labrie F, Luu-The V, Pelletier G. J Histochem Cytochem. 1991;39:1403–1407. doi: 10.1177/39.10.1940311. [DOI] [PubMed] [Google Scholar]
- 14.Ghersevich S A, Poutanen M H, Martikainen H K, Vihko K. J Endocrinol. 1994;143:139–150. doi: 10.1677/joe.0.1430139. [DOI] [PubMed] [Google Scholar]
- 15.Mäentausta O, Boman K, Isomaa V, Stendahl U, Backstrom T, Vihko R. Cancer. 1992;70:1551–1555. doi: 10.1002/1097-0142(19920915)70:6<1551::aid-cncr2820700618>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 16.Poutanen M, Isomaa V, Lehto V P, Vihko R K. Int J Cancer. 1992;50:386–390. doi: 10.1002/ijc.2910500310. [DOI] [PubMed] [Google Scholar]
- 17.Poutanen M, Isomaa V, Peltoketo H, Vihko R. J Steroid Biochem Mol Biol. 1995;55:525–532. doi: 10.1016/0960-0760(95)00201-4. [DOI] [PubMed] [Google Scholar]
- 18.James V H T, McNeil J M, Beranek P A, Bonney R C, Reed M J. J Steroid Biochem. 1986;25:787–790. doi: 10.1016/0022-4731(86)90309-2. [DOI] [PubMed] [Google Scholar]
- 19.Adams E F, Newton C J, Tait G H, Braunsberg H, Reed M J, James V H T. Int J Cancer. 1988;42:119–122. doi: 10.1002/ijc.2910420122. [DOI] [PubMed] [Google Scholar]
- 20.James V H T, Reed M J, Coldham N G. Ann NY Acad Sci. 1990;595:227–235. doi: 10.1111/j.1749-6632.1990.tb34296.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghosh D, Pletnev V Z, Zhu D W, Wawrzak Z, Duax W L, Pangborn W, Labrie F, Lin S X. Structure. 1995;3:503–513. doi: 10.1016/s0969-2126(01)00183-6. [DOI] [PubMed] [Google Scholar]
- 22.Azzi A, Rehse P H, Zhu D W, Campbell R L, Labrie F, Lin S X. Nat Struct Biol. 1996;8:665–668. doi: 10.1038/nsb0896-665. [DOI] [PubMed] [Google Scholar]
- 23.Breton R, Housset D, Mazza C, Fontecilla-Camps J C. Structure. 1996;4:905–915. doi: 10.1016/s0969-2126(96)00098-6. [DOI] [PubMed] [Google Scholar]
- 24.Mazza C, Breton R, Housset D, Fontecilla-Camps J C. J Biol Chem. 1998;273:8145–8152. doi: 10.1074/jbc.273.14.8145. [DOI] [PubMed] [Google Scholar]
- 25.Jörnvall H, Persson B, Krook M, Atrian S, Gonzalez-Duarte R, Jeffrey J, Ghosh D. Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh D, Wawrzak Z, Weeks C M, Duax W L, Erman M. Structure. 1994;2:629–640. doi: 10.1016/s0969-2126(00)00064-2. [DOI] [PubMed] [Google Scholar]
- 27.Miettinen M M, Mustonen M V J, Poutanen M H, Isomaa V V, Vihko R K. Biochem J. 1996;314:839–845. doi: 10.1042/bj3140839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Otwinowski Z, Minor W. The hkl Manual. New Haven, CT: Yale Univ. Press; 1995. [Google Scholar]
- 29.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 30.Brunger A T. xplor version 3.1: A System for X-Ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. [Google Scholar]
- 31.Jones T A. J Appl Crystallogr. 1978;11:268–272. [Google Scholar]
- 32.Kleywegyt G J, Brunger A T. Structure. 1996;4:897–904. doi: 10.1016/s0969-2126(96)00097-4. [DOI] [PubMed] [Google Scholar]
- 33.Lin S X, Yang F, Jin J Z, Breton R, Zhu D W, Luu-The V, Labrie F. J Biol Chem. 1992;267:16182–16187. [PubMed] [Google Scholar]
- 34.Busetta B, Courseille C, Hospital M. Acta Crystallogr B. 1973;29:298–313. [Google Scholar]
- 35.Sawicki, M. W., Li, N. & Ghosh, D. (1999) Acta Crystallogr. C., in press. [DOI] [PubMed]
- 36.Duax W L, Griffin J F, Ghosh D. Curr Opin Struct Biol. 1996;6:813–823. doi: 10.1016/s0959-440x(96)80012-1. [DOI] [PubMed] [Google Scholar]
- 37.IUPAC-IUB Joint Commission on Biomedical Nomenclature. Eur J Biochem. 1983;131:9–15. doi: 10.1111/j.1432-1033.1983.tb07225.x. [DOI] [PubMed] [Google Scholar]
- 38.Eden J A, Wren B G. Cancer Treat Rev. 1996;22:335–343. doi: 10.1016/s0305-7372(96)90006-7. [DOI] [PubMed] [Google Scholar]
- 39.Willis D B. Maturitas. 1997;27:105–108. doi: 10.1016/s0378-5122(97)00028-5. [DOI] [PubMed] [Google Scholar]
- 40.Hunt K, Vessey M, McPherson K. Br J Obstet Gynaecol. 1990;97:1080–1086. doi: 10.1111/j.1471-0528.1990.tb02494.x. [DOI] [PubMed] [Google Scholar]
- 41.Evans S V. J Mol Graphics. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]