Abstract
Multisubunit tethering complexes may contribute to the specificity of membrane fusion events by linking transport vesicles to their target membrane in an initial recognition event that promotes SNARE assembly. However, the interactions that link tethering factors to the other components of the vesicle fusion machinery are still largely unknown. We have previously identified three subunits of a Golgi-localized complex (the Vps52/53/54 complex) that is required for retrograde transport to the late Golgi. This complex interacts with a Rab and a SNARE protein found at the late Golgi and is related to two other multisubunit tethering complexes: the COG complex and the exocyst. Here we show that the Vps52/53/54 complex has an additional subunit, Vps51p. All four members of this tetrameric GARP (Golgi-associated retrograde protein) complex are required for two distinct retrograde transport pathways, from both early and late endosomes, back to the TGN. vps51 mutants exhibit a distinct phenotype suggestive of a regulatory role. Indeed, we find that Vps51p mediates the interaction between Vps52/53/54 and the t-SNARE Tlg1p. The binding of this small, coiled-coil protein to the conserved N-terminal domain of the t-SNARE therefore provides a crucial link between components of the tethering and the fusion machinery.
INTRODUCTION
Vesicular traffic between different organelles in the secretory and endocytic pathways involves the highly regulated docking and fusion of a transport vesicle with a specific target membrane. Current models suggest that at least some of the specificity of the docking and fusion process is provided by the recognition and pairing of small membrane-anchored proteins called SNAREs (Chen and Scheller, 2001). As a single v-SNARE on the vesicle membrane assembles with three t-SNAREs on the target membrane, opposing membranes are drawn together, resulting in bilayer mixing and fusion. However, additional factors implicated in vesicle docking such as Rab GTPases and multisubunit tethering complexes may act at a step that precedes SNARE pairing and assembly (Whyte and Munro, 2002). Although much debate has centered on which of these factors mediate the primary membrane recognition event, specificity may be determined not by a single component, but instead by a combination of SNAREs, Rabs, and tethers that together uniquely define a given transport step. Defining the nature of these interactions remains a major challenge.
The tethering factors identified to date are a diverse collection of long coiled-coil proteins and multisubunit complexes that are presumed to link vesicles to their target membrane in a step that promotes subsequent SNARE complex formation (Whyte and Munro, 2002). Many if not all of these tethering factors interact with specific Rab proteins and therefore may couple membrane recognition to the activation of the Rab GTPase. What is less clear is whether tethering factors promote fusion simply by increasing the local concentration of vesicles at the correct target membrane, or if they act directly on the SNAREs to activate their assembly.
SNARE proteins have a membrane proximal coiled-coil SNARE motif that assembles to form a stable four-stranded helical bundle referred to as the core complex (Fernandez et al., 1998). In addition, some SNARE proteins have an independently folded N-terminal domain that binds the C-terminal SNARE motif, thus preventing its assembly with other SNAREs (Dulubova et al., 1999). Removal of the N-terminal domain of syntaxin accelerates SNARE complex formation in vitro (Parlati et al., 1999). Therefore, an attractive model is that regulatory factors interact with the N-terminal domain of syntaxin-like t-SNAREs to modulate fusion.
Sec1p inhibits assembly of SNAREs at the plasma membrane by binding to syntaxin and holding it in the closed conformation (Dulubova et al., 1999). Other Sec1 family members bind to the N-terminal regions of SNARE proteins but do not prevent SNARE formation and instead may contribute to its specificity (Peng and Gallwitz, 2002; Dulubova et al., 2002; Yamaguchi et al., 2002). Furthermore, the N-terminal domains of many SNARE proteins are structurally related to that of syntaxin but not all of these form closed conformations in vitro (Dulubova et al., 2001, 2002; Antonin et al., 2002). Understanding the precise role of SNARE N-terminal domains in the fusion process will require a more detailed picture of their interactions with regulatory proteins.
