Abstract
The organization of multiple mitochondrial DNA (mtDNA) molecules in discrete protein-DNA complexes called nucleoids is well studied in Saccharomyces cerevisiae. Similar structures have recently been observed in human cells by the colocalization of a Twinkle-GFP fusion protein with mtDNA. However, nucleoids in mammalian cells are poorly characterized and are often thought of as relatively simple structures, despite the yeast paradigm. In this article we have used immunocytochemistry and biochemical isolation procedures to characterize the composition of human mitochondrial nucleoids. The results show that both the mitochondrial transcription factor TFAM and mitochondrial single-stranded DNA-binding protein colocalize with Twinkle in intramitochondrial foci defined as nucleoids by the specific incorporation of bromodeoxyuridine. Furthermore, mtDNA polymerase POLG and various other as yet unidentified proteins copurify with mtDNA nucleoids using a biochemical isolation procedure, as does TFAM. The results demonstrated that mtDNA in mammalian cells is organized in discrete protein-rich structures within the mitochondrial network. In vivo time-lapse imaging of nucleoids show they are dynamic structures able to divide and redistribute in the mitochondrial network and suggest that nucleoids are the mitochondrial units of inheritance. Nucleoids did not colocalize with dynamin-related protein 1, Drp1, a protein of the mitochondrial fission machinery.
INTRODUCTION
Mammalian mitochondrial DNA (mtDNA) is a 16.5-kb circular double-stranded DNA (Anderson et al., 1981; Bibb et al., 1981) present in one to several thousand of copies per cell (e.g., Takamatsu et al., 2002). All proteins involved in mtDNA maintenance are encoded by the nuclear genome. These are traditional proteins in replication and repair such as the mtDNA polymerase POLG, but also include proteins directly or indirectly involved in e.g., segregation.
MtDNA in the yeast Saccharomyces cerevisiae and to a lesser extent in a few other species, appears to be organized in discrete foci within mitochondria called nucleoids (Miyakawa et al., 1984). These have been inferred to be the units of inheritance each containing several copies of yeast mtDNA (Jacobs et al., 2000; MacAlpine et al., 2000 and references therein). Biochemical purification and protein analysis have defined several of the constituents of yeast nucleoids (Miyakawa et al., 1995; Newman et al., 1996; Kaufman et al., 2000). One of the core components is the Abf2 protein (Abf2p), an orthologue of the human mitochondrial transcription factor TFAM. The function of Abf2p in yeast is essential for mtDNA maintenance by providing a mtDNA-packaging function. It also modestly stimulates yeast transcription in in vitro assays (Parisi et al., 1993). A mouse TFAM knockout shows embryonic lethality with complete loss of mtDNA (Larsson et al., 1998), but this is generally believed to be the result of impairment of transcription initiation that would generate primers for mtDNA replication.
Other yeast nucleoid components include Rim1p, the yeast single-stranded DNA-binding protein, and also several proteins with separate roles believed to be unrelated to nucleoid maintenance. Such proteins include α-ketoglutarate dehydrogenase subunits and aconitase (Kaufman et al., 2000). It seems likely that these dual-function nucleus-encoded enzymes that nevertheless have conserved catalytic cores have randomly acquired additional functions and coevolved with mtDNA in various eukaryotic lineages. This suggests that a great variability in the types of proteins associated with nucleoids in different species can be expected.
Although mtDNA inheritance is significantly different between budding yeast and mammalians, there is both genetic and cell biological evidence to believe that also in mammals mtDNA is organized in nucleoid-like structures (Jacobs et al., 2000; Lehtinen et al., 2000). Nevertheless, only our recent demonstration of specific colocalization of Twinkle-EGFP with mtDNA in punctate intramitochondrial structures for the first time clearly visualized the existence of discrete mtDNA-protein complexes in human mitochondria (Spelbrink et al., 2001). Twinkle shows similarity to the bacteriophage T7 primase/helicase gene 4 protein. Mutations in the gene for Twinkle, C10orf2, are associated with forms of a late onset autosomal dominant disorder, progressive external opthalmoplegia, characterized at the molecular level by the accumulation of multiple mtDNA deletions.
Mitochondria are maintained as a highly active and flexible network. This is accomplished by mitochondrial growth, fusion, and division, and active movement to sites of energy demand. This ensures proper number and distribution of mitochondria. The distribution and inheritance of mtDNA during these processes has been relatively underexplored, especially in mammalian cells, because of a lack of appropriate markers to directly visualize mtDNA in vivo. Nevertheless, in recent years, many of the proteins involved in the mitochondrial division and fusion machinery have been identified. Drosophila, yeast, and mammalian Fzo/Mitofusin proteins are essential for mitochondrial fusion (Hales and Fuller, 1997; Hermann et al., 1998; Santel and Fuller, 2001). They are transmembrane GTPases with the catalytic domain exposed to the cytoplasm.
The first factor described to be responsible for mitochondrial division is the Caenorhabditis elegans and mammalian dynamin-related protein Drp1 (also known as Dlp1, Dymple; Kamimoto et al., 1998; Smirnova et al., 1998; Labrousse et al., 1999; Pitts et al., 1999) and its yeast orthologue Dnm1 (Bleazard et al., 1999; Sesaki and Jensen, 1999). Mutations in this protein cause mitochondria to collapse into perinuclear aggregates consisting of long interconnected tubules. This is probably the consequence of a shift in the balance between fission and fusion (Smirnova et al., 2001). Genetics screens in yeast point at MDV1 and FIS1 as Drp1(Dnm1) partners (Fekkes et al., 2000; Mozdy et al., 2000; Tieu and Nunnari, 2000).
MATERIALS AND METHODS
Cell Culture and Transfection
HEK293EBNA, 143B osteosarcoma, A549 adenocarcinoma cells and primary human fibroblasts were cultured as described (Spelbrink et al., 2000). Cells were seeded in six-well plates 1–2 d before transfection at 40–70% density. Transfection used 10 μl lipofectamine (Life Technologies, Paisley, United Kingdom) for 293 cells, A549 cells and fibroblasts diluted in 1 ml Optimem (Life Technologies) according to the manufacturer's protocol. Five hours after transfection, we added 2 ml fresh medium and replaced the medium 24 h after transfection. For 143B cell transfection, we used FuGENE 6 (Roche, Mannhiem, Germany) as per the manufacturer's specifications.
Expression Constructs
Twinkle-EGFP, Twinkle-MycHis, and GFP-Drp1 constructs were as described before (Smirnova et al., 2001; Spelbrink et al., 2001)
Immunocytochemistry plus Confocal Microscopy and Time-lapse Epifluorescence Microscopy
Mitotracker Red (Molecular Probes, Eugene, Oregon) staining and cell preparation were as described elsewhere (Spelbrink et al., 2000). Cells were fixed for 20 min in PBS/3.7% formaldehyde/5% sucrose at 37°C, washed twice with PBS and mounted using Vectashield mounting medium (Vector Laboratories, Burlingame, CA). For ICC, fixed cells were processed by lysis for 15 min in PBS/0.5% Triton X-100 at room temperature, washed twice in PBS, and blocked in PBS/3% BSA. For colocalization studies of Drp1 and mtSSB we dehydrated and rehydrated samples before ICC as described below for (5-bromo-2-deoxy-uridine) BrdU labeling. Antibodies were diluted in PBS/3% BSA. Coverslips were incubated with the antibody solution on slides for 1 h at room temperature. Dilutions were as follows: mouse anti–c-Myc mAb 9E10 (Roche) 1:1000 dilution of 5 mg/ml stock; rabbit anti-human TFAM (kind gift of Dr R. Wiesner) 1:200; rabbit anti-mtSSB (kind gift of Dr M. Zeviani) 1:100; rabbit anti-POLG (Santa Cruz, Santa Cruz, CA, sc-5930) 1:50; and rabbit anti-POLG2 (kind gift of Dr. P. Lestienne) 1:400. Slides were washed three times with PBS and incubated with a 1:200 dilution of secondary biotinylated antibodies (anti-mouse IgG or anti-rabbit IgG, Vector Laboratories, Inc.; anti-goat IgG, DAKO, Glostrup, Denmark), washed as above, and incubated with a 1:200 dilution of either streptavidin-Texas Red/Rhodamine or streptavidin-Fluorescein (Vector Laboratories Inc.) All fixed samples were examined by confocal scanning laser microscopy, using a Perkin Elmer-Cetus/Wallac UltraView LCI system (Wellesly, MA). Confocal images were processed by the UltraView 4.0 software and further handled using Microsoft Photo Editor 3.01 and Adobe Photoshop 6.0 to obtain appropriate sections with best contrast/brightness and resolution. Individual images were assembled using Microsoft Powerpoint and further handled with Adobe Photoshop 6.0 again to obtain best possible resolution for final printing.
