Abstract
After invasion of erythrocytes, the human malaria parasite Plasmodium falciparum resides within a parasitophorous vacuole and develops from morphologically and metabolically distinct ring to trophozoite stages. During these developmental phases, major structural changes occur within the erythrocyte, but neither the molecular events governing this development nor the molecular composition of the parasitophorous vacuole membrane (PVM) is well known. Herein, we describe a new family of highly cationic proteins from P. falciparum termed early transcribed membrane proteins (ETRAMPs). Thirteen members were identified sharing a conserved structure, of which six were found only during ring stages as judged from Northern and Western analysis. Other members showed different stage-specific expression patterns. Furthermore, ETRAMPs were associated with the membrane fractions in Western blots, and colocalization and selective permeabilization studies demonstrated that ETRAMPs were located in the PVM. This was confirmed by immunoelectron microscopy where the PVM and tubovesicular extensions of the PVM were labeled. Early expressed ETRAMPs clearly defined separate PVM domains compared with the negatively charged integral PVM protein EXP-1, suggesting functionally different domains in the PVM with an oppositely charged surface coat. We also show that the dynamic change of ETRAMP composition in the PVM coincides with the morphological changes during development. The P. falciparum PVM is an important structure for parasite survival, and its analysis might provide better understanding of the requirements of intracellular parasites.
INTRODUCTION
Plasmodium falciparum, the causative agent of the most dangerous form of human malaria, provokes >200 million clinical attacks and kills 1–3 million people each year. A vaccine against malaria is still not available and resistance to drugs is widespread. Improvement of this situation is unlikely to occur without an increased understanding of many aspects of P. falciparum biology that could reveal new intervention targets.
The symptoms of malaria are caused by the asexual development of the parasite within red blood cells (RBCs). Encompassed in a parasitophorous vacuolar membrane (PVM), the parasites develop from ring stages (0–22 h postinvasion [hpi]) to trophozoites (22–36 hpi) and finally to schizonts (36–48 hpi). Rupture of schizonts releases up to 24 merozoites into the bloodstream, which initiate a new round of schizogony. Human erythrocytes are highly specialized cells devoid of internal organelles and a functional protein-trafficking system. This metabolically inert cell allows the parasite to hide from the immune system. As a trade-off, the parasite needs to refurbish the host cell to import nutrients, dispose of waste products, and export proteins across its plasma membrane (PPM), the surrounding PVM, and the erythrocyte cytosol and plasma membrane. Parasite-induced modifications in the host cell are believed to mediate these tasks. A tubovesicular network extends from the PVM into the cytoplasm of trophozoite-infected RBCs (Elmendorf and Haldar, 1993, 1994). In addition, flattened vesicular structures (Maurer's clefts) normally running parallel to and just beneath the red cell membrane occur in the host cell cytosol of late ring stage-infected erythrocytes (Langreth et al., 1978). Maurer's clefts were proposed to be involved in transport of parasite proteins to the RBC plasma membrane (Barnwell, 1990; Hinterberg et al., 1994) and are associated with P. falciparum homologs of proteins involved in vesicle transport (Albano et al., 1999a; Adisa et al., 2001). Recently, they also have been shown to be involved in transport and possibly assembly of proteins forming knobs (Wickham et al., 2001). New permeation pathways occur at >15 hpi in the host cell membrane (Staines et al., 2001), and several parasite proteins become associated with the RBC cytoskeleton (reviewed in Cooke et al., 2001). Appearance of these modifications at the late ring to early trophozoite stage coincides with onset of rapid parasite growth and sequestration in postcapillary venules in vivo. To date, little is known about the molecular events taking place in the preceding ring stage. Compared with later stage parasites, ring stages are characterized by low metabolic and biosynthetic activity (Zolg et al., 1984; de Rojas and Wasserman, 1985; ter Kuile et al., 1993) and little change in size and morphology. However, it is unlikely that the parasite lies dormant during half of its asexual development in RBCs, and we assumed that this “lag”-phase serves the parasite to induce the elaborate host cell modifications apparent in later stages. This initial host cell refurbishment is necessary for growth and survival and must include a protein-trafficking system to deliver the required components beyond the parasite's boundaries into different locations of the host cell. Apart from the intriguing cell biological aspects, this unique situation most probably demands unusual processes essential for parasite survival. These might be sufficiently different from host processes to present new targets for interventions that would leave the host unaffected.
We have used suppression subtractive hybridization to clone genes exclusively transcribed during the P. falciparum ring stage (Spielmann and Beck, 2000). In contrast to genes originating from a trophozoite-specific library, few of the identified ring-specific genes showed homologies to known genes of other organisms, which is in accordance with the unique nature of the molecular events in early stages. One of these genes has previously been shown to code for a protein located in Maurer's clefts and was proposed to bind the erythrocyte scaffold (Blisnick et al., 2000). Among the other ring stage-specific genes, we identified three members of a new gene family coding for highly charged putative membrane proteins we referred to as early transcribed membrane proteins (etramp) (Spielmann and Beck, 2000). Herein, we report on the complete identification of 13 different etramps. Expression of six ETRAMPs was highly ring stage specific, whereas the others showed a different developmental regulation. ETRAMPs localized to the PVM, defining domains distinct to the distribution of the PVM protein EXP-1. Furthermore, we show that the composition of individual ETRAMP members in the PVM changes in a developmentally regulated manner. This change occurs at the transition from ring stage to trophozoite and correlates with appearance of parasite proteins in the RBC cytoplasm and onset of rapid parasite growth. Our findings provide new insights about the Plasmodium PVM and cell biology of intracellular pathogens in general. We suggest that ETRAMPs play an important role in parasite survival and might represent new targets for drug-mediated interventions.
MATERIALS AND METHODS
Identification of etramps and Sequence Analysis
The RsaI cDNA fragments of three different etramps (clones R4b, R1.10, and R20; Spielmann and Beck, 2000) were used to deduce the complete open reading frames (ORFs) from the P. falciparum genome project with the program BlastN (Altschul et al., 1990; http://www.tigr.org; http://www.sanger.ac.uk/Projects/P_falciparum/; http://sequence-www.stanford.edu/group/malaria/). The predicted amino acid (aa) sequences were used to identify additional related sequences in the P. falciparum genome by using tBLASTN on the same Web sites and on the National Center for Biotechnology Information custom BLAST server (http://www.ncbi.nlm.gov/Malaria/plasmodiumblcus.html). Chromosomal organization of etramps was determined using PlasmoDB (http://plasmodb.org/, release 3.2, 30.11.01; Bahl et al. 2002). We thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P. falciparum (3D7) public before publication of the completed sequence. The Sanger Center (Cambridge, United Kingdom) provided sequence for chromosomes 1, 3–9, and 13, with financial support from the Wellcome Trust. A consortium composed of The Institute for Genome Research, along with the Naval Medical Research Center (Baltimore, MD), sequenced chromosomes 2, 10, 11, and 14, with support from National Institute of Allergy and Infectious Diseases/National Institutes of Health, the Burroughs Wellcome Fund, and the Department of Defense. The Stanford Genome Technology Center (Palo Alto, CA) sequenced chromosome 12, with support from the Burroughs Wellcome Fund. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania (Philadelphia, PA) and Monash University (Melbourne, Australia), supported by the Burroughs Wellcome Fund.
The following programs available at http://www.expasy.ch were used for etramp sequence analysis: compute pI/MW tool (Bjellqvist et al., 1993), SignalP version 2.0 (Nielsen et al., 1997), TMHMM (Krogh et al., 2001; Moller et al., 2001), ClustalX at EBI European Bioinformatics Institute (Oxford, United Kingdom) (Thompson et al., 1994), Coils (Lupas et al., 1991), Paircoil (Berger et al., 1995), and PESTfind (Rogers et al., 1986). PC gene version 6.7 was used to plot a dendrogram.
Parasite Culture
P. falciparum strain 3D7 was cultured as described previously (Trager and Jensen, 1978) by using 0.5% AlbuMAX (Invitrogen, Groningen, Switzerland) as a substitute of human serum (Dorn et al., 1995). Parasites were synchronized with 5% sorbitol (Lambros and Vanderberg, 1979). One cycle before harvest, already synchronously growing parasites were synchronized with two sorbitol treatments 12 h apart to obtain highly synchronous parasites.
Northern Analysis
Templates to prepare specific probes were generated by polymerase chain reaction (PCR) on 3D7 DNA with primers listed in Table 1, except for etrampBLOB.1 and 11.2. For etrampBLOB.1, clone R10 (Spielmann and Beck, 2000) was used as a template. For etramp11.2, a clone obtained from a pool of four different ETRAMP sequences generated with degenerate primers (5′-ATGAAARTYWCAARGATYTYRTWTTTY-3′ and 5′-GCWAMASCNGARGCWAYRGMAGWR-3′) was used as a template. PCR products were purified using NucleoSpin columns (Macherey-Nagel, Oesingen, Switzerland) and labeled using [α-32P]dCTP and the HighPrime system (Roche Diagnostics, Rotkreuz, Switzerland).
Table 1.
Accession numbers and primers used for amplification of etramps
| etramp name | Accession no. | PlasmoDB v4 identification | Oligonucleotides used (5′-3′) |
|---|---|---|---|
| etramp2 | D71622a | PFB0120w | fw ATTTACATAAAAATAACAAGGGAGACb |
| rv TTAGGCTCTGTACATGGTACTTGAb | |||
| etramp4 | AJ420674c | PFD1120c | fw CTTAGTAAATCTAATAAAAAACCAGAAGb |
| NP_702882a | rv AACAGTAGTGGGTACTGCTGTAGb | ||
| etrampBLOB.1 | N98151d | PFE1590w | |
| AJ290927 | |||
| CAD51676a | |||
| etrampBLOB.2 | CAD51079ah | MAL8P1.6h | fw CTTGTACGTGTAAAAAGGATTATGG |
| rv AAGCCAACTTTACATCTGTATTTCC | |||
| etramp10.1 | AJ420670e | Pf10_0019 | fw TATAAGAATCATAAATCCGATAACAAAGb |
| AJ420672c | rv AGCTTTAGCAGTTAAAAGTGGTGTGCb | ||
| NP_700493a | |||
| etramp10.2 | AJ420673c | Pf10_0323 | fw TATATGAAAAAGAAAAATGTTGACTCTGb |
| CAD51676a | rv TTCTTTTCTGACTCTTGGTGTGGb | ||
| etramp10.3 | NP_700638ai | Pf10_0164i | fw TTAGGATGTGTTTTAACTCTTAACC |
| rv CCAGCTCTTATATCCTAAACTTCC | |||
| etramp11.1 | AJ290926f | Pf11_0039 | fw ATGAAAGTCACAAGGATTTTATATTTC |
| rv CTATGCTGGAGTTTTCTTTACTACTTGTC | |||
| etramp11.2 | AJ420671e | Pf11_0040 | |
| NP_700905a | |||
| etramp12 | NP_701751a | PFL1945c | fw AGTGTTTGTAACGAAAATGTTGAGG |
| rv TTAAGGTGAGGTGATCTGGAAGC | |||
| etramp13 | CAB93662g | Pf13_0012 | fw TATGGATATAAAAAATCGCGAGAAGb |
| CAB93664g | rv GCATATATTCCATGTTATTTACATGAGb | ||
| etramp14.1 | AJ290924f | Pf14_0016 | fw ATGAAAGTTTCTAATATTTTATTC |
| rv TTAAGCTGTTGTGGCTGGG | |||
| etramp14.2 | NP_702618ak | Pf14_0729k | fw GGAAATGTATGCAATGAATTAAAGG |
| rv TCTATATCCTTACCAACTTCATCC |
Published in Gardner et al. (1998, 2002).