In a few instances, tethering complexes unrelated to Sec1p have been shown to interact directly with t-SNARE N-terminal domains, and it may be that more such associations will be found as more of these complexes are identified and characterized. Whereas Rabs, SNAREs, and Sec1-like proteins are members of conserved families, tethering factors are somewhat more diverse and have been identified in functional studies rather than by sequence similarity. Candidate tethering factors exist for many but not all fusion events, and the subunit structure/components of these complexes are still being identified. The COG (conserved oligomeric Golgi) complex, also known as the Sec34/35 complex, is localized to the cis-Golgi where it has been shown to have a tethering function in an in vitro assay (Morsomme and Riezman, 2002; Ungar et al., 2002). The recent identification of six new subunits of the COG complex led to the recognition that at least some of these components are related at the sequence level to components of two other large, multisubunit tethering factors: the exocyst and Vps52/53/54 complexes (Whyte and Munro, 2001, 2002). COG has now been shown to be an effector of the Rab protein Ypt1p and to bind the cis-Golgi syntaxin Sed5p (Suvorova et al., 2002). The exocyst, an eight-subunit complex that directs the fusion of secretory vesicles with the plasma membrane, is the best studied of the three. An effector of the Sec4p Rab protein, the exocyst localizes to sites of polarized secretion where exocytosis takes place (Guo et al., 1999), though no interaction with t-SNAREs has yet been reported.
Although the exocyst and COG complexes each have eight subunits, only three components of the Vps52/53/54 complex have previously been identified (Conibear and Stevens, 2000). Vps52p is related to the exocyst component Sec3p, whereas Vps53p and Vps54p both contain an N-terminal domain that is shared by subunits of the other two docking complexes (Whyte and Munro, 2001). Vps52p, Vps53p, and Vps54p are large coiled-coil containing proteins that are tightly associated in a stoichiometric complex localized to the late Golgi. The sorting defects of these mutants suggest that the Vps52/53/54 complex is required for the retrograde trafficking of vesicles from the late endosomal/prevacuolar compartment back to the late Golgi. The Vps52/53/54 complex has been identified as an effector of the Rab protein Ypt6p and also binds the N-terminal region of the TGN t-SNARE Tlg1p, further supporting the idea that this complex is a tethering factor for vesicle fusion at the TGN (Siniossoglou and Pelham, 2001). Sequence relationships between components of these multisubunit tethering complexes suggest that all three may share a common mechanism of action. Therefore, understanding the interactions that link tethering complexes to SNARE proteins will be important for understanding both the mechanism of vesicle docking and the regulation of SNARE complex formation.
Here, we identify a fourth component of the Vps52/53/53 complex, Vps51p, that regulates its association with the N-terminal domain of the t-SNARE Tlg1p. In addition to a role in retrograde transport from the late endosomal/prevacuolar compartment, we find that all four subunits of this complex are also required for trafficking on a distinct transport pathway, from early endosomes back to the late Golgi. We have named this tetrameric complex the GARP (Golgi-associated retrograde protein) complex to reflect its role in the docking and fusion of multiple classes of endosome-derived vesicles with the late Golgi membrane.
MATERIALS AND METHODS
Enzymes, antibodies, and general molecular biology methods were as described previously (Conibear and Stevens, 2000, 2002).
Plasmid and Strain Construction
Identification of the transposon insertion site in IRS4 by recovery and sequencing of yeast genomic DNA containing the Tn3-LacZ insertion cassette was as described previously (Conibear and Stevens, 2000). A 2.7-kb PCR fragment containing IRS4 was subcloned into BamHI-NotI cut pRS316 and Yep352 to create pLC17 and pLC18, respectively. A 2.6-kb BamHI-XbaI fragment of pLC30 containing YKR020w was subcloned into BamHI-XbaI cut pLC17 to create the vps51-1 complementing plasmid pLC31. Sequences encoding IRS4 were subsequently removed by digestion and religation of SacI-cut pLC31 to create pLC35. To make the YKR020w knockout plasmid pLC36, the HindIII-XbaI fragment from pLC30 was first subcloned into KS+ and digested with EcoRI-MfeI, and TRP1 was inserted on an EcoRI fragment derived from pJJ246.