For time-lapse imaging, COS-7 cells were grown in DMEM supplemented with 10% fetal calf serum. Cells were seeded onto glass-bottom dishes (Mat-Tek, Ashland, MA) for in vivo observation and transfected by using FuGENE transfection reagent (Roche) according to the manufacturers' procedures. Before observation, cells were incubated with 0.1 μM MitoTracker Red for 10 min at 37°C. The images were acquired 18 h after the start of the transfection by a cooled CCD camera (PXL; Photometrics) powered by Isee Inovision Corporation software. Time-lapse images were collected every 15 s using Plan 100×/1.25 NA oil immersion objective at room temperature. Individual time-lapse frames were imported to the UltraView 4.0 software, to obtain images of “regions of interest,” these were again exported and handled further as above. For the measurement of mitochondrial and Twinkle-GFP movements, all the individual frames were imported into NIH Image 1.62. For each chosen mitochondrion, one of its tips was followed through the frames acquired every 15 s for >7 min. The average velocity was calculated from the measured distances, including the points when the mitochondrion has not moved. The results are expressed as a mean value ± SE.
Metabolic Labeling of mtDNA using BrdU
BrdU and an mAb specific for BrdU in single-stranded DNA were from Roche. One or 2 d before BrdU labeling 143B osteosarcoma cells deficient for cytoplasmic thymidine kinase (TK−) were seeded at low density on coverslips. BrdU was added to a final concentration of 10 μM and cells were incubated for 2–24 h. Cells were subsequently fixed and lysed as described above for immunocytochemistry. This was followed by dehydration using serial washes of 70, 90, and 100% ethanol. Cells were rehydrated using several PBS washes. DNA was denatured as described (Davis and Clayton, 1996). Before antibody incubations the coverslips were blocked for 15 min using 3% BSA in PBS. The anti-BrdU mAb was used as a 1:50 dilution in PBS/1% BSA. Horse anti-mouse–Fluorescein IgG was used as a secondary antibody at a 1:200 dilution in PBS/3% BSA. Slides were subsequently used for ICC or directly mounted. For the detection of Myc-tagged Twinkle using the anti-Myc monoclonal in ICC, BrdU-ICC slides were postfixed for 10 min in PBS/3.7% formaldehyde/5% sucrose at 25°C and washes three times in PBS before Myc-ICC. After the anti-Myc mAb incubation, slides were incubated with 1:200 biotinylated anti-mouse IgG as secondary antibody followed by streptavidin-Texas Red. This resulted in very little or no cross-reactivity of the biotinylated anti-mouse IgG with the BrdU mAb that was presumably inaccessible due to cross-linked horse anti-mouse-Fluorescein IgG (compare Figures 3, D and E, lower left corner, and unpublished data).
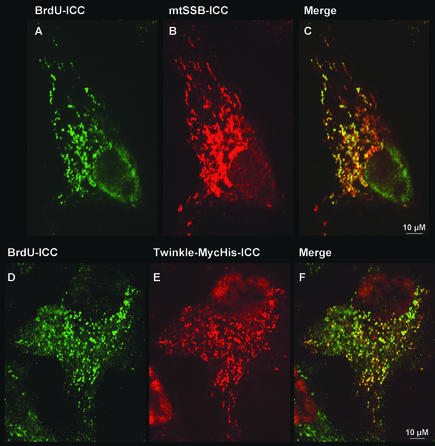
Figure 3.
Intramitochondrial foci that are positive for mtSSB or Twinkle are sites of BrdU incorporation. MtDNA was labeled with BrdU for 24 h (A–C) or 2 h (D–F) in 143B(TK−) cells seeded at low density 24–48 h before labeling. BrdU incorporation was detected by a BrdU-specific mAb (A and D). The cells shown in A were subsequently processed for mtSSB-ICC (B), whereas the cells shown in D, transfected with a Twinkle-MycHis construct 24 h before BrdU labeling, were processed for Twinkle-MycHis-ICC (E). (C and F) The merged images for A+B and D+E, respectively. Results show an abundant and specific BrdU labeling in most foci that also specifically stain for either mtSSB or Twinkle, demonstrating that these foci are nucleoids.
Immunoelectron Microscopy
For immunoelectron microscopy HEK 293 cells were grown on Termanox plastic dishes, fixed with 0.01% glutaraldehyde, and 3.5% paraformaldehyde in 0.1 M phosphate buffer, pH 7.2, and embedded in Lowicryl HM 20 (Agar Scientific, Stamsted, United Kingdom). Thin sections were stained by indirect immunogold labeling according to standard methods with the rabbit anti-human TFAM antibody (1:400) and 5- or 10-nm protein A-gold and poststained with uranyl acetate and lead citrate.
Isolation of Mitochondria
HEK293EBNA cells were disrupted to isolate mitochondria in a chilled 5 ml Dounce homogenizer by 25 strokes at medium speed, essentially as described (Spelbrink et al., 2000).
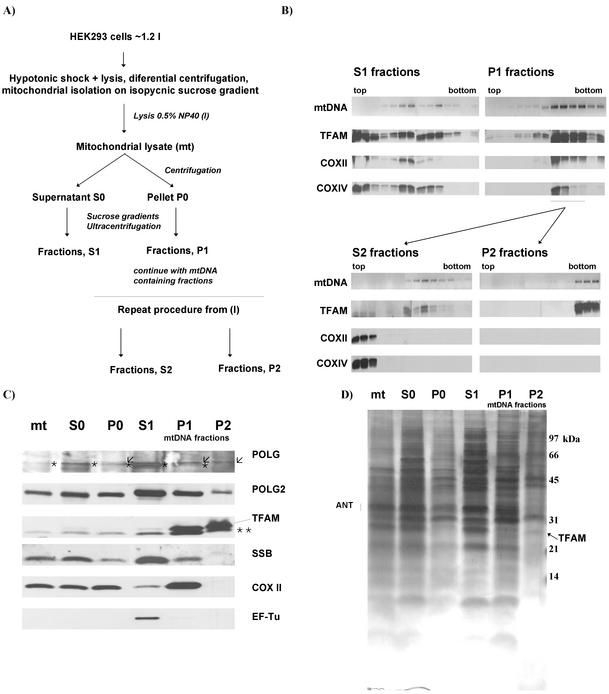
Mitochondria were further purified by isopycnic gradient centrifugation: the pellet was resuspended in 1 mM EDTA, 0.1% BSA, and 10 mM Tris-HCl, pH 7.5, containing 0.8 M sucrose, layered on top of a continuous sucrose gradient from 1 to 2 M sucrose in the same buffer and centrifuged at 80,000 × g for 2 h. Mitochondria were collected and diluted with 2 vol of 1 mM EDTA, 10 mM Tris-HCl, pH 7.4 and pelleted at 16,000 × g for 15 min. The purified mitochondrial pellet was stored at −80°C.