Used to generate constructs for recombiant GST fusion protein expression.
gDNA containing partial ORF.
Published as antigen 22 (Horii et al., 1988).
cDNA containing partial ORF PCR amplified with degenerate primers.
cDNA containing complete ORF.
Published as OrfP (Sallicandro et al, 2000).
This annotation differs from our prediction in a second exon at the N-terminus leading to absence of a signal peptide.
This annotation differs from our prediction in a second small exon.
This annotation differs from our prediction in seven amino acids missing at the N-terminus, resulting in a disruption of the signal peptide.
Northern blotting was done as described previously (Spielmann and Beck, 2000). Total RNA was prepared using TRIzol reagent (Invitrogen) and separated on 1.4% agarose gels containing 5 mM guanidinium isothiocyanate (Goda and Minton, 1995; Kyes et al., 2000). Equal loading (∼5 μg/lane) and quality of RNA were checked visually. The gel was soaked in 10× SSC for 30 min and transferred to Hybond XL membranes (Amersham Biosciences, Dübendorf, Switzerland) for 3 h with 10× SSC containing 10 mM NaOH with a vacuum blotter (Appligene Oncor, Basel, Switzerland). Hybridization was carried out at 42°C in UltraHyb (Ambion, Lugano, Switzerland) overnight. Filters were washed with high stringency (0.1× SSC/0.1%SDS) at 50°C. Autoradiography was done at −70°C by using a Transcreen HE enhancer and MS films (Eastman Kodak, Rochester, NY).
Expression of Recombinant ETRAMP-GST Fusion Proteins in Escherichia coli and Immunization
The sequences coding for the C terminus of etramp2 (nucleotide [nt] 211–318), etramp4 (nt 226–426), etramp10.1 (nt 211–321), etramp10.2 (nt 232-1065), and etramp13 (nt 250–555, partial C terminus only) were PCR amplified using primers listed in Table 1 and cloned into the SmaI site 3′ of the glutathione S-transferase (GST) in pGEX-6P-2 (Amersham Biosciences) by cycle restriction ligation (Push et al., 1997). Constructs were confirmed by sequencing. GST fusion proteins were expressed in E. coli BL21 cells and purified using glutathione-Sepharose (Amersham Biosciences).
Mice were immunized with a total of three injections 10–14 d apart, each containing 10 μg of recombinant protein in RIBI adjuvant (Corixa, Seattle, WA).
Preparation of Parasite Protein Extracts
For total parasite extracts, parasites were released from RBCs by lysis with 0.03% saponin for 20 min on ice, washed in phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4), and resuspended in Laemmli sample buffer.
Triton X-114 phase separation (Bordier, 1981) was performed according to Smythe et al. (1988) with modifications. Ten milliliters of parasite culture (10% parasitemia) was saponin treated to release parasites from RBCs, washed in human tonicity phosphate-buffered saline (HTPBS; 137 mM NaCl, 2.7 mM KCl, 8.1 mM sodium phosphate, pH 7.2), resuspended in 0.7 ml of 1× HTPBS containing 0.5% Triton X-114 and 1 mM phenylmethylsulfonyl fluoride, and stored at −20°C until use. After thawing, the extract was kept on ice for 30 min with intermittent mixing. Twenty microliters was removed and Laemmli sample buffer was added. The remaining extract was spun at 15,000 × g for 20 min to pellet insoluble material. The insoluble fraction was washed three times in HTPBS containing 0.5% Triton X-114 and resuspended in Laemmli sample buffer. The supernatant, containing the detergent-soluble proteins, was layered over a 0.5-ml ice-cold sucrose cushion (6% sucrose, 0.06% Triton X-114), incubated at 37°C for 5 min, and centrifuged at 500 × g for 5 min at room temperature. The resulting three phases were treated as follows: 1) the detergent-depleted upper layer was collected, Triton X-114 added to 0.5%, and the depletion step repeated; 2) the sucrose cushion was discarded; and 3) the detergent pellet, containing the membrane proteins, was resuspended in 0.5 ml of HTPBS, the purification over a sucrose cushion was repeated, and the pellet resuspended in 0.5 ml of HTPBS. Both the final detergent-depleted fraction (1) and the final detergent fraction (3) were precipitated with trichloroacetic acid and analyzed by SDS-PAGE (Laemmli, 1970).
Saponin-lysed and trypsinized infected red blood cells (IRBCs) were obtained as described by Ansorge et al. (1997). Five milliliters of saponin-lysed parasite culture (5–10% parasitemia) was washed and resuspended in 500 μl of RPMI 1640 medium either with or without 1 mg/ml trypsin and incubated at 37°C for 30 min. Cells were spun at 2000 × g and resuspended in PBS containing 2 mg/ml trypsin inhibitor and incubated for 15 min at 37°C. Cells were washed, resuspended in 10 mM Tris-HCl pH 8.0, and resolved by SDS-PAGE.
Western Analysis
Western analysis was carried out according to standard procedures (Ausubel et al., 1989) with the following modifications. Methanol was omitted from all buffers because of the sensitivity of the epitope. Proteins were transferred to a porablot 0.2-μm polyvinylidene difluoride membrane (Macherey-Nagel, Oesingen, Switzerland) in 10 mM CAPS pH 10.8 for 3 h by using a Trans-Blot semidry electroblotter (Bio-Rad, Reinach, Switzerland). Blots were washed with 1× PBS, and antibody incubation and blocking were done in 1× PBS containing 1% bovine serum albumin (BSA). Dilutions of mouse antisera (containing 50% glycerol) were as follows: ETRAMP2, 1/300; ETRAMP10.1, 1/350; ETRAMP4, 1/400; ETRAMP10.2, 1/250; monoclonal anti-GAPDH antibody, 1/120; and anti-rabbit EXP-1 serum, 1/400. The EXP-1 serum (raised against the C terminus of EXP-1) was a kind gift of Dr. Lingelbach (Phillips-Universität, Marburg, Germany). The monoclonal antibody against GAPDH (purified B-cell supernatant) was a kind gift of Dr. Daubenberger (Swiss Tropical Institute, Basel, Switzerland). Alkaline phosphatase-conjugated anti-mouse IgG (Sigma, Buchs, Switzerland) diluted 1/2000 was used as secondary antibody.
Indirect Immunofluorescence Assay
Ten-well, 8-mm slides (Bio-Microtech, Bolton, ON, Canada) were coated for 30 min with 20 μl of concanavalin A (0.5 mg/ml in distilled H2O; Sigma), and IRBCs in PBS were added to the wells for 20 min. Unbound IRBCs were washed off. For selective permeabilization, 0.01% saponin or 0.1% Triton X-100 was added to the wells for 30 min at 4°C either in PBS (no fixation), in PBS/0.1% formaldehyde (mild fixation), or in PBS/0.5% formaldehyde (increased fixation). For streptolysin O (SLO; Sigma) treatment 20 U of SLO was added in PBS and incubated for 15 min, and then wells were washed three times and fixed if required. Control wells were treated with PBS, 0.1 or 0.5% formaldehyde in PBS only. For complete fixation, bound IRBCs were air-dried and subsequently fixed with 1% formaldehyde or 100% acetone for 15 min at room temperature, followed by three washes with PBS. Wells were blocked for 15 min in PBS/1% BSA, and primary antibody was added in blocking solution for 1 h. After five washes with PBS/1% BSA, second antibody was added in PBS/1% BSA for 1 h and washed again five times. Mouse sera containing 50% glycerol were used at 1/120 (anti-ETRAMP2), 1/150 (anti-ETRAMP10.1), 1/200 (anti-ETRAMP4), and 1/75 (anti-ETRAMP10.2). Rabbit sera were kindly provided by Dr. Lingelbach and used at 1/400 (anti-aldolase and anti-SERP) and 1/200 (anti-EXP-1C). Fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (0.5 mg/ml; KPL, Gaithersburg, MD) was diluted 1/300, Cy3-conjugated goat anti-rabbit IgG (Jackson Immunoresearch Laboratories, West Groove, PA) 1/500, and Texas Red-conjugated goat anti-rabbit IgG (Southern Biotechnology Associates, Birmingham, AL) 1/400. Parasite nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) (5 μg/ml) added to the secondary antibody solution. Slides were analyzed by light microscopy or with a TCS SP confocal laser scanning microscope (Leica, Wetzlar, Germany).
Secondary antibody alone and uninfected RBCs were used as negative controls and were always unlabeled. There was also no signal with preimmune sera or with a serum raised against GST alone. Competition immunofluorescence assays (IFAs) were done as described above, except that 15 min before addition to a slide, primary antisera were diluted in blocking solution containing 1 mg/ml recombinant ETRAMP-GST fusion protein or GST alone.
Immunoelectron Microscopy
A preembedding protocol was used. Unsynchronized infected red blood cells were saponin-permeablized and then resuspended in the primary antibody (either anti-ETRAMP2 or anti-ETRAMP4 serum containing 50% glycerol) with a dilution of 1:40 in PBS for 1 h. The samples were then washed in buffer and resuspended in anti-mouse Ig conjugated to 5-nm gold with a dilution of 1:50 in PBS for 1 h. After washing, the samples were fixed in 2% glutaraldehyde and processed for electron microscopy by routine techniques. This involved postfixation in osmium tetroxide, dehydration in ethanol, treatment with propylene oxide, and embedding in Spurrs's epoxy resin. Thin sections were stained with uranyl acetate and lead citrate before examination in a 1200EX electron microscope (JOEL, Tokyo, Japan).