Isolation and Characterization of mtDNA Nucleoids
We modified a published isolation procedure for mtDNA nucleoids from yeast (Newman et al., 1996). Briefly, purified mitochondria were thawed on ice, resuspended in NE2 buffer (0.25 M sucrose, 20 mM Tris-HCl, pH 7.6, 2 mM EDTA, 7 mM β-mercaptoethanol), and diluted with equal volume of 0.5× NE2 buffer to a final concentration of 5–7 mg mitochondrial protein/ml. Spermidine (1.0 M) was added to a final concentration of 3 mM, and mitochondria were lysed by adding 20% NP40 to a final concentration of 0.5%. After 15 min with gentle stirring, the lysate was fractionated at 12,000 × g for 20 min into supernatant (S0) and pellet (P0) fractions. The pellet fraction was resuspended as above.
Next, supernatant and pellet fractions were layered on top of step gradients comprised of 3.5 ml 20%/2.5 ml 40%/1.8 ml 60%/0.9 ml 75% sucrose in gradient buffer (20 mM Tris-HCl, pH 7.6, 1 mM EDTA, 1 mM spermidine, 7 mM β-mercaptoethanol, 1 mM PMSF) and centrifuged at 111,000 × g for 75 min. Gradients were fractioned and analyzed for distribution of mtDNA and protein. mtDNA containing samples derived from the S and P fractions from an initial NP40 extraction are hereafter referred to as S-1 and P-1, respectively. P-1 sample was collected, diluted with 2 vol ice cold gradient buffer, treated again with 0.5% NP40 for 15 min, and centrifuged through a second step gradient at 49,000 × g for 3 h, to yield S-2 and P-2.
Sucrose gradient samples were dialyzed at 4°C for several hours against NE2 buffer in order to reduce the sucrose concentration before analysis by SDS-PAGE
Nucleoid Analyses
mtDNA distribution in gradients was determined by PCR. After proteinase K treatment mtDNA was extracted by ethanol precipitation and amplified with specific primers FR6 5′ GGTGCAGCCGCTATTAAAGGTCG 3′ and FR7 5′ CCGATCAGGGCGTAGTTTG 3′, amplifying a 685-base pair fragment of human mtDNA corresponding to base pairs 3013–3698 of the Cambridge Reference Sequence (as described, Spelbrink et al., 2000).
Protein distribution was analyzed by SDS-PAGE on 10% or 14% polyacrylamide gels according to Laemmli (1970) or according Schägger and Von Jagow (1987). PAGE markers were from Bio-Rad (Hercules, CA). Before loading all samples were heated at 95°C for 5 min in SDS sample buffer (50 mM Tris-HCl, pH 6.8, 12% glycerol, 4% SDS, 0.01% Serva Blue G, 0.1 M DTT). After electrophoresis, gels were stained with 0.1% Coomassie Brilliant Blue in 40% methanol, 10% acetic acid for 30 min. Destaining was carried out in 40% methanol, 10% acetic acid followed by 10% methanol, 10% acetic acid. Coomassie-stained gels were next silver-stained (Morrissey, 1981). Immunoblot analysis was essentially done as described (Spelbrink et al., 2000). Primary antibodies and dilutions used were as follows: mouse anti–c-Myc mAb 9E10 (Roche) 1:15,000; mouse anti-COXII and mouse anti-COXIV monoclonal (kind gift of Dr R. Capaldi) 1:10,000; rabbit anti-human TFAM 1:10,000; rabbit anti-mtSSB 1:5000; rabbit anti-POLG (Santa Cruz, sc-5930) 1:2000; rabbit anti-POLG2 1:10,000; mouse monoclonal anti–EF-Tu (kind gift of Dr. F. Henkler) 1:500; and goat antihistone H2A (Santa Cruz, sc-8648) 1:500. The POLG blocking peptide (Santa Cruz, sc-5930p) was used at 10 times excess of the antibody. Blots were further processed as previously described (Spelbrink et al., 2000). For larger 13-cm gels as shown in Figure 4, C and D, all samples were run on the same gel, but because of gel size it was sliced in two, each half being blotted to a separate membrane. Both membranes were always incubated with antibody together.
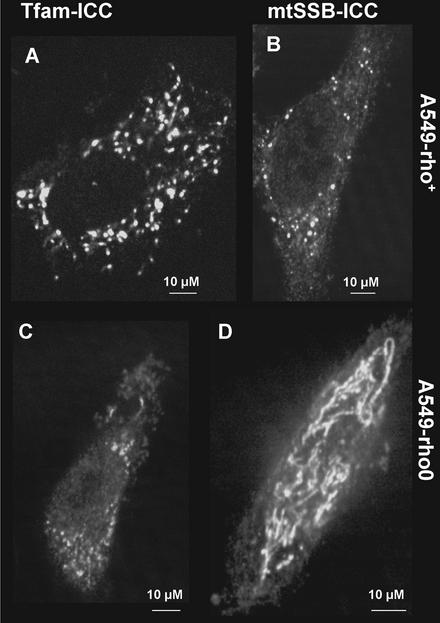
Figure 4.
Distribution and expression levels of endogenous TFAM and mtSSB in rho-zero cells. (A) ICC for endogenous TFAM in A549 rho+ and rho-zero (C) cells. (B) ICC for endogenous mtSSB in A549 rho+ and rho-zero (D) cells. The results show a significant reduction in detectable TFAM protein levels. Note that the exposure and image enhancement for C is different from that in A in order to obtain the most informative image in each case.
RESULTS
Although mtDNA-protein complexes were isolated from mammalian cells many years ago (see e.g., Albring et al., 1977), these complexes remain poorly characterized. Twinkle, a protein with similarity to the phage T7 gene 4 protein (T7 gp4), is the first protein that was found to specifically colocalize in vivo with mtDNA in mammalian cells (Spelbrink et al., 2001), thus corroborating the existence of highly organized nucleoids in mammals. A punctate submitochondrial localization had until then only been observed for proteins of the mitochondrial fusion and fission apparatus such as dynamin-related protein 1 (Drp1; Smirnova et al., 2001), although these proteins so far have not been shown to colocalize with mtDNA. Other proteins, such as the mitochondrial transcription factor TFAM and the mitochondrial polymerase POLG seem to be uniformly distributed within mitochondria (Spelbrink et al., 2000, and unpublished observations), but these results are based only on GFP reporter assays.
Endogenous TFAM and mtSSB, But Not POLG and POLG2 Specifically Colocalize with Twinkle in Nucleoids in Human Mitochondria
As a first approach to further determine nucleoid protein composition we examined the in vivo localization of various endogenous mitochondrial proteins with a known function in mtDNA maintenance. To this end we used immunocytochemistry (ICC) with antibodies against TFAM, the mitochondrial single-stranded DNA-binding protein mtSSB, POLG and its accessory subunit POLG2 (also known as the beta subunit). Both untransfected cells or cells transiently transfected with a Twinkle-GFP construct were studied. All antibodies, except the POLG antibody, have been shown to be highly specific and show little background staining on Western blots with isolated mitochondria (see Spelbrink et al., 2000, and unpublished data).
Figure 1 shows that both endogenous TFAM and mtSSB have a punctate mitochondrial staining pattern, untypical for the overall mitochondrial network structure in these cells seen with counterstaining with Mitotracker Red. The combined images (Figure 1, C and F) show that both TFAM and mtSSB are concentrated in defined foci within mitochondria, reminiscent to those observed with Twinkle-GFP. This was true for various human cell lines such as osteosarcoma and lung carcinoma cells as well as primary or transformed fibroblasts and was also observed in mouse 3T3 cells (unpublished data) and COS7 cells (see below). Especially TFAM, though concentrated in punctate structures, also showed weak, more uniform mitochondrial fluorescence, revealing the mitochondrial network as observed after Mitotracker staining (see also Figure 2A).