RESULTS
Identification and General Structure of etramps
Using three previously identified etramp sequences (accessions AJ290924, AJ290926, and AJ290927; Spielmann and Beck, 2000), we identified 10 additional sequences from the P. falciparum genome project with similarities to the original etramps. Thus, we have identified a new gene family comprising 13 related open reading frames of 91–355 amino acids, hereafter referred to as “etramp ” with the extension giving the chromosome number they were found on (Table 1). Part of etrampBLOB1 has previously been published as antigen22 (Horii et al., 1988), and part of etramp13 has been published as OrfP (Sallicandro et al., 2000).
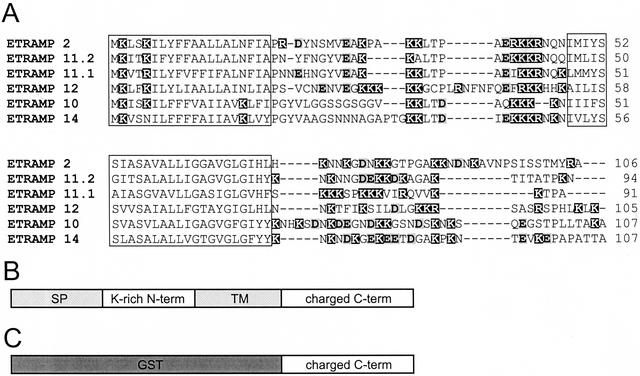
All etramps were predicted to be encoded by a single exon. All members of the ETRAMP family share the same general structure (Figure 1B) with a predicted N-terminal signal peptide, a short (∼20 aa) cationic domain followed by a predicted transmembrane domain (TM), and a highly charged C-terminal domain of variable length (22–280 aa). Amino acid sequence alignment of the six most similar ETRAMPs is shown in Figure 1A. The large number of positively charged residues in etramp coding regions results in a calculated pI of 9–10.4, except for ETRAMPBLOB.1, which is rich in negative residues with a calculated pI of 5.3. This sequence is unique also in an ∼50 aa insertion between the signal peptide and the transmembrane segment, consisting mainly of serines and prolines but otherwise seeming to resemble ETRAMPs. Several ETRAMPs contained regions of 20–30 aa in length, flanking the TM N and/or C terminally, that were predicted to have a tendency to form coiled coils.
Figure 1.
Conserved structure of ETRAMPs. (A) Alignment of the predicted aa sequence of the six most closely related ETRAMPs. The two hydrophobic regions containing the predicted signal peptide and TM are shown in boxes. Amino acids with positively charged side chains are shadowed in black; aa with negatively charged side chains are boldface and shaded in gray. The high number of charged residues is evident. (B) General structure of ETRAMPs. (C) Schematic view of GST-ETRAMP fusion used for recombinant expression. SP, signal peptide.
To test whether ETRAMP features were not just common features of many P. falciparum integral membrane proteins, we compared all ETRAMPs with 21 predicted integral membrane proteins containing a single TM domain that were randomly selected from published sequences of chromosomes 2 and 3 (Gardner et al., 1998; Bowman et al., 1999). Apart from some similarities in the signal peptides and transmembrane domains, all selected sequences were clearly distinct from ETRAMPs (our unpublished data).
Analysis of the chromosomal location of etramps by using PlasmoDB revealed that the six most closely related etramps are located within 20–60 kb to the subtelomeric rifins, except etramp12, which lies within 10 kb from the chromosome internal var cluster. etramps11.1 and 11.2 are linked tail to tail within 3.2 kb, suggesting that they may have evolved from a sequence duplication event.
Limited sequencing of etramps from different isolates gave no evidence of sequence polymorphism (our unpublished data). None of the etramp coding regions showed significant sequence homologies to known proteins of other organisms in a BLAST search against a nonredundant protein database. But searches using National Center for Biotechnology Information custom BLAST and PlasmoDB revealed homologous sequences sharing the same overall structure in P. vivax, P. berghei, and P. yoelii. A preliminary analysis showed that at least nine of the P. yoelii etramps homologs code for proteins that share much higher homology to each other than to P. falciparum ETRAMPs, and are more conserved than the six most closely related P. falciparum ETRAMPs (our unpublished data).
Stage-specific Transcription of etramps
To test whether all etramps were ring stage-specifically transcribed and to analyze the transcription pattern across the intraerythrocytic cell cycle, we performed Northern blots with total RNA of highly synchronized Plasmodium cultures. PCR-amplified etramp fragments (Table 1) were used to generate labeled probes. The low sequence homology among etramps ensured specificity of the probes.
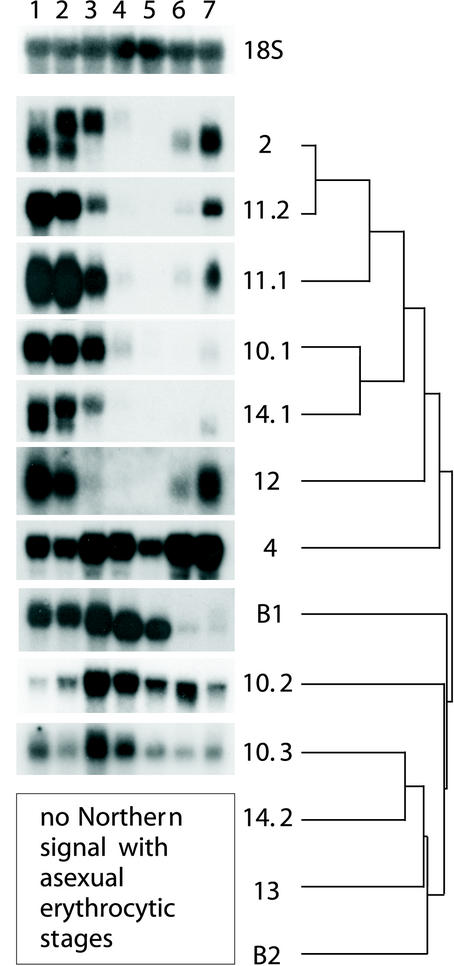
Northern blot results are shown in Figure 2. As indicated with the dendrogram, the most closely related etramps (2, 10.1, 11.1, 11.2, 12, and 14) showed similar, tightly regulated ring stage-specific transcription, which is shut down before 18 hpi, corresponding to the late ring stage (Figure 2, lanes 1–3). As apparent in lane 7, etramps2, 11.1, 11.2, and 12 are already transcribed in late schizonts or very early ring stages (0–4 hpi). Transcription levels occurred to be high, reflected in short exposure times (10–15 min). etramps2, 14, and BLOB.1 are each encoded by two transcripts of different sizes (∼1.4 and 1.8 kb; ∼1.5 and 2.1 kb, and ∼2.0 and 2.3 kb, respectively) that partly overlap during different time points. The large transcript sizes compared with the size of the coding sequences observed for etramp2, 14.1, and BLOB.1 indicates the presence of extensive untranslated regions. This feature, often observed with P. falciparum mRNAs, was also seen with etramp10.1 (∼1.5 kb), etramp11.1 (∼1.4 kb), etramp11.2 (∼2.4 kb), and etramp12 (∼1 kb).
Figure 2.
Developmental regulation of etramp transcription. Northern analysis was performed with filters containing total RNA of highly synchronized asexual blood stage cultures harvested at 0–6, 6–12, 12–18, 18–24, 24–30, 33–39, and 40–4 hpi (lanes 1–7). etramps used as probes are shown on the right side, including a dendrogram to show the degree of relative aa sequence similarities between different ETRAMPs. The six ring-transcribed etramps show the highest degree of similarity. A second group contains only one actively transcribed family member (etramp10.3). Two identical filters that were stripped and reprobed were used for all experiments. A 18s rRNA gene probe was used to demonstrate equal loading and transfer.
In contrast to the six most closely related etramps, etramp4 (∼2.4 kb) was transcribed throughout the cycle, with peaks at the transition from ring stage to trophozoites and at the schizont stage. etrampBLOB.1 was transcribed from ring stage to mid-trophozoites, and etramp10.2 (∼3.8 kb) and 10.3 (∼2.4 kb) were mainly transcribed at the transition from ring to trophozoite stage. Transcriptional levels for etramp10.2 and 10.3 seemed to be much lower (8- and 16-h exposure time, respectively). etramp4, BLOB1, 10.2, and 10.3 define a much less consistent group but share a prominent increase in transcription at the transition from ring to trophozoite stage (Figure 2, lanes 3 and 4). This transcriptional activity coincides with the repression of the six ring-transcribed etramps.
We were unable to detect a signal with etramp13, 14.2, and BLOB.2, even after prolonged exposure. These sequences comprise a group of etramps most distinct from the others (Figure 2, dendrogram). Within this group, only etramp10.3 gave a signal on Northern blots containing RNA from asexual blood stages. Northern analysis was confirmed with independently isolated RNA covering the asexual cycle >5 time points (our unpublished data). Furthermore, Northern analysis with blots containing RNA of the P. falciparum strains HB3 and ITG2F6 showed transcripts of all etramps tested (etramps2, 4, 10.1, 11.2, 14.1, and BLOB.1; our unpublished data).
Characterization of anti-ETRAMP Sera
The C termini of five ETRAMPs were expressed as GST-fusion proteins in E. coli (Figure 1C). We chose two ring stage-transcribed ETRAMPs (ETRAMP2, aa 71–106 and ETRAMP10, aa 71–107) and three ETRAMPs that differed in transcription pattern (ETRAMP4, aa 76–136; ETRAMP10.2, aa 78–355; and ETRAMP13, aa 84–185). To analyze antigenicity, all recombinant proteins were tested on Western blots with a serum pool from semiimmune individuals from Papua New Guinea. All recombinant fusion proteins but ETRAMP13 were recognized as shown in Figure 3A. This confirmed results from Northern blots in which no transcript of ETRAMP13 could be detected, indicating that this protein is not (or at very low levels) expressed in the human host. We therefore omitted ETRAMP13 from further studies.
Figure 3.
Characterization of anti-ETRAMP sera. (A) ETRAMPs are recognized by natural antibodies. Western blot containing recombinant GST (lane 1), MSP-2 (lane 2), ETRAMP2-GST (lane 3), ETRAMP4-GST (lane 4), ETRAMP10.1-GST (lane 5), ETRAMP13-GST (lane 6), and ETRAMP10.2-GST (lane 7) were probed with a serum pool of individuals living in a high endemicity malaria region. ETRAMP10.2 shows some degradation in E. coli. MSP-2 served as a positive control, and GST alone as negative control. (B) Western blots of total protein extracts generated from uninfected RBCs (lanes 1) or parasites released by saponin lysis (lanes 2) were probed with mouse antisera against ETRAMP10.1, 2, 4, and 10.2 (shown on top of corresponding boxes). The faint signal in the RBC lane is due to red staining originating from hemoglobin. Single bands for ETRAMP10.1, 2, and 4 show specificity of the corresponding antisera. ETRAMP10.2 seems to be processed into a soluble (lane s) and a membrane portion (lane m). Molecular mass markers (in kilodaltons) are shown on the right of each filter.
The four asexual stage ETRAMPs were recognized with a comparable strength as the merozoite surface protein-2 (Figure 3A, lane 2), and no signal was obtained with GST alone. Likewise, only the ETRAMP domain was recognized when the fusion protein was cleaved with PreScission protease (our unpublished data). Sera from infants of endemic areas or returning travelers with an acute P. falciparum infection readily recognized the fusion proteins, but no signal was obtained with sera from never-exposed individuals (our unpublished data). This demonstrates that ETRAMPs are expressed in vivo and that the recombinant protein is recognized by naturally occurring antibodies, and suggests a strong immunogenicity of these proteins.
We raised antibodies in mice against the four ETRAMP-GST fusion proteins. Sera were tested on Western blots of total parasite protein or RBC proteins separated by SDS-PAGE. None of the sera reacted with RBC extracts (Figure 3B). Sera against ETRAMPs10.1, 2, and 4 recognized single bands in the total parasite protein preparation (Figure 3B). Bands were ∼20% larger than the predicted sizes (including the signal peptide) of 11.3 kDa for ETRAMP10.1, 11.5 kDa for ETRAMP2, and 14.9 kDa for ETRAMP4. As often observed with cationic proteins, bands migrate slower than predicted. This is explained by nonuniform binding of SDS to highly charged regions (Coppel et al., 1994). It is noteworthy that a signal was only obtained when methanol was omitted from the Western transfer buffer. Due to the low protein concentration of ring stages (these stages contain up to 30 times less protein than mature parasites), P. falciparum cultures had to be enriched for ring stages to obtain a signal with antisera against ETRAMP10.1 and 2.
Sera raised against ETRAMP10.2 reacted with two bands of 28 and 18 kDa in total parasite extracts. Together, this accounts for 46 kDa, which corresponds to the predicted size for ETRAMP10.2 (38.9 kDa), including 20% due to the high positive charge of this protein. ETRAMP10.2 contains a PEST sequence within the C terminus (aa 210–227, with a PESTfind prediction score of 15). Such sequences are known to target proteins for proteolysis (Rogers et al., 1986; Rechsteiner and Rogers, 1996). Assuming a transmembrane location of ETRAMP10.2, processing would result in the presence of one domain in the membrane fraction, whereas the other domain would be found soluble. This was confirmed on Western blots with the detergent-depleted fraction and the membrane fraction (Figure 3, lanes s and m). Identical stage-specific presence of both bands further indicated that they originated from the same polypeptide (our unpublished data).
ETRAMPs Are Stage-specifically Expressed Membrane Proteins
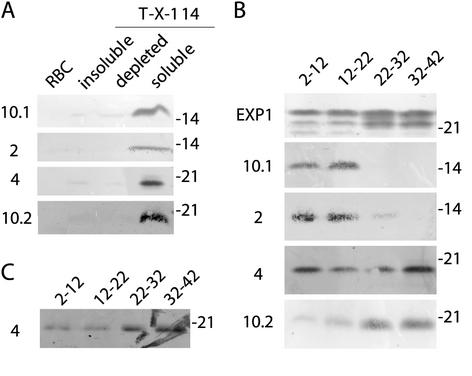
Western analysis using fractions generated by Triton X-114 phase separation of parasite proteins verified the predicted association of ETRAMPs with the membrane fraction. Apart from the soluble part of ETRAMP10.2 (Figure 3B), no ETRAMP could be detected in the Triton X-114–insoluble or the Triton X-114–depleted fraction (Figure 4A).
Figure 4.
ETRAMPs are detected in the membrane fraction only and are developmentally expressed. (A) Western analysis with filters containing insoluble, soluble Triton X-114–depleted and Triton X-114–soluble parasite protein. All ETRAMPs were found in the membrane (Triton X-114–soluble) fraction only except for a 28-kDa ETRAMP10.2 signal in the detergent-depleted fraction (our unpublished data). (B) Western analysis with filters containing membrane protein harvested at four time points of synchronized P. falciparum in vitro cultures. Lanes 1 (2–12 hpi) and lane 2 (12–22 hpi) correspond to early and late ring stage, respectively; lane 3 (22–32 hpi) trophozoite stage; lane 4 (32–42 hpi) schizont stage. ETRAMP10.1 and 2 are only expressed in ring stages, whereas ETRAMP4 is found throughout the cycle. ETRAMP10.2 shows the highest expression after the ring stage. An antiserum against EXP-1 was used to confirm equal loading. (C) Formaldehyde (1%) cross-linking before extraction of membrane protein greatly diminished ETRAMP4 signal in rings. Antisera used are shown on the left; molecular mass is indicated in kilodaltons on the right of each filter.
We then used membrane extracts of synchronized parasites harvested during four consecutive time points of asexual development to analyze the expression pattern. The results shown in Figure 4B are consistent with the transcriptional analysis. Ring-transcribed ETRAMP10.1 and 2 were both only detected in extracts of ring stage parasites (Figure 4B, 2–22 hpi). ETRAMP4 was continuously found with a marked increase in schizonts and a moderate increase in ring stage parasites, which is in accordance with the transcription levels during the schizont stage. Levels of ETRAMP10.2 increased after the ring stage, immediately after the time point when an increase of transcription was observed in Northern blots. Thereafter, the protein persisted until the end of schizogony. Similar results with all four ETRAMPs were obtained with Western blots of total protein extracts (in HTPBS/0.5% Triton X-114) of synchronized parasites (our unpublished data).
Western analysis with rabbit anti EXP-1 sera and silver staining of SDS-PAGE gels confirmed equal loading of membrane extracts (Figure 4B; our unpublished data).
ETRAMPs Localize to Parasite Periphery
When IRBCs from asynchronous cultures were immobilized on concanavalin A-coated slides, dried, and fixed either with 1% formaldehyde or acetone, ETRAMP antisera gave a positive IFA signal. No signal was obtained when slides were fixed with methanol or ethanol.
Both sera against the ring stage expressed ETRAMP2 and 10.1 labeled a complete or partial ring (our unpublished data; Figure 5E with cells that were not dried). IFAs with synchronized parasites revealed that ETRAMP2 and 10.1 labeled all ring stage parasites, but no fluorescence was found in later stage parasites (our unpublished data). This is accordance with the results obtained in Western blots.
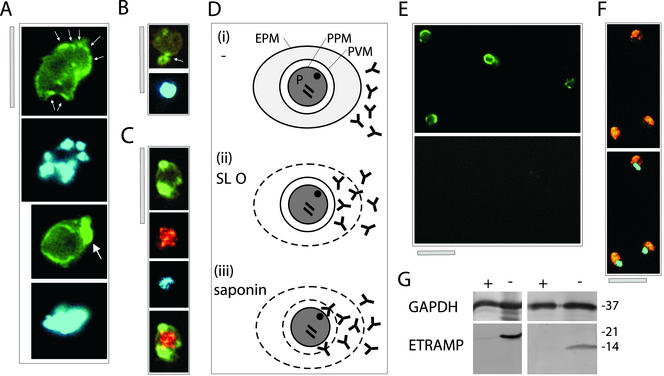
Figure 5.
ETRAMPs are located in the PVM with their C termini facing the RBC cytosol. (A–C) Distribution of ETRAMP4 around the parasite periphery in fixed cells. Top two panels of A depict ETRAMP4 staining (FITC) and nuclear staining (DAPI) of a schizont stage parasite; spots of more intense staining are indicated by small arrows. Bottom two panels of A show ETRAMP4 (FITC) and nuclear staining (DAPI) of a late trophozoite stage parasite. The single patch of strong staining is indicated by a large arrow. (B) Extensions from the parasite body are labeled by anti-ETRAMP4 serum, indicating localization in blebbing vesicles or the TVM. The top panel shows ETRAMP4 (FITC) detected in a young trophozoite; below the nucleus is shown (DAPI). The extension is indicated by an arrow. (C) Costaining with a rabbit anti-aldolase serum verifies that the periphery of the parasite is decorated by ETRAMP4 antiserum; the four panels represent the same section depicting a young trophozoite. From top to bottom, ETRAMP4 (FITC), aldolase (Cy3), nucleus (DAPI), and aldolase and ETRAMP4 merged. Bars, 5 μm. (D) Representation of the selective permeabilization scheme: 1) an untreated IRBC (P, parasite; EPM, erythrocyte plasma membrane; PPM, parasite plasma membrane); 2) a cell permeabilized with SLO, enabling access of antibodies through the perforated RBC membrane, resulting in a fluorescence signal if the corresponding antigen is located in the PVM facing the RBC cytosol; 3) a cell permeabilized with saponin, which allows to detect antigens facing the outer side of the PPM or both sides of the PVM. (E) Formaldehyde (1%)-fixed and saponin-permeabilized ring stage parasites showed a signal for ETRAMP10.1 (green, FITC, top), but not for aldolase (red, Cy3, bottom). Both panels show the same section. (F) Aldolase is strongly detected under similar fixation conditions and treatment with Triton X-100, indicating that aldolase is well detectable in ring stages. (G) Western analysis with extracts of saponin-released parasites that were either treated with (+) or without (−) trypsin before protein extraction, shows that the ETRAMP4 (left) and ETRAMP10.1 (right) C termini were digested, but not the cytoplasmic parasite enzyme GAPDH. Molecular mass (in kilodaltons) is indicated on the right.