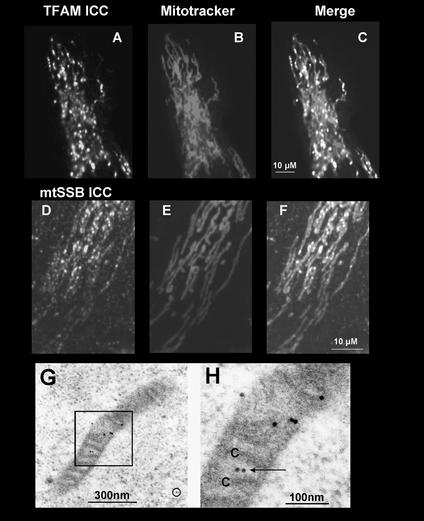
Figure 1.
Localization of endogenous TFAM and mtSSB in intramitochondrial foci. 143B osteosarcoma cells grown on coverslips for 1–2 d were stained with Mitotracker Red (B and E). Detection of TFAM (A) and mtSSB (D) by ICC used polyclonal rabbit antibodies and a secondary fluorescein labeled antibody. (C; TFAM, F; SSB) Merged fluorescent images. (G) Immunolabeling for TFAM in a HEK293 cell mitochondrion. Seven gold particles were concentrated along a stretch of 300 nm of the mitochondrion. Very little background labeling was observed outside mitochondria (circle). (H) Blow-up of the boxed area in G, showing TFAM label (arrow) in the mitochondrial matrix near cross-sectioned cristae (C).
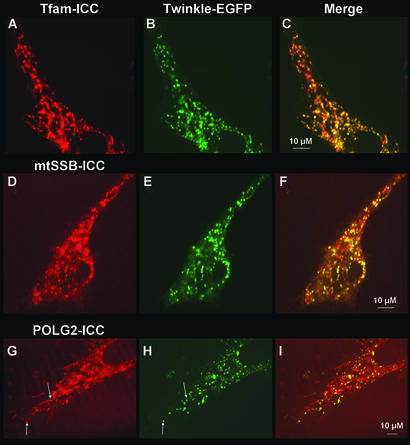
Figure 2.
Colocalization of Twinkle-EGFP with endogenous TFAM, mtSSB, and POLG2. 143B osteosarcoma cells (or A549 lung-carcinoma cells) were grown on coverslips and transfected with a Twinkle-EGFP constructs. One to 2 d after transfection ICC for the detection of TFAM, mtSSB, or POLG2 was performed. Secondary antibodies had rhodamine or Texas Red fluorescent groups. (A, D, and G) ICC for TFAM, mtSSB, and POLG2, respectively. (B, E, and H) Same cells as for A, D, and G showing the expression of Twinkle-EGFP. (C, F, and I) Merged fluorescent images showing colocalization of Twinkle-EGFP with TFAM, SSB, and POLG2, respectively. Some but not all foci containing the apparent highest POLG2 concentrations also contained Twinkle-EGFP as indicated by arrows in G and H, but overall POLG2 fluorescence was more uniform than TFAM or mtSSB fluorescence.
In contrast to TFAM and mtSSB, endogenous POLG and POLG2 showed an almost uniform mitochondrial fluorescence very similar to the Mitotracker staining. POLG2 did show some apparent foci with higher concentrations inside mitochondria (see Figure 2 and below). It was difficult to detect possible foci with higher concentrations of POLG because the antibody gave only weak mitochondrial fluorescence with some background in ICC (unpublished data).
As an additional approach to study nucleoid protein composition and to localize nucleoids at higher resolution, we used immunoelectron microscopy. After staining with the TFAM antibody, immunogold label was often found concentrated at regions within the mitochondrial matrix (Figure 1, G and H). Most mitochondria showed patches with several gold particles close together and long stretches devoid of label. On several occasions gold particles in cristae-free areas were associated with filamentous structures possibly containing nucleic acid (unpublished data).
To establish more precisely the localization of TFAM and mtSSB, we transfected osteosarcoma cells with Twinkle-GFP and subjected the cells to ICC 2 d after transfection. The results show a clear colocalization of Twinkle-GFP both with TFAM and with mtSSB (Figure 2, A–F) strongly suggesting that endogenous TFAM and SSB are preferentially present in mitochondrial nucleoids. Foci with apparent highest concentrations of POLG2 appeared also to colocalize with Twinkle-GFP (Figure 2, G–I, indicated by arrows).
We had previously used ethidium bromide staining to look for colocalization of Twinkle with mtDNA (Spelbrink et al., 2001), but this staining was not as discrete as the Twinkle localization, possibly because of costaining of mitochondrial RNA. We now used BrdU metabolic labeling of mtDNA, followed by immunodetection of denatured DNA containing BrdU, and ICC for mtSSB or ICC for cotransfected Twinkle with a MycHis epitope tag. BrdU is a thymidine analog that can be specifically incorporated in DNA once it has become phosphorylated by thymidine kinase. In cells that are deficient in their cytoplasmic thymidine kinase (TK−) while retaining the mitochondrial isozyme, specific mtDNA staining can be easily detected (see Davis and Clayton, 1996 and references therein). However, a disadvantage of BrdU labeling could be that incorporation of this analog in mtDNA was previously shown to mainly take place in perinuclear mitochondria, suggestive of a spatial regulation of mtDNA replication (Davis and Clayton, 1996). This was hypothesized to be the result of a limitation in one or several factors encoded by the nucleus that needed to be imported by the mitochondria. To test BrdU labeling in 143B(TK−) cells, we labeled cells for 2 and 24 h and detected BrdU incorporation using a monoclonal BrdU-specific antibody. We found that BrdU incorporation was very efficient, even after a 2-h labeling period, with labeling of mtDNA more or less uniform throughout the cell (see also Figure 3). This meant that BrdU labeling could be used as a highly specific DNA “stain” to demonstrate specific incorporation in foci that we have so far tentatively termed nucleoids. The results of these experiments are shown in Figure 3. In this particular experiment, mtSSB ICC showed somewhat more uniform mitochondrial fluorescence, but foci could still be spotted easily. This more uniform staining was within the normal range of variation that we observed with this antibody (unpublished data). There was, nevertheless, very good colocalization of mtSSB and of Twinkle-MycHis with sites of BrdU incorporation, unequivocally demonstrating that Twinkle and mtSSB colocalize with mtDNA. Similar results were obtained for Tfam (unpublished data). Because labeling with BrdU is metabolic, it is not surprising that some foci stain positive for Twinkle or mtSSB but not for BrdU. The number of foci that were positive for mtSSB or Twinkle and negative for BrdU after 24 h of labeling was generally smaller than after 2 h of labeling. Nevertheless, even after 2 h most mtSSB or Twinkle-positive foci were already BrdU positive in a considerable proportion of cells of the culture. An example of this is shown in Figure 3, D–F, for Twinkle-transfected cells. No BrdU incorporation was observed in rho-zero cells (unpublished data). Because also after a 24-h labeling period most BrdU incorporation sites still colocalized with mtSSB or Twinkle, the results strongly suggest that nucleoid associated proteins are not merely DNA synthesis/repair factories that dissociate after replication and/or repair. Further BrdU labeling studies under well controlled experimental conditions could prove very useful to unravel dynamics of mtDNA synthesis and the maintenance machinery.
Nucleoid Proteins No Longer Form Nucleoid-like Structures in rho-zero Cells
Nucleoids can be expected to be held together both by protein–protein and protein–DNA interactions. In cells without mtDNA (rho-zero cells) one might therefore expect a disruption of nucleoid structure. The various proteins tested by ICC gave different results as a consequence of the absence of mtDNA (Figure 4). TFAM levels appeared severely reduced (Figure 4, A and C, compare rho+ and rho-zero cells of the same genetic background), although the remaining staining did appear somewhat punctate. It did not strictly colocalize with Twinkle-GFP (unpublished data), even though most remaining TFAM staining was mitochondrial. Mitochondrial SSB was easily detectable by ICC. The staining in rho-zero cells (compare Figure 4, B and D), in sharp contrast with staining in rho+ cells, showed a much more uniform mitochondrial fluorescence revealing a mitochondrial network similar to that observed after Mitotracker staining (unpublished data). Both POLG and POLG2 showed uniform mitochondrial fluorescence and did not appear to be down- or upregulated compared with cells with mtDNA (unpublished data).
Biochemical Purification of Nucleoids from Cultured Human Cells Demonstrates Partial Copurification of TFAM, mtSSB, POLG, and POLG2 with mtDNA
The ICC results have defined some of the proteins that should be present in purified mitochondrial nucleoids. To biochemically purify nucleoids we used isopycnic-gradient purified mitochondria followed by two sequential NP40 lysis and sucrose-density gradient centrifugation steps (see Figure 5A). Initial purification steps and gradient fractions were monitored using PCR amplification for mtDNA, and TFAM served as a nucleoid-protein marker on Western blots. Cytochrome c oxidase subunit II (COXII) and IV (COXIV) were used as markers for possible inner membrane contamination. A typical result from the first lysis step onward showing most sucrose gradient fractions is shown in Figure 5B. More extensive subsequent analysis by Western blotting and total protein staining was done, using representative sucrose gradient fractions only, on large SDS-PAGE Schägger-von Jagow gels. Apart from TFAM and COXII, we checked for purification of mtSSB, POLG, POLG2, and mitochondrial elongation factor Tu (mtEF-Tu) as another negative control because it is involved in translation and as far as we know not in mtDNA replication or repair. It was therefore not surprising to find mtEF-Tu mainly in the S1 fraction, which also nicely illustrated the differential enrichment we observed for the various proteins. As previously shown for yeast nucleoid isolation procedures from which our procedure was adapted, the sucrose gradient fractions containing most mtDNA and copurifying proteins are derived from the insoluble pellet fraction after lysis of mitochondria using 0.5% NP40. The soluble NP40 fractions that contained mtDNA consistently purified in the gradient at lower density. Also, as observed for yeast nucleoids, the first NP40 pellet gradient still showed a considerable amount of COXII and COXIV copurifying with mtDNA and TFAM, suggesting contamination with membrane protein complexes. After a second NP40 treatment of these mtDNA-containing fractions and a second sucrose-density gradient of the resulting NP40 pellet, very little or no COXII and COXIV was left in the mtDNA fractions but were now detected in the soluble fraction. In addition, various proteins such as what we assume is an adenine nucleotide translocator isoform (ANT) are still present at high concentrations in the first “pellet”-gradient P1 (Figure 5D). These were no longer detectable after the second lysis and gradient step, consistent with a further purification of the nucleoids and loss of nonspecific proteins. In sharp contrast, hardly any TFAM was lost from the nucleoid fraction by the second NP40 lysis step (Figure 5, C and D), showing that this protein in contrast to COXII, ANT, and other proteins was strongly associated with mtDNA and specifically retained.
Figure 5.
Biochemical isolation of nucleoids from cultured human cells. HEK 293 Cells were grown to ∼80% confluence and isolated by low-speed centrifugation. The mitochondrial isolation procedure and subsequent steps of the nucleoid isolation procedure is depicted in A. Abbreviations for the various fractions as used in the other panels are also indicated. (B) Analysis of sucrose gradients from the various purification steps as indicated in A. (C) A more thorough analysis of fractions from each step of the isolation procedure, using various specific antibodies against mitochondrial proteins. The S1 fraction used in this case was from the top of the sucrose gradient. Please note that a major cross-reacting non-POLG protein (indicated by asterisk) was observed with slightly higher mobility than POLG itself (indicated by open arrow). The cross-reacting species is prominent in fractions mt, S0, P0, and S1, whereas it is only faintly visible in P1 and no longer visible in P2. POLG was very faintly visible in P0 while becoming more prominent in P1 and P2, indicating its enrichment. Two asterisks indicate a TFAM breakdown product (see main text for further explanations). (D) The same samples as used for C, but the gel was Coomassie and subsequently silver-stained to reveal all proteins. Protein markers are indicated on the right in kDa. We tentatively assigned the TFAM protein by comparison of the gel with the immunoblots shown in C and the adenine nucleotide translocator (ANT) on the basis of its molecular mass and abundance. Note also that because of gel loading limitations total protein concentration in the final P2 fraction is significantly less than, for example, that in P1, Also note that bands in P2 are slightly upshifted possibly because of incomplete dialysis to remove excess sucrose.
The TFAM antibody that we used recognized two TFAM species, the lower of the two being partially degraded because it was hardly detected when we included a cocktail of protease inhibitors during the purification procedure (unpublished data). In addition, TFAM copurifying with mtDNA was mostly full length, in contrast to TFAM found in some of the other fractions. DNAseI treatment of mitochondrial lysates resulted in increased sensitivity of TFAM to degradation and furthermore resulted in the formation of an additional lower molecular weight species (unpublished data). MtSSB was mostly lost from the nucleoid fraction after the second lysis and gradient step.
Despite the ICC findings, the results also showed that not only TFAM copurifies with mtDNA nucleoids to the very last step but also small amounts of POLG2. Interestingly, POLG almost exclusively copurified with nucleoids contradicting the ICC results for this protein. However, the commercial POLG antibody appeared to cross-react with several other proteins that were present in our mitochondrial fractions at much higher concentrations than POLG (see Figure 5C). Nevertheless, recognition of only one protein with the expected size could be specifically inhibited on Western blots by the POLG peptide used for immunization (unpublished data), making it possible to positively identify POLG (indicated by an arrow in Figure 5C, see also figure legend).
Finally, as a control for the purity of our nucleoid preparations we examined the possibility of contamination with nuclear chromatin proteins. Using a commercial polyclonal antibody we could not detect any human histone protein 2A (H2A) in our mitochondrial preparations, whereas the protein was clearly detected in a nuclear extract (unpublished data). Second, a mitochondrial preparation that we fixed after the isopycnic gradient purification showed DAPI-stained intramitochondrial spots, supposedly nucleoids, but not any extramitochondrial DNA material that would indicate the presence of chromatin.