ETRAMP4 and 10.2 were also localized to the periphery of the parasite (shown for ETRAMP4 in Figure 5A), with much weaker staining of ETRAMP10.2. Usually, the distribution of both ETRAMPs around the cell was not uniform as seen in Figure 5A showing a schizont (top two panels) and a trophozoite stage parasite (bottom panels). Most parasites had either an additional single patch of fluorescence (Figure 5A, thick arrow), intermittent regions of stronger fluorescence (Figure 5A, small arrows), or blebbing extensions from the parasite (Figure 5B). Costaining with Cy3-labeled anti-aldolase showed that the parasite cytoplasm is encircled by the ETRAMP signal (Figure 5C). In contrast to the ring stage-expressed ETRAMPs, we observed fluorescence only in later stage parasites (our unpublished data). This is in contrast to results from Western and Northern blots obtained for ETRAMP4 and may indicate that the protein is masked in ring stages. This is further supported by the almost complete loss of signal on Western blots by using stage-specific protein extracts from parasites that were cross-linked with 1% formaldehyde before protein extraction (Figure 4C). No labeling was observed with segmented schizonts.
To exclude that the similar fluorescence pattern was due to cross-reactivity between ETRAMPs, we preincubated sera against ETRAMP with an excess of each expressed ETRAMP. Immunofluorescence was only abolished with the corresponding GST-fusion protein, but neither with any of the other ETRAMP-fusion proteins or with GST alone (our unpublished data). No fluorescence was observed with sera against GST (our unpublished data).
ETRAMPs Are Located in PVM
Peripheral labeling obtained with all antisera suggested a localization of ETRAMPs in the PPM or the PVM. But the close association of these membranes does not allow assignment of the signal to one of these compartments. To determine the exact location of ETRAMPs, we applied the selective permeabilization procedure for Western analysis (Ansorge et al., 1997) to IFAs. A schematic representation of the procedure is shown in Figure 5D. IRBCs were either treated with saponin, which permeabilizes the RBC membrane and the PVM but not the PPM (Figure 5D, ii), or with SLO, which permeabilizes the RBC membrane only but leaves intact the PVM and the PPM (Figure 5D, iii). Anti-ETRAMP sera were added to IRBCs treated with one of these two agents, and the cells were analyzed by immunofluorescence. Depending on the location of ETRAMPs, a fluorescence signal is only observed if the permeabilizing agent created access to the antigen. In all experiments, a rabbit serum specific for parasite aldolase was added together with the mouse anti-ETRAMP serum to confirm that antibodies had no access to the parasite cytosol after selective permeabilization. As a positive control, IRBCs were dried or treated with Triton X-100 to permeabilize all membranes.
In mildly or unfixed cells, permeabilized with saponin or SLO, fluorescence was only observed with sera against the ring expressed ETRAMP10.1 and 2, whereas aldolase staining was only visible in Triton X-100–permeabilized cells (Table 2 and Figure 5, E and F). We were not able to detect ETRAMPs after treatment of cells with Triton X-100, but dried and fixed cells showed a signal with all sera, indicating the presence of all antigens (Table 2). Because sera were raised against the C-terminal domain of ETRAMPs, the signal with saponin-permeabilized IRBCs demonstrated that the C termini of both ring-expressed ETRAMPs were located outside of the parasite PPM. Furthermore, immunofluorescence with SLO-treated IRBCs showed that ETRAMP10.1 and 2 could only be located in the PVM, with the C terminus facing the RBC cytoplasm.
Table 2.
Fluorescence signal with selectively permeabilized IRBCs
| Fixation Treatment antisera | no/0.1%
Formaldehyde
|
0.5% Formaldehyde
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| None | SLO | Saponin | T-X-100 | None | SLO | Saponin | T-X-100 | Dried | |
| 10.1 | − | + | + | − | ±a | + | + | − | + |
| 2 | − | + | + | − | ±a | + | + | − | + |
| 4 | − | − | − | − | ±a | + | + | − | + |
| 10.2 | − | − | − | − | ±a | + | + | − | + |
| Aldolase | − | − | − | + | − | ±b | ±b | + | + |
T-X-100, Triton-X-100.
Only few of the IRBCs were labeled.
Very poor signal.
To obtain a signal with sera against ETRAMP4 and 10.2, the formaldehyde concentration had to be raised to 0.5% (Table 2), resulting in some loss of cell integrity. This was evident in a low proportion of nonpermeabilized cells with an ETRAMP signal (Table 2) and in permeabilized cells with a faint staining of aldolase. Therefore, we could not completely exclude that ETRAMP4 and 10.2 are located in the PPM, although the extensions from the parasite periphery suggest PVM location (Figure 5B).
To corroborate further localization of ETRAMP4 in the PVM, we performed Western blots with proteins from saponin-treated and subsequently trypsinized cells. In the proposed classical vesicular transport pathway inside plasmodial cells (Albano et al., 1999b), a type I integral PPM protein would face the parasite cytoplasm with the C terminus. After permeabilization and trypsination, we would expect ETRAMP4 to be truncated to the TM and the C terminus. But no truncated protein was detected on Western blots and the ETRAMP4 signal was lost completely (Figure 5G). This is in accordance with a localization of ETRAMP4 in the PVM and was also shown for ETRAMP10.1. In contrast, PfGAPDH was still detected with a monoclonal antibody in trypsinized and control fractions of the same extracts, demonstrating that trypsin was unable to enter the cytoplasm of the parasite.
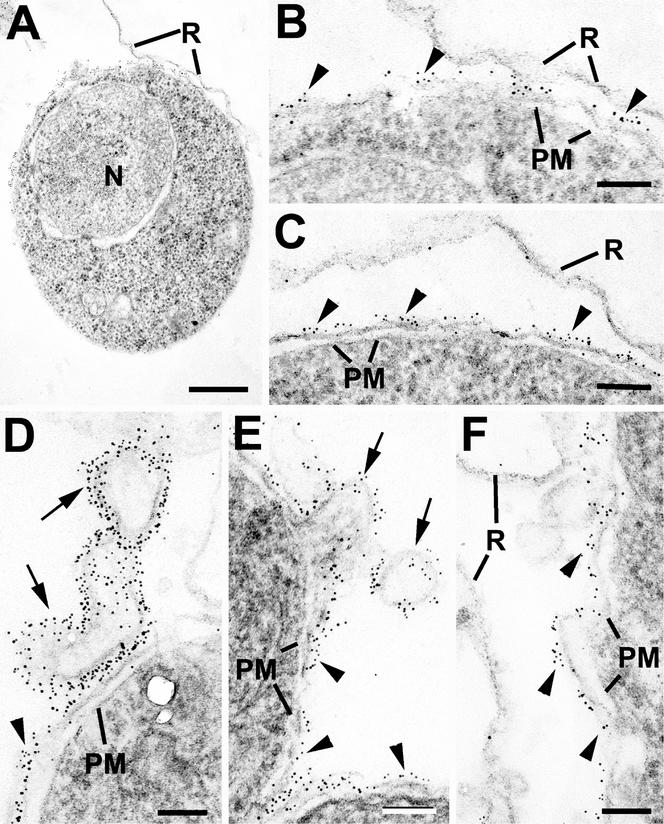
By immunoelectron microscopy, it was observed that infected cells containing young trophozoites and stained for ETRAMP2 showed labeling of the outer aspect of the PVM (Figure 6, A–C). The red cell membrane, Maurer's clefts and PPM were unlabeled. It was also noted that certain extensions of the PVM and tubovesicular blebs were also strongly labeled (Figure 6, D and E). In samples stained for ETRAMP4, the level of staining was lower but a similar pattern was observed with labeling of the PVM of certain cells containing more mature multinucleate trophozoites (Figure 6F).
Figure 6.
Immunoelectron microscopy of permeablized red blood cells infected with P. falciparum and labeled with anti-ETRAMP2 (A–E) or anti-ETRAMP4 (F) and visualized with 5-nm gold particles. (A) Low-power image showing an intact early trophozoite with a single nucleus (N). (B) Detail form the periphery of A in which numerous gold particles are associated with the PVM (arrowheads). Note the lack of staining of the red cell membrane (R). (C) Similar section to B showing that the labeling is limited to the PVM (arrowheads). (D and E) Details of the periphery of intracellular parasites showing strong labeling of the PVM (arrowheads) and tubovesicular extensions of this membrane (arrows). (F) Section of the periphery of a large trophozoite stained with anti-ETRAMP4 in which the labeling is limited to the PVM (arrowheads). PM, parasite plasma membrane; R, red cell membrane. Bars, 0.5 μm (A) and 100 nm (B–F).
ETRAMP4 and 10.2 Colocalize with Markers of Parasitophorous Vacuole (PV) and PVM
We performed colocalization studies with dried and acetone-fixed cells by using rabbit antisera against EXP-1, a PVM membrane protein, and SERP, a soluble protein found in the PV of late stage parasites
Colocalization by confocal microscopy with serum against SERP revealed only limited overlap in stage specificity with ETRAMP4 and 10.2. These ETRAMPs occurred earlier in the cycle than SERP and were not detectable in the late schizont. However, when both proteins were present, cells clearly showed similar peripheral labeling for both antigens, although distribution around the cell differed slightly (shown for ETRAMP4 in Figure 7A). The expanded peripheral ETRAMP4 location (arrow, Figure 7A) might be due to an artificial detachment of the PVM due to drying and fixing.
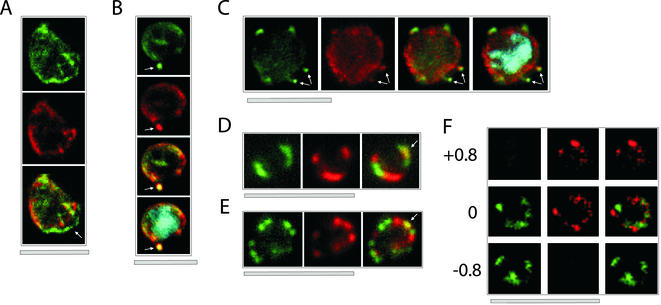
Figure 7.