Nucleoid Dynamics in Live Cells
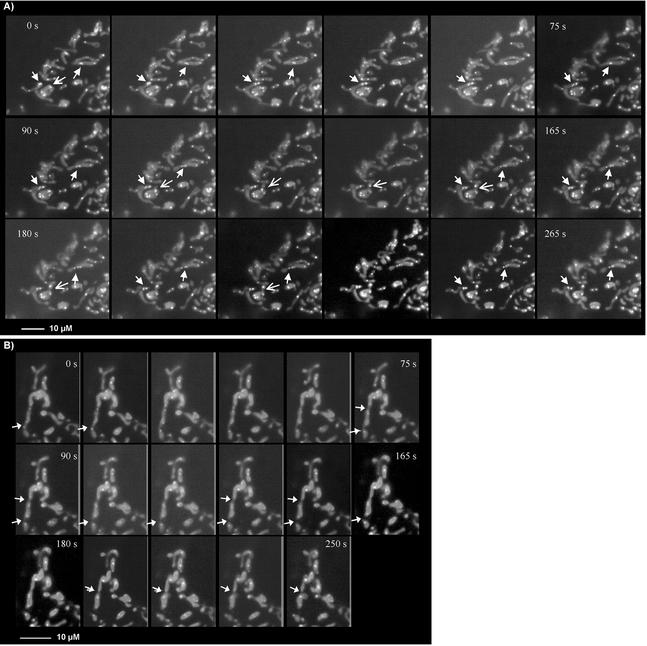
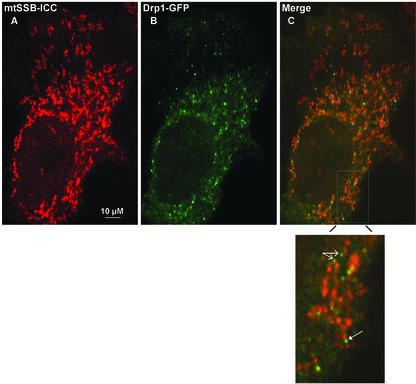
Being able for the first time to visualize mammalian nucleoids using GFP-tagged Twinkle, we tried to address basic questions of nucleoid dynamics in living cells by performing time-lapse fluorescence microscopy. The time-lapse results first show that Twinkle-GFP punctae are not static but mobile, following the mitochondrial dynamics. When observed at room temperature, most mitochondria (93%) exhibited short oscillatory movement. This movement was not confined to a single direction, but rather consisted of multiple trajectories. It also manifested itself as a change of mitochondrial shape and/or stretching and contraction of some of its parts. The calculated velocity of mitochondrial displacement in any direction in our experimental system was 0.01 ± 0.0007 μm/s (7 mitochondria, 149 time points). A small fraction (7 ± 0.8%) of mitochondria moved faster than 0.05 μm/s, mostly in saltatory unidirectional movements. There were ∼2.3 ± 0.4 Twinkle-GFP spots per mitochondrion (n = 58). Their movements were similar to those of mitochondria both quantitatively and qualitatively. Both nondividing and dividing Twinkle-GFP foci oscillated within the mitochondrion, and the velocity of their displacement was 0.01 ± 0.0008 μm/s (5 Twinkle-GFP spots, 110 time points), similar to the overall mitochondrial velocity. These results strongly suggest a membrane association of nucleoids, consistent with previous suggestions of membrane association of mtDNA via the D-loop region (see also Albring et al., 1977). With Twinkle-GFP and ICC, we also observed what appears to be a minimum size for Twinkle, TFAM, and mtSSB containing spots of ∼0.1–0.3 μm in diameter. The largest Twinkle containing spots were typically up to 0.8–1.0 μm in diameter and were often asymmetrical. Most importantly, these larger sized structures were on several occasions observed to break up in several smaller sized Twinkle containing spots (indicated by solid arrows in Figure 6A), suggesting nucleoid division events. Although we did on occasion see smaller spots apparently join to form larger spots, this usually was an apparent rejoining of nucleoid elements that had been together in a larger assembly before (see also Figure 6A). A third observation was that nucleoid division can take place without division of mitochondria (see also solid arrows Figure 6A). Nevertheless, in 67% of the dividing mitochondria, nucleoids appear to be positioned at or near a site of mitochondrial division and could show apparent concomitant division or redistribution to a daughter mitochondrion (open arrow in Figure 6A). Alternatively, at the cell periphery we did observe clear mitochondrial fission events without division of the nucleoids that were distributed over the entire length of the mitochondrion so that each daughter mitochondrion would have one or more nucleoids (Figure 6B, arrows indicate fission events). The combined results show that upon mitochondrial fusion or fission, mtDNA is distributed or redistributed to ensure proper inheritance. Even though the cells that we used for the time-lapse imaging are effectively two-dimensional at the cell periphery (L.G. and A.M.vdB., unpublished data), we tried to further examine nucleoid positioning to possible sites of mitochondrial fission by covisualization of one of the components of the mitochondrial division machinery, Drp1, with mtSSB. In this case we used GFP-Drp1, whereas mtSSB was visualized using ICC. This experiment, shown in Figure 7, clearly demonstrated there is essentially no colocalization of mtSSB-nucleoid foci with Drp1. Drp1-GFP could be observed at sites of what appear to be constrictions of mitochondria with mtSSB foci at either site of the constriction (indicated with a solid arrow in the enlargement of a section from Figure 7C). In addition, Drp1 was also frequently observed at tips of mitochondria of what appear as sites of recent fission events (an example is indicated with open arrows), very similar to the results reported for Drp1 in C. elegans (Labrousse et al., 1999). The same observations were made using cotransfection of GFP-Drp1 and Twinkle with a c-Myc epitope tag that could be detected by ICC and with Tfam ICC and by transfection of Twinkle-GFP combined with detection of endogenous Drp1 using a polyclonal Drp1-specific antibody (unpublished results). Time-lapse experiments using different fluorescently labeled Drp1 and Twinkle should shed light on the exact sequence of events of nucleoid and mitochondrial division.
Figure 6.
Nucleoids and mitochondrial dynamics. Nucleoid dynamics were studied in MitoTracker Red–stained and Twinkle-GFP–transfected COS7 cells. Mitotracker shows as uniform mitochondrial staining, whereas Twinkle-GFP appears as brighter intramitochondrial spots. (A) A section of a cell with several nucleoid divisions, indicated by solid arrows. In addition it shows an apparent redistribution of a nucleoid moving into what appears to be a daughter mitochondrion about to be split of (indicated by an open arrow). Both in A and B, frames of 15-s intervals are shown going from left to right and top to bottom. (B) Two mitochondrial division events (solid arrows) in which nucleoids are positioned so that each daughter mitochondrion has at least one new nucleoid element. Although nucleoids appear close to the tips after division, they do not appear to actively participate in the division event itself.
Figure 7.
Drp1 does not colocalize with mitochondrial nucleoids. To further demonstrate that nucleoids were excluded from sites of mitochondrial fission we looked at colocalization, in human osteosarcoma cells, of nucleoids stained by mtSSB ICC (A), with the mitochondrial fission protein Drp1, used here as a GFP fusion protein (B). The merged image (C) shows a general lack of colocalization of Drp1-GFP with mtSSB foci. Indicated in an enlarged section of the cell shown in C is a mitochondrial constriction site containing Drp1-GFP with mtSSB foci on either site of this putative future fission site (solid arrow). Indicated by open arrows are two faint Drp1-GFP foci at mitochondrial tips, suggestive of a recent fission event.
DISCUSSION
The results presented in this article provide many valuable insights into the organization and dynamics of mtDNA nucleoids in cultured human cells. It is for the first time clearly shown, both by in situ cell biological methods and biochemical methods, that nucleoids in mammalian cells are stable assemblies of multiple mitochondrial proteins with mtDNA. Furthermore, the results show that nucleoids are dynamic structures that divide and follow the dynamics of the mitochondrial network, presumably to ensure the transmission of mtDNA to daughter mitochondria during mitochondrial growth and division. MtSSB did not colocalize with Drp1, which is often found concentrated at sites of past or future mitochondrial fission. In addition to basic considerations of mtDNA organization in mammals, the study of nucleoid structure and dynamics should also help us to better understand important aspects of mtDNA maintenance, such as mtDNA segregation and complementation, in human mitochondrial disease.
Mitochondrial Nucleoids in Human Cells Are Assemblies of Multiple Proteins with mtDNA
The first suggestions that nucleoids are discrete structures containing mtDNA and proteins has come from studies mostly involving the yeast S. cerevisiae. Evidence for a similar organization in mammalian cells had until recently only been suggested by DAPI staining (Satoh and Kuroiwa, 1991). Twinkle was the first protein to be positively identified as a human mitochondrial protein associated with mtDNA nucleoids in vivo (Spelbrink et al., 2001).