Colocalization of ETRAMPs with EXP-1 and SERP. The first image in each set shows ETRAMP labeling (green, FITC), the second Texas-Red-labeled SERP (A) or EXP-1 (B–F), and the third image represents the merged picture. The fourth panel in B and C depicts the merged picture, including nuclear staining (DAPI). (A) Confocal microscopy analysis of SERP and ETRAMP4 colocalization in schizont stage parasites (dried and 1% formaldehyde fixed). The ETRAMP4 signal seems partially detached from the PV content (arrow). (B and C) ETRAMP4 and EXP-1 localize to dots, probably representing vesicular structures that are detached from the PVM (E) or seem connected to the PVM (F). Dots are indicated by arrows. (D and E) Fluorescent microscopic analysis of ETRAMP and EXP-1 costaining of a ring-stage parasite displaying an even circular pattern (D, 0.1% formaldehyde-fixed IRBCs, ETRAMP10.1) or a beads on a string pattern (E, unfixed IRBCs, ETRAMP2). Yellow color shows regions of overlap (arrows). This indicated that ETRAMPs and EXP-1 localize to different regions of the PVM. (F) Three sections through a ring-stage parasite (dried and fixed with 1% formaldehyde), costained with ETRAMP10.1 and EXP-1, were analyzed by confocal microscopy. The planes of the first and the third row are 0.8 μm apart from the plane shown in the central row. The top plane contains only EXP-1 signal, whereas the bottom plane displays only ETRAMP signal. Bars, 5 μm.
Analysis of ETRAMP4 and EXP-1 colocalization by using fluorescent microscopy showed overlapping staining around the periphery of the cell. EXP-1 was found occasionally as small fluorescent dots inside the RBC cytosol, often close to the PVM, which might reflect the known association with vesicular structures in the RBC cytoplasm (Simmons et al., 1987; Kara et al., 1988a,b). ETRAMP4 colocalized also to these dots that occurred either disconnected from the PVM (Figure 7B, arrows) or connected to the PVM (Figure 7C, arrows). When the focus was adjusted to the dots, the parasite body was slightly out of focus and peripheral staining was less prominent. Occasionally, there were also dots or extensions from the PVM showing only one of the two antigens (our unpublished data). These results further corroborate a PVM location of ETRAMP4.
Ring Stage-expressed ETRAMPs Define Discrete Domains in PVM
Colocalization of ring-expressed ETRAMPs (2 and 10.2) and EXP-1 was analyzed with saponin-permeabilized IRBCs. For both EXP-1 and ETRAMPs, fluorescence around the cell seemed concentrated at discrete sites. Together, the two antigens gave a ring-formed distribution around the parasite but did clearly not colocalize (shown for ETRAMP10.1 in Figure 7D). A similar distribution was also seen with ring stage-infected cells showing a bead on a string fluorescence, a pattern we observed frequently in unfixed cells, and that has been described for proteins in the PV detected in unfixed ring stages (Waller et al., 2000; Wickham et al., 2001). Again, beads showed either green (ETRAMP) or red (EXP-1) labeling (shown for ETRAMP2 in Figure 7E) with occasional overlaps (Figure 7, D and E, arrows). Similar results were obtained with both fixed and unfixed IRBCs.
To analyze the three-dimensional distribution of ring stage-specific ETRAMPs and EXP-1, dried and formaldehyde-fixed IRBCs were analyzed by confocal microscopy. Focus was adjusted to the center of a parasite, and optical planes 0, +0.8, and −0.8 μm were recorded for EXP-1 and ETRAMP10.1. As shown in Figure 7F, the two signals varied considerably in their distribution, e.g., EXP-1 was only present in the top and central optical section, and ETRAMP10.1 only in the central and lower plane. Optical sections ±1.6 μm showed no signal, demonstrating that the observed staining derived from a single cell. Analysis of several cells showed similar results, with optical planes often showing only one signal or both with clear spatial separation. These results clearly indicate spatial separation of ETRAMPs and EXP-1 in different domains of the PVM.
DISCUSSION
The P. falciparum ring stage takes up the first half of asexual intraerythrocytic development, and our understanding of the molecular events during this stage is very limited. Herein, we have described and characterized a new P. falciparum gene family called etramp, coding for highly charged, small membrane proteins. Using etramps isolated from a ring stage-specific cDNA library (Spielmann and Beck, 2000), we identified a total of 13 etramps from the P. falciparum genome. Six members belonged to a coherent group expressed exclusively in the ring stage, whereas other members showed a different developmental regulation. Some ETRAMPs were not transcribed at all in asexual blood stage parasites. One of these, ETRAMPs (ETRAMP13), shows 31% aa identity to a P. yoelii protein (Py22) with typical features of an ETRAMP. Py22 is transcribed in salivary gland sporozoites (Matuschewski et al., 2002), indicating that expression of the ETRAMP protein family is not restricted to blood stages. ETRAMP13 was previously published as OrfP and detected by reverse transcription-PCR but not by Northern analysis of asexual blood stage RNA (Sallicandro et al., 2000). We also failed to detect the corresponding mRNA on Northern blots, and we did not find natural antibodies against the recombinant GST-fusion protein. Because we have only analyzed asexual blood stages, ETRAMP13 might be expressed in other life cycle stages.
We used colocalization and selective permeabilization IFAs to determine the location of four ETRAMPs in IRBCs. Whereas both ring-expressed ETRAMPs could be unequivocally localized to the PVM, ETRAMP4 and 10.2 were only detectable in selectively permeabilized cells fixed with 0.5% formaldehyde. The peripheral staining with antisera against these ETRAMPs suggested that they were either located in the PPM, or fixing was required to detect them in the PVM. There are several lines of evidence for the latter possibility: 1) with 0.5% formaldehyde, aldolase signal was very faint, indicating that access to the cytosol of the parasite was limited, but anti-ETRAMP signal was intense. Moreover, in unfixed or mildly fixed cells, the antiserum against the C terminus of EXP-1 was only reactive with the PVM of ring stage parasites, whereas all parasites were recognized with 0.5% formaldehyde (our unpublished data), pointing to a general stage-specific effect. 2) Western analysis with extracts obtained from saponin-treated and trypsinized IRBCs indicated PVM localization. 3) Extensions from the cell periphery, including dots in the RBC cytoplasm, were decorated by the anti-ETRAMP sera, and colocalized with the PVM protein EXP-1. Such structures have previously been shown to contain EXP-1 and were attributed to vesicles in the RBC cytoplasm and the TVN originating from the PVM (Simmons et al., 1987; Kara et al., 1988a,b). Further corroborating these findings, immunoelectron microscopy of saponin-permeabilized IRBCs with sera against ETRAMP2 and 4 showed specific staining of the PVM facing the RBC cytosol as well as PVM extensions and proximate vesicles. Together, these results strongly indicate PVM localization of all four ETRAMPs tested and suggest a similar localization for all other ETRAMPs expressed during the asexual blood stage. Furthermore, the SLO and immunoelectron microscopy experiments showed that ETRAMP C termini face the RBC cytoplasm, whereas the N terminus protrudes into the PV. This orientation is similar to that of EXP-1 (Ansorge et al., 1997), the only well characterized P. falciparum integral PVM protein.
In the related apicomplexan Toxoplasma gondii, the rhoptry proteins ROP2, 3, 4, and 7 are inserted into the PVM (Beckers et al., 1994) in a process concomitant with host cell invasion and PVM formation. In P. falciparum, proteins derived from merozoite apical organelles were also found to be associated with the PVM after invasion (Bushell et al., 1988; Sam-Yellowe et al., 1988; Trager et al., 1992) but in contrast to T. gondii, these structures seem to be lost after invasion. We cannot exclude that some ETRAMPs are already inserted into the PVM upon invasion. However, different ETRAMPs are synthesized at various stages of the asexual blood cycle, demonstrating transport to the PVM throughout intraerythrocytic development. Furthermore, we did not detect ETRAMPs in late schizonts, which would be a prerequisite for PVM insertion upon invasion. Therefore, ETRAMP transport is comparable with secretion from the dense granules in T. gondii, which occurs after invasion (Carruthers and Sibley, 1997). The orientation of ETRAMPs in the PVM supports a continuous vesicle fusion-budding protein transport to the PVM originally proposed on the basis of EXP-1 orientation (Günther et al., 1991; Ansorge et al., 1997; Lingelbach and Joiner, 1998). This contrasts the T. gondii dense granule protein GRA5, which is secreted as a soluble protein and is subsequently inserted into the PVM as a transmembrane protein in the opposite orientation (Lecordier et al., 1999).
It seems that in P. falciparum, the default secretory route for soluble proteins targets them to the PV (Waller et al., 2000; Wickham et al., 2001) or possibly into the RBC cytosol (Burghaus and Lingelbach, 2001). It is not known whether integral membrane proteins are also exported by default and whether there are specific PVM retention motifs. Targeting signals are often present in the C-terminal cytoplasmic domains (Urbe et al., 1997). However, no obvious common aa sequence element was found in ETRAMP C termini. Either the targeting information lies in the more conserved N terminus, is very variable, or no specific signal is required for transport and retention in the PVM. Posttranslational modifications could also mediate targeting. EXP-1 was shown to be myristilated (Kara et al., 1988a). We cannot exclude that the observed larger size of ETRAMPs on Western blots of parasite extracts is due to such modifications.
We showed that in the ring stage, the PVM contains subdomains in terms of protein composition. The cationic ring-expressed ETRAMPs define discrete regions compared with the anionic PVM protein EXP-1. This implies that two functionally distinct domains exist in the PVM. Nonuniform distribution of EXP-1 around the cell has already been observed (Behari and Haldar, 1994). But in contrast to EXP-1, the ETRAMP composition of the PVM changes with development. The ring-expressed ETRAMPs disappear and are replaced by other ETRAMPs at the transition from ring to trophozoite stage. There is also a peak in transcription for ETRAMP4 and B1 during this transition stage, suggesting increased synthesis of these proteins. Therefore, the host cell–parasite interface changes its properties concomitant with morphological changes, a first peak of increased protein synthesis (de Rojas and Wasserman, 1985), appearance of parasite-induced host cell modifications, and subsequent rapid parasite growth. The early appearance of parasite proteins at the parasite–host cell interface is in accordance with the need to create a connection to the host cell to acquire nutrients and to establish protein transport into the RBCs. The ring-expressed ETRAMPs define this stage and are part of a set of early transcribed genes lacking homologies to known proteins (Spielmann and Beck, 2000). This is in accordance with a unique nature of this process in cell biology. We propose that the loss of ring-expressed ETRAMPs defines a switch in development seen in the morphological transition from ring to trophozoite stage. This makes ETRAMPs excellent markers for this stage both at the RNA and the protein level.
Highly charged molecules such as ETRAMPs could interact with structural proteins to fix the PVM in the RBCs or could interact with soluble proteins in the parasite cytoplasm to act as carriers out of the parasite. However, we were unsuccessful to detect interaction partners (from parasite and RBC extracts) with the recombinant ETRAMP C termini fused to GST by using GST pull-down and Far Western techniques (our unpublished data).
ETRAMPs could also interact with each other, requiring a membrane environment and the predicted coiled coil domains on both sides of the TM. Such multimers could form a channel that is responsible for the molecular sieve properties of the PVM (Desai et al., 1993; Desai and Rosenberg, 1997) or might be involved in protein transport through the PVM (Ansorge et al., 1996). However, it has to be noted that coiled coil predictions of lysine-rich sequences have to be interpreted with care (Berger et al., 1995).
There are striking analogies between ETRAMPs and integral PVM proteins found in other intracellular pathogens, indicating a common scheme to equip this compartment. Similar to ETRAMPs, the T. gondii rhoptry proteins ROP2, 4, 7, and 8 have a pI > 9; share sequence homology among each other; contain a TM; are found in the PVM early after invasion; and show punctuate, nonuniform localization around the cell periphery (Beckers et al., 1994). Chlamydia spp. modify their PVM by insertion of Inc proteins. This family contains many charged proteins and shares a characteristic hydrophobic domain of >40 aa, but otherwise exhibits no sequence similarity. Inc proteins are small proteins expressed early after infection (Scidmore-Carlson et al., 1999), and some are unevenly distributed around the cell (Bannantine et al., 1998; Scidmore-Carlson et al., 1999). Neither ETRAMPs nor Incs nor ROP proteins show sequence homology to known proteins. To date, little is known about the function of PVM-located proteins. ROP2 was recently shown to mediate association of host cell organelles to the T. gondii PVM (Sinai and Joiner, 2001), and IncA seems to be involved in fusion of different Chlamydia inclusions (Hackstadt et al., 1999). Although such functions are not required for Plasmodium blood stages, the ability of these proteins to mediate interaction or fusion with membranes might be relevant for ETRAMPs and a common property of PVM proteins. The domain responsible for ROP2 insertion into lipid bilayers is highly positively charged, a feature also typical for ETRAMPs. High positive charge is a characteristic of proteins capable to directly traverse membranes (Schwarze and Dowdy, 2000) and of peptides capable to disrupt lipid bilayers (Bechinger, 1997). If ETRAMPs had membrane interacting/penetrating capabilities, they could be required for the proposed junction between the TVN and the RBC membrane to acquire nutrients (Lauer et al., 1997), to obtain lipids from the RBC membrane, for vesicular transport processes at the PVM, or to anchor the PVM to the RBC membrane. Different membrane interaction tasks would explain the presence of a number of related proteins and the need for different proteins during different life cycle stages. Furthermore, the biophysical properties required for such a function, as binding negative lipid head groups or insertion into lipid bilayers, could explain the technical difficulties encountered to detect ETRAMPs when methanol or Triton X-100 was used.
We were unable to demonstrate any sequence difference between etramps of different strains, and all parasites expressed all ETRAMPs they were analyzed for. The presence of etramp transcripts in strains other than 3D7 indicates that this gene family is expressed in all P. falciparum strains. Furthermore, ETRAMPs were readily recognized by sera of malaria patients, indicating expression in vivo. The strong antigenicity of ETRAMPs is notable in the light of the protective immune response against a PVM located antigen of liver stages (Charoenvit et al., 1999).
The presence of ETRAMP homologs in other Plasmodium species indicates that this family is involved in processes common to all Plasmodia. Intracellular pathogens residing in a PVM have to maintain this barrier and must be equipped to allow controlled cross talk and to modify it according to the needs of each the life cycle stage. ETRAMP composition in the P. falciparum PVM is subjected to a dynamic change concomitant with parasite development. Furthermore, ring expressed ETRAMPs define subdomains in the PVM. This indicates spatial- and stage-dependent functional heterogeneity of this compartment. Therefore, we believe that ETRAMPs are important mediators of the Plasmodium–host cell interaction and will reveal significant insight into the biology of Plasmodium and the properties of host–parasite interfaces of intracellular pathogens in general.
ACKNOWLEDGMENTS
We thank Drs. T. Voss, K. Lingelbach, and J. Burckhardt for critically reading the manuscript and comments. We are grateful to Drs. K. Lingelbach and S. Baumeister for antisera against aldolase, EXP-1, and SERP and to Dr. C. Daubenberger for antibodies to GAPDH. We also thank Dr. H. Reichert for providing access to the confocal microscope and S. Sprecher and L. Kammermeier for expert help with this instrument. We are grateful for Dr. R. Brun and S. Scheurer of the protozoology group for providing culture facilities and consumables. We also thank I. Endriss and K. Gysin for doing the mouse work. We thank the scientists and funding agencies comprising the international Malaria Genome Project for making sequence data from the genome of P. falciparum (3D7) public before publication of the completed sequence. D.J.P.F. was supported by an equipment grant of the Wellcome Trust. The Sanger Center provided sequence for chromosomes 1, 3–9, and 13, with financial support from the Wellcome Trust. A consortium composed of The Institute for Genome Research, along with the Naval Medical Research Center, sequenced chromosomes 2, 10, 11, and 14, with support from National Institute of Allergy and Infectious Diseases/National Institutes of Health, the Burroughs Wellcome Fund, and the Department of Defense. The Stanford Genome Technology Center sequenced chromosome 12, with support from the Burroughs Wellcome Fund. The Plasmodium Genome Database is a collaborative effort of investigators at the University of Pennsylvania and Monash University, supported by the Burroughs Wellcome Fund.
Abbreviations used:
- aa
amino acid(s)
- IRBC
infected red blood cell
- hpi
hours postinvasion
- PPM
parasite plasma membrane
- PV
parasitophorous vacuole
- PVM
PV membrane
- RBC
red blood cell
- TVN
tubovesicular network
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–04–0240. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–04–0240.
REFERENCES
- Adisa A, Albano FR, Reeder J, Foley M, Tilley L. Evidence for a role for a Plasmodium falciparumhomologue of Sec31p in the export of proteins to the surface of malaria parasite-infected erythrocytes. J Cell Sci. 2001;114:3377–3386. doi: 10.1242/jcs.114.18.3377. [DOI] [PubMed] [Google Scholar]
- Albano FR, Berman A, La Greca N, Hibbs AR, Wickham M, Foley M, Tilley L. A homologue of Sar1p localizes to a novel trafficking pathway in malaria-infected erythrocytes. Eur J Cell Biol. 1999a;78:453–462. doi: 10.1016/S0171-9335(99)80072-7. [DOI] [PubMed] [Google Scholar]
- Albano FR, Foley M, Tilley L. Export of parasite proteins to the erythrocyte cytoplasm: secretory machinery and traffic signals. Novartis Found Symp. 1999b;226:157–172. doi: 10.1002/9780470515730.ch12. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ansorge I, Benting J, Bhakdi S, Lingelbach K. Protein sorting in Plasmodium falciparum-infected red blood cells permeabilized with the pore-forming protein streptolysin O. Biochem J. 1996;315:307–314. doi: 10.1042/bj3150307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansorge I, Paprotka K, Bhakdi S, Lingelbach K. Permeabilization of the erythrocyte membrane with streptolysin O allows access to the vacuolar membrane of Plasmodium falciparumand a molecular analysis of membrane topology. Mol Biochem Parasitol. 1997;84:259–261. doi: 10.1016/s0166-6851(96)02806-x. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidmann JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1989. [Google Scholar]
- Bahl A, et al. PlasmoDB: the Plasmodiumgenome resource. An integrated database providing tools for accessing, analyzing and mapping expression and sequence data (both finished and unfinished) Nucleic Acids Res. 2002;30:87–90. doi: 10.1093/nar/30.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine JP, Rockey DD, Hackstadt T. Tandem genes of Chlamydia psittacithat encode proteins localized to the inclusion membrane. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- Barnwell JW. Vesicle-mediated transport of membrane and proteins in malaria-infected erythrocytes. Blood Cells. 1990;16:379–395. [PubMed] [Google Scholar]
- Bechinger B. Structure and functions of channel-forming peptides: magainins, cecropins, melittin and alamethicin. J Membr Biol. 1997;156:197–211. doi: 10.1007/s002329900201. [DOI] [PubMed] [Google Scholar]
- Beckers CJM, Dubremetz JF, Mercereaupuijalon O, Joiner KA. The Toxoplasma gondiirhoptry protein Rop-2 is inserted into the parasitophorous vacuole membrane, surrounding the intracellular parasite, and is exposed to the host-cell cytoplasm. J Cell Biol. 1994;127:947–961. doi: 10.1083/jcb.127.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behari R, Haldar K. Plasmodium falciparum: protein localization along a novel, lipid-rich tubovesicular membrane network in infected erythrocytes. Exp Parasitol. 1994;79:250–259. doi: 10.1006/expr.1994.1088. [DOI] [PubMed] [Google Scholar]
- Berger B, Wilson DB, Wolf E, Tonchev T, Milla M, Kim PS. Predicting coiled coils by use Sf pairwise residue correlations. Proc Natl Acad Sci USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino-acid-sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- Blisnick T, Eugenia M, Betoulle M, Barale JC, Uzureau P, Berry L, Desroses S, Fujioka H, Mattei D, Breton CB. Pfsbp 1, a Maurer's cleft Plasmodium falciparumprotein, is associated with the erythrocyte skeleton. Mol Biochem Parasitol. 2000;111:107–121. doi: 10.1016/s0166-6851(00)00301-7. [DOI] [PubMed] [Google Scholar]
- Bordier C. Phase-separation of integral membrane-proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Bowman S, et al. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- Burghaus PA, Lingelbach K. Luciferase, when fused to an N-terminal signal peptide, is secreted from transfected Plasmodium falciparumand transported to the cytosol of infected erythrocytes. J Biol Chem. 2001;276:26838–26845. doi: 10.1074/jbc.M100111200. [DOI] [PubMed] [Google Scholar]