On the basis of localization studies of human mitochondrial replication proteins such as POLG, POLG2, and TFAM using GFP tagging, we previously concluded that none of these proteins specifically colocalize with mtDNA (Spelbrink et al., 2000, and J.N.S., unpublished observations). However, we questioned especially the result for TFAM for various reasons. First, the GFP methodology usually highly overexpresses the protein of interest, whereas the GFP tag could mask regions of the protein involved in specific nucleoid interaction or interfere with proper protein folding. Second, the yeast TFAM orthologue Abf2p has convincingly been demonstrated to be a yeast mitochondrial nucleoid protein (Newman et al., 1996; Kaufman et al., 2000). Third, both yeast Abf2p and human TFAM have low binding specificity for DNA but high binding affinity and have similarity with the bacterial histone like protein HU and eukaryotic high mobility group proteins (Diffley and Stillman, 1991, 1992; Fisher et al., 1992; Newman et al., 1996), suggesting a function in DNA packaging similar to nuclear histone proteins. Fourth, TFAM protein levels in mammalian mitochondria have been recently suggested to be much higher than previously believed, being sufficient to fully wrap mtDNA assuming TFAM binding at intervals of 30 base pairs (Takamatsu et al., 2002). Finally, two related mammalian mitochondrial transcription factors, similar to the S. cerevisiae mitochondrial RNA polymerase specificity factor, Mtf1p, were recently identified and shown to greatly stimulate TFAM-dependent transcription activation in in vitro assays (McCulloch et al., 2002; Falkenberg et al., 2002). It was shown that the novel transcription factors interact with the mitochondrial RNA polymerase, and further experiments suggested them to be important for transcription initiation (Falkenberg et al., 2002), similar to Mtf1p (Karlok et al., 2002). These findings for the first time show the apparent necessity of transcription factors other then TFAM in mammalian mtDNA transcription. However, also packaging of mtDNA by TFAM is likely to be important for transcription and replication, similar to the regulation of nuclear genes by histones, which are themselves regulated by protein modification (see e.g., Marmorstein, 2001).
Thus, we demonstrate for the first time colocalization of endogenous TFAM and mtSSB with Twinkle in intramitochondrial foci, which also contain mtDNA. Both TFAM and mtSSB are also concentrated in foci in the absence of transgenically expressed Twinkle-EGFP suggesting they are intrinsically part of nucleoids. This is also compatible with the concentrated localization of TFAM shown by our immunoelectron microscopy results. Furthermore, the size of the immunolabeled area in the mitochondria fits the reported size of nucleoids in yeast (Miyakawa et al., 1987).
Both TFAM and SSB are well characterized as DNA-binding proteins (see above for TFAM and, e.g., Curth et al., 1994) and copurify, albeit to varying extend, with mtDNA in our nucleoid preparations. TFAM was specifically retained in both the first and the second gradient purification step, whereas other proteins that we have considered contaminations were lost in the second purification step. SSB was less well retained and was hardly detectable in our final nucleoid prep. Because mtSSB is a binding protein with specificity for single-stranded DNA, its in situ presence in nucleoids is likely to be a consequence of single-stranded DNA regions such as the D-loop (Zeviani et al., 1995). Loss of mtSSB might be a consequence of the partial loss of these single-stranded structures during the purification. It is interesting to note that based on ICC most, if not all, nucleoids contain both TFAM, mtSSB and Twinkle, suggesting that all three proteins are structural components of nucleoids, presumably also stabilizing and protecting mtDNA.
The results for POLG and POLG2 were somewhat less straightforward. POLG2 ICC showed mostly uniform fluorescence, but also some foci with apparently higher POLG2 concentrations. Most but not all of these foci also contained Twinkle. An attractive hypothesis is that the POLG2 foci represent actively replicating mitochondrial genomes. The ICC results in this case are consistent with the biochemical purification data that suggest that most of POLG2 is not present within the nucleoid fraction. In contrast, POLG ICC showed uniform mitochondrial fluorescence, whereas the biochemical purification showed significant POLG enrichment in the nucleoid fractions. This inconsistency could be explained by low-affinity nonspecific binding of the POLG antibody, presumably recognizing at least one nonnucleoid mitochondrial protein also during the ICC. Despite the enrichment in nucleoids, POLG concentration is probably low because a protein of the correct size could not clearly be distinguished in the total-protein stain after SDS-PAGE of nucleoid fractions. These data could point to POLG, like POLG2, being present mainly at sites of ongoing DNA synthesis or repair. Because POLG and POLG2 have been shown to physically interact and usually copurify (Wang et al., 1997), the results also point to a large excess of POLG2. This could be a safeguarding mechanism to assure that POLG always finds its partner that has been shown to increase the fidelity and processivity of the polymerase enzyme (Carrodeguas et al., 1999; Lim et al., 1999). It might also suggest an additional function for POLG2 inside mitochondria, apart from its role as a processivity factor. Although it should be no surprise that POLG and POLG2 at least partially copurify with mtDNA because they have been clearly implicated in mtDNA maintenance (Foury, 1989; Iyengar et al., 1999, 2002; Spelbrink et al., 2000), the data we present here do not support the hypothesis that these are structural nucleoid proteins.
In the absence of mtDNA, nucleoid integrity is lost, showing that the interaction of Twinkle, TFAM, and SSB with mtDNA is essential for their colocalization in discrete foci within mitochondria. The most clear demonstration of this comes from the almost uniform mitochondrial distribution of mtSSB in rho-zero cells. TFAM levels, in contrast with mtSSB or POLG and POLG2 levels, were severely reduced. This agrees with previous studies showing a strong correlation between TFAM protein levels and mtDNA levels, using rho-zero cell lines, cell lines partially depleted by dideoxycytidine treatment, material from patients suffering from mtDNA depletion and heterozygous TFAM knockout mice (Larsson et al., 1994; Poulton et al., 1994; Larsson et al., 1998). This strongly suggested that TFAM protein stability and turnover depends on its interaction with mtDNA and is supported by our observation of an apparent dependence of the occurrence of TFAM degradation products on a lack of DNA binding.
Nucleoid Dynamics
In yeast mitochondrial genetics, it has been demonstrated that the number of segregating units is clearly less than the number of mtDNA molecules, but more or less equal to the number of mtDNA-nucleoid structures as observed by DAPI staining. This not only demonstrated that nucleoids contain more than one mtDNA molecule, but clearly implicated nucleoids as the units of mtDNA inheritance.
Here we show in vivo behavior and distribution of Twinkle-GFP suggesting that Twinkle containing elements in human mitochondria, which we here showed to contain mtDNA, are the heritable elements of mtDNA. Most of the mitochondria contain multiple Twinkle-GFP punctae that appeared to be distributed among the “daughter” mitochondria upon division.
It is tempting to speculate that the minimal size nucleoids we observed are protein-DNA structures containing single copies of mtDNA. Interestingly, it was recently concluded based on mtDNA-specific FISH experiments that multiple copies of mtDNA are often clustered inside the mitochondrial network (Margineantu et al., 2002). Also in this case, single small spots possibly consisting of individual mtDNA molecules could be seen. Nevertheless, at this point neither the previously published results nor our results can exclude the possibility that the minimum visible structures are actually the most minimal size of nucleoids but still containing multiple mtDNA copies.
As observed with the FISH experiments and also visible in our previous published pictures (Spelbrink et al., 2001; Margineantu et al., 2002), mtDNA nucleoids are frequently localized at or near the tips or constriction sites on the mitochondria, suggesting a possible association with Drp1. Drp1 cycles on and off mitochondria, but when on the mitochondrial outer membrane, it localizes at the spots where past and future scissions occur (van der Bliek, 2000). We did not notice any significant colocalization between SSB (or Twinkle) and Drp1. This indicates that nucleoid proteins are not active participants of the division machinery. Nevertheless, Twinkle follows mitochondrial dynamics and could play an important role in the active segregation of mtDNA.
ACKNOWLEDGMENTS
J.N.S. thanks Howy Jacobs, Ian Holt, and Anu Wartiovaara for many useful comments and discussions. This work was supported in part by The Academy of Finland Center of Excellence program, the Academy of Finland Life2000 program, and the Academy of Finland grants 80939 and 80936 to J.N.S., by a grant from the Sigrid Jusélius Foundation to J.W., and by a postdoctoral fellowship by the American Heart Association to L.G.