- Bushell GR, Ingram LT, Fardoulys CA, Cooper JA. An antigenic complex in the rhoptries of Plasmodium falciparum. Mol Biochem Parasitol. 1988;28:105–112. doi: 10.1016/0166-6851(88)90057-6. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Sibley LD. Sequential protein secretion from three distinct organelles of Toxoplasma gondiiaccompanies invasion of human fibroblasts. Eur J Cell Biol. 1997;73:114–123. [PubMed] [Google Scholar]
- Charoenvit Y, Majam VF, Corradin G, Sacci JB, Wang RB, Doolan DL, Jones TR, Abot E, Patarroyo ME, Guzman F, Hoffman SL. CD4(+) T-cell- and γ interferon-dependent protection against murine malaria by immunization with linear synthetic peptides from a Plasmodium yoelii17-kilodalton hepatocyte erythrocyte protein. Infect Immun. 1999;67:5604–5614. doi: 10.1128/iai.67.11.5604-5614.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke BM, Mohandas N, Coppel RL. The malaria-infected red blood cell: structural and functional changes. Adv Parasitol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppel RL, Davern KM, McConville MJ. Immunochemistry of parasite antigens. In: van Oss CJ, van Regenmortel MHV, editors. Immunochemistry. New York: Marcel Dekker; 1994. pp. 475–532. [Google Scholar]
- de Rojas MO, Wasserman M. Temporal relationships on macromolecular synthesis during the asexual cell cycle of Plasmodium falciparum. Trans R Soc Trop Med Hyg. 1985;79:792–796. doi: 10.1016/0035-9203(85)90119-1. [DOI] [PubMed] [Google Scholar]
- Desai SA, Krogstad DJ, McCleskey EW. A nutrient-permeable channel on the intraerythrocytic malaria parasite. Nature. 1993;362:643–646. doi: 10.1038/362643a0. [DOI] [PubMed] [Google Scholar]
- Desai SA, Rosenberg RL. Pore size of the malaria parasite's nutrient channel. Proc Natl Acad Sci USA. 1997;94:2045–2049. doi: 10.1073/pnas.94.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn A, Stoffel R, Matile H, Bubendorf A, Ridley RG. Malarial hemozoin β-hematin supports heme polymerization in the absence of protein. Nature. 1995;374:269–271. doi: 10.1038/374269a0. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Haldar K. Secretory transport in Plasmodium. Parasitol Today. 1993;9:98–102. doi: 10.1016/0169-4758(93)90216-3. [DOI] [PubMed] [Google Scholar]
- Elmendorf HG, Haldar K. Plasmodium falciparumexports the Golgi marker sphingomyelin synthase into a tubovesicular network in the cytoplasm of mature erythrocytes. J Cell Biol. 1994;124:449–462. doi: 10.1083/jcb.124.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, et al. Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science. 1998;282:1126–1132. doi: 10.1126/science.282.5391.1126. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda SK, Minton NP. A simple procedure for gel-electrophoresis and Northern blotting of RNA. Nucleic Acids Res. 1995;23:3357–3358. doi: 10.1093/nar/23.16.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther K, Tummler M, Arnold HH, Ridley R, Goman M, Scaife JG, Lingelbach K. An exported protein of Plasmodium falciparumis synthesized as an integral membrane protein. Mol Biochem Parasitol. 1991;46:149–157. doi: 10.1016/0166-6851(91)90208-n. [DOI] [PubMed] [Google Scholar]
- Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. The Chlamydia trachomatisIncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1999;1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- Hinterberg K, Scherf A, Gysin J, Toyoshima T, Aikawa M, Mazie JC, da Silva LP, Mattei D. Plasmodium falciparum: the Pf332 antigen is secreted from the parasite by a brefeldin A-dependent pathway and is translocated to the erythrocyte membrane via the Maurer's clefts. Exp Parasitol. 1994;79:279–291. doi: 10.1006/expr.1994.1091. [DOI] [PubMed] [Google Scholar]
- Horii T, Bzik DJ, Inselburg J. Characterization of antigen-expressing Plasmodium falciparumcDNA clones that are reactive with parasite inhibitory antibodies. Mol Biochem Parasitol. 1988;30:9–18. doi: 10.1016/0166-6851(88)90127-2. [DOI] [PubMed] [Google Scholar]
- Kara UAK, Stenzel DJ, Ingram LT, Bushell GR, Lopez JA, Kidson C. Inhibitory monoclonal-antibody against a (myristylated) small-molecular-weight antigen from Plasmodium falciparumassociated with the parasitophorous vacuole membrane. Infect Immun. 1988a;56:903–909. doi: 10.1128/iai.56.4.903-909.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara UAK, Stenzel DJ, Ingram LT, Kidson C. The parasitophorous vacuole membrane of Plasmodium falciparum: demonstration of vesicle formation using an immunoprobe. Eur J Cell Biol. 1988b;46:9–17. [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of head of bacteriophage-T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparumerythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Langreth SG, Jensen JB, Reese RT, Trager W. Fine structure of human malaria in vitro. J Protozool. 1978;25:443–452. doi: 10.1111/j.1550-7408.1978.tb04167.x. [DOI] [PubMed] [Google Scholar]
- Lauer SA, Rathod PK, Ghori N, Haldar K. A membrane network for nutrient import in red cells infected with the malaria parasite. Science. 1997;276:1122–1125. doi: 10.1126/science.276.5315.1122. [DOI] [PubMed] [Google Scholar]
- Lecordier L, Mercier C, Sibley LD, Cesbron-Delauw MF. Transmembrane insertion of the Toxoplasma gondiiGRA5 protein occurs after soluble secretion into the host cell. Mol Biol Cell. 1999;10:1277–1287. doi: 10.1091/mbc.10.4.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingelbach K, Joiner KA. The parasitophorous vacuole membrane surrounding Plasmodium and Toxoplasma: an unusual compartment in infected cells. J Cell Sci. 1998;111:1467–1475. doi: 10.1242/jcs.111.11.1467. [DOI] [PubMed] [Google Scholar]
- Lupas A, Vandyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Matuschewski K, Ross J, Brown SM, Kaiser K, Nussenzweig V, Kappe SH. Infectivity-associated changes in the transcriptional repertoire of the malaria parasite sporozoite stage. J Biol Chem. 2002;277:41948–41953. doi: 10.1074/jbc.M207315200. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, vonHeijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Push C, Schmitt H, Blin N. Increased cloning efficiency by cycle restriction-ligation (CRL) Technical Tips Online. 1997;1:T40071. [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- Rogers S, Wells R, Rechsteiner M. Amino-acid-sequences common to rapidly degraded proteins: the pest hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- Sallicandro P, Paglia MG, Hashim SO, Silvestrini F, Picci L, Gentile M, Mulaa F, Alano P. Repetitive sequences upstream of the pfg27/25 gene determine polymorphism in laboratory and natural lines of Plasmodium falciparum. Mol Biochem Parasitol. 2000;110:247–257. doi: 10.1016/s0166-6851(00)00274-7. [DOI] [PubMed] [Google Scholar]
- Sam-Yellowe TY, Shio H, Perkins ME. Secretion of Plasmodium flaciparumrhoptry protein into the plasma-membrane of host erythrocytes. J Cell Biol. 1988;106:1507–1513. doi: 10.1083/jcb.106.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarze SR, Dowdy SF. In vivo protein transduction: intracellular delivery of biologically active proteins, compounds and DNA. Trends Pharmacol Sci. 2000;21:45–48. doi: 10.1016/s0165-6147(99)01429-7. [DOI] [PubMed] [Google Scholar]
- Scidmore-Carlson MA, Shaw EI, Dooley CA, Fischer ER, Hackstadt T. Identification and characterization of a Chlamydia trachomatisearly operon encoding four novel inclusion membrane proteins. Mol Microbiol. 1999;33:753–765. doi: 10.1046/j.1365-2958.1999.01523.x. [DOI] [PubMed] [Google Scholar]
- Simmons D, Woollett G, Bergin-Cartwright M, Kay D, Scaife J. A malaria protein exported into a new compartment within the host erythrocyte. EMBO J. 1987;6:485–491. doi: 10.1002/j.1460-2075.1987.tb04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinai AP, Joiner KA. The Toxoplasma gondiiprotein ROP2 mediates host organelle association with the parasitophorous vacuole membrane. J Cell Biol. 2001;154:95–108. doi: 10.1083/jcb.200101073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe JA, Coppel RL, Brown GV, Ramasamy R, Kemp DJ, Anders RF. Identification of 2 integral membrane-proteins of Plasmodium falciparum. Proc Natl Acad Sci USA. 1988;85:5195–5199. doi: 10.1073/pnas.85.14.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmann T, Beck HP. Analysis of stage-specific transcription in Plasmodium falciparumreveals a set of genes exclusively transcribed in ring stage parasites. Mol Biochem Parasitol. 2000;111:453–458. doi: 10.1016/s0166-6851(00)00333-9. [DOI] [PubMed] [Google Scholar]
- Staines HM, Ellory JC, Kirk K. Perturbation of the pump-leak balance for Na(+) and K(+) in malaria-infected erythrocytes. Am J Physiol Cell Physiol. 2001;280:C1576–C1587. doi: 10.1152/ajpcell.2001.280.6.C1576. [DOI] [PubMed] [Google Scholar]
- ter Kuile F, White NJ, Holloway P, Pasvol G, Krishna S. Plasmodium falciparum: in vitro studies of the pharmacodynamic properties of drugs used for the treatment of severe malaria. Exp Parasitol. 1993;76:85–95. doi: 10.1006/expr.1993.1010. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal-W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific Gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, Jensen JB. Cultivation of malarial parasites. Nature. 1978;273:621–622. doi: 10.1038/273621a0. [DOI] [PubMed] [Google Scholar]
- Trager W, Rozario C, Shio H, Williams J, Perkins ME. Transfer of a dense granule protein of Plasmodium falciparumto the membrane of ring stages and isolation of dense granules. Infect Immun. 1992;60:4656–4661. doi: 10.1128/iai.60.11.4656-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbe S, Tooze SA, Barr FA. Formation of secretory vesicles in the biosynthetic pathway. Biochim Biophys Acta. 1997;1358:6–22. doi: 10.1016/s0167-4889(97)00050-5. [DOI] [PubMed] [Google Scholar]
- Waller RF, Reed MB, Cowman AF, McFadden GI. Protein trafficking to the plastid of Plasmodium falciparumis via the secretory pathway. EMBO J. 2000;19:1794–1802. doi: 10.1093/emboj/19.8.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham ME, Rug M, Ralph SA, Klonis N, McFadden GI, Tilley L, Cowman AF. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20:5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolg JW, Macleod AJ, Scaife JG, Beaudoin RL. The accumulation of lactic acid and its influence on the growth of Plasmodium falciparumin synchronized cultures. In Vitro. 1984;20:205–215. doi: 10.1007/BF02618189. [DOI] [PubMed] [Google Scholar]