Abbreviations used:
- Drp1
dynamin-related protein 1
- BrdU
5-bromo-2-deoxy-uridine
- (E)GFP
(enhanced) green fluorescent protein
- ICC
immunocytochemistry
- MtSSB/SSB
mitochondrial single-stranded DNA-binding protein
- POLG
polymerase gamma
- POLG2
accessory subunit of POLG
- TFAM
transcription factor A of mitochondria
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0399. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0399.
REFERENCES
- Albring M, Griffith J, Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci USA. 1977;74:1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Van Etten RA, Wright CT, Walberg MW, Clayton DA. Sequence and gene organization of mouse mitochondrial DNA. Cell. 1981;26:167–180. doi: 10.1016/0092-8674(81)90300-7. [DOI] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrodeguas JA, Kobayashi R, Lim SE, Copeland WC, Bogenhagen DF. The accessory subunit of Xenopus laevis mitochondrial DNA polymerase gamma increases processivity of the catalytic subunit of human DNA polymerase gamma and is related to class II aminoacyl-tRNA synthetases. Mol Cell Biol. 1999;19:4039–4046. doi: 10.1128/mcb.19.6.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curth U, Urbanke C, Greipel J, Gerberding H, Tiranti V, Zeviani M. Single-stranded-DNA-binding proteins from human mitochondria and Escherichia coli have analogous physicochemical properties. Eur J Biochem. 1994;221:435–443. doi: 10.1111/j.1432-1033.1994.tb18756.x. [DOI] [PubMed] [Google Scholar]
- Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. J Cell Biol. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. A close relative of the nuclear, chromosomal high-mobility group protein HMG1 in yeast mitochondria. Proc Natl Acad Sci USA. 1991;88:7864–7868. doi: 10.1073/pnas.88.17.7864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Stillman B. DNA binding properties of an HMG1-related protein from yeast mitochondria. J Biol Chem. 1992;267:3368–3374. [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1, and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- Fekkes P, Shepard KA, Yaffe MP. Gag3p, an outer membrane protein required for fission of mitochondrial tubules. J Cell Biol. 2000;151:333–340. doi: 10.1083/jcb.151.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Lisowsky T, Parisi MA, Clayton DA. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP 1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989;264:20552–20560. [PubMed] [Google Scholar]
- Hales KG, Fuller MT. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 1997;90:121–129. doi: 10.1016/s0092-8674(00)80319-0. [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–373. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B, Luo N, Farr CL, Kaguni LS, Campos AR. The accessory subunit of DNA polymerase gamma is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc Natl Acad Sci USA. 2002;99:4483–4488. doi: 10.1073/pnas.072664899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B, Roote J, Campos AR. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-gamma125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs HT, Lehtinen SK, Spelbrink JN. No sex please, we're mitochondria: a hypothesis on the somatic unit of inheritance of mammalian mtDNA. Bioessays. 2000;22:564–572. doi: 10.1002/(SICI)1521-1878(200006)22:6<564::AID-BIES9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Kamimoto T, Nagai Y, Onogi H, Muro Y, Wakabayashi T, Hagiwara M. Dymple, a novel dynamin-like high molecular weight GTPase lacking a proline-rich carboxyl-terminal domain in mammalian cells. J Biol Chem. 1998;273:1044–1051. doi: 10.1074/jbc.273.2.1044. [DOI] [PubMed] [Google Scholar]
- Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc Natl Acad Sci USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlok MA, Jang S-H, Jaehning JA. Mutations in the yeast mitochondrial RNA polymerase specificity factor, Mtf1, verify an essential role in promoter utilization. J Biol Chem. 2002;277:28143–28149. doi: 10.1074/jbc.M204123200. [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Oldfors A, Holme E, Clayton DA. Low levels of mitochondrial transcription factor A in mitochondrial DNA depletion. Biochem Biophys Res Commun. 1994;200:1374–1381. doi: 10.1006/bbrc.1994.1603. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lehtinen SK, Hance N, El Meziane A, Juhola MK, Juhola KM, Karhu R, Spelbrink JN, Holt IJ, Jacobs HT. Genotypic stability, segregation and selection in heteroplasmic human cell lines containing np 3243 mutant mtDNA. Genetics. 2000;154:363–380. doi: 10.1093/genetics/154.1.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SE, Longley MJ, Copeland WC. The mitochondrial p55 accessory subunit of human DNA polymerase gamma enhances DNA binding, promotes processive DNA synthesis, and confers N-ethylmaleimide resistance. J Biol Chem. 1999;274:38197–38203. doi: 10.1074/jbc.274.53.38197. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Perlman PS, Butow RA. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 2000;19:767–775. doi: 10.1093/emboj/19.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margineantu DH, Cox WG, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmorstein R. Protein modules that manipulate histone tails for chromatin regulation. Nat Rev Mol Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Aoi H, Sando N, Kuroiwa T. Fluorescence microscopic studies of mitochondrial nucleoids during meiosis and sporulation in the yeast. Saccharomyces cerevisiae. J Cell Sci. 1984;66:21–38. doi: 10.1242/jcs.66.1.21. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Sando N, Kawano S, Nakamura S, Kuroiwa T. Isolation of morphologically intact mitochondrial nucleoids from the yeast. Saccharomyces cerevisiae. J Cell Sci. 1987;88:431–439. doi: 10.1242/jcs.88.4.431. [DOI] [PubMed] [Google Scholar]
- Miyakawa I, Fumoto S, Kuroiwa T, Sando N. Characterization of DNA-binding proteins involved in the assembly of mitochondrial nucleoids in the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 1995;36:1179–1188. [PubMed] [Google Scholar]
- Morrissey JH. Silverstain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;117:307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SM, Zelenaya-Troitskaya O, Perlman PS, Butow RA. Analysis of mitochondrial DNA nucleoids in wild-type and a mutant strain of Saccharomyces cerevisiae that lacks the mitochondrial HMG box protein Abf2p. Nucleic Acids Res. 1996;24:386–393. doi: 10.1093/nar/24.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi MA, Xu B, Clayton DA. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol Cell Biol. 1993;13:1951–1961. doi: 10.1128/mcb.13.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol Biol Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J, et al. Deficiency of the human mitochondrial transcription factor h-mtTFA in infantile mitochondrial myopathy is associated with mtDNA depletion. Hum Mol Genet. 1994;3:1763–1769. doi: 10.1093/hmg/3.10.1763. [DOI] [PubMed] [Google Scholar]
- Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–874. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- Satoh M, Kuroiwa T. Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell. Exp Cell Res. 1991;196:137–140. doi: 10.1016/0014-4827(91)90467-9. [DOI] [PubMed] [Google Scholar]
- Schägger H, Von Jagow G. Tricine-sodium dodecylsulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1–100 kD. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Ryazantsev SN, van der Bliek AM. A human dynamin-related protein controls the distribution of mitochondria. J Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelbrink JN, et al. In vivo functional analysis of the human mitochondrial DNA polymerase POLG expressed in cultured human cells. J Biol Chem. 2000;275:24818–24828. doi: 10.1074/jbc.M000559200. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- Takamatsu C, Umeda S, Ohsato T, Ohno T, Abe Y, Fukuoh A, Shinagawa H, Hamasaki N, Kang D. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 2002;18:451–456. doi: 10.1093/embo-reports/kvf099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–366. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Bliek AM. A mitochondrial division apparatus takes shape. J Cell Biol. 2000;151:F1–F4. doi: 10.1083/jcb.151.2.f1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Farr CL, Kaguni LS. Accessory subunit of mitochondrial DNA polymerase from Drosophila embryos. Cloning, molecular analysis, and association in the native enzyme. J Biol Chem. 1997;272:13640–13646. doi: 10.1074/jbc.272.21.13640. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Amati P, Comi G, Fratta G, Mariotti C, Tiranti V. Searching for genes affecting the structural integrity of the mitochondrial genome. Biochim Biophys Acta. 1995;1271:153–158. doi: 10.1016/0925-4439(95)00022-v. [DOI] [PubMed] [Google Scholar]