Yeast two-hybrid bait plasmids pLC133 and pLC134, containing VPS51 and VPS54, respectively, were generated by PCR amplification of genomic DNA and insertion into pGBD-C1 (James et al., 1996). A plasmid expressing the GALAD-Tlg1p fusion was created by PCR and in vivo homologous recombination in pOAD. Plasmids pGBD-TLG2 and pGAD-TLG2 were gifts from Lucy Robinson (Louisiana State University Health Science Center, Shreveport, LA). To create vectors for the expression of GST-Tlg1p fusion proteins in Escherichia coli, sequences corresponding to the Tlg1p cytosolic domain (M1 to C206), N-terminal domain (M1 to E106), and C-terminal domain (P129 to C206) were amplified by PCR and subcloned as a SacI-BamHI fragment into NotI-BamHI cut pGEX-5X-1 (Amersham Pharmacia Biotech, Piscataway, NJ) after filling in SacI and NotI sites to create compatible blunt ends, resulting in pLC158 (GST-full length), pLC159 (GST-Nterm), and pLC160 (GST-Cterm).
Table 1 describes the yeast strains used in this study. A 6.2-kb BlgII fragment of genomic DNA containing the transposon insertion was rescued from a backcrossed vps51-1 strain after transformation with pRSQ305, creating pLC30. pLC30 was cut with BglII and used to introduce vps51-1 into SEY6210. To delete YKR020w (VPS51), yeast were transformed with the HindIII-XbaI fragment from pLC36. A tlg1::TRP1 deletion cassette together with flanking genomic sequences were PCR-amplified from yeast strain JHY016 (Holthuis et al., 1998) and used for single-step gene replacements. Absence of Tlg1 protein in disrupted strains was confirmed by Western blotting. A plasmid for the integration of GFP-Snc1p (Lewis et al., 2000) was digested with StuI to target it to URA3 locus, and transformants expressing low levels of the fusion protein were selected.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SNY36-9B | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 | Nothwehr et al. (1993) |
| LCY131 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 vps51::tn3-URA3 | This study, 1993 |
| LCY159 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 irs4Δ::LEU2 | This study |
| LCY222 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 vps52Δ::kanr | Conibear and Stevens (2000) |
| LCY221 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 vps53::Tn3URA3 | Conibear and Stevens (2000) |
| LCY200 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 vps54Δ::TRP1 | Conibear and Stevens (2000) |
| LCY271 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 vps51Δ::TRP1 | This study |
| LCY226 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS52-HA | Conibear and Stevens (2000) |
| LCY320 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51-myc::HIS5 | This study |
| LCY325 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS52-HA::URA3::vps52 VPS51-myc::HIS5 | This study |
| LCY227 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS52-HA vps53::TRP1::VPS53-myc | Conibear and Stevens (2000) |
| LCY228 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 vps53::TRP1::VPS53-myc | Conibear and Stevens (2000) |
| LCY330 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51::3HAkanr | This study |
| LCY326 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS53-HA::TRP1::vps53 VPS51-myc::HIS5 | This study |
| LCY300 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 tlg1Δ::TRP1 | This study |
| LCY328 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51-myc::HIS5+ OCH1-HA::URA3::och1 | This study |
| LCY329 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51-myc::HIS5+ KEX2-HA::URA3::kex2 | This study |
| LCY321 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS52-HA vps51Δ::TRP1 | This study |
| LCY421 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51-myc::HIS5 vps52::tn3-URA3 | This study |
| LCY423 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS51-myc::HIS5 tlg1Δ::TRP1 | This study |
| LCY425 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS52-HA tlg1Δ::TRP1 | This study |
| LCY390 | MATα ura3Δ leu2Δ his3Δ200 trp1-901 lys2Δ vps52::URA3::VPS52-HA | This study |
| LCY392 | MATα ura3Δ leu2Δ his3Δ200 trp1-901 lys2Δ vps52::URA3::VPS52-HA ypt6Δ::kanr | This study |
| LCY327 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-901 lys2-801 suc2-Δ9 pho8Δ::ADE2 VPS54-HA VPS51-myc::HIS5 | This study |
Epitope-tagging of VPS51 was accomplished by integration of 13xmyc-HIS3 or 3xHA-kanr cassettes at the VPS51 genomic locus using a PCR-based gene targeting method (Longtine et al., 1998). Epitope-tagging of VPS52, VPS53, VPS54, and OCH1 and the construction of vps52 and vps53 mutant strains have been described previously (Conibear and Stevens, 2000). The KEX2-HA integration plasmid pLC137 was constructed by subcloning the C-terminal region of KEX2-HA from pSN222 into the BamHI-XhoI site of pRS306. Integration of HpaI-cut pLC137 created a functional HA-tagged copy of KEX2 at the genomic locus. The ability of all epitope-tagged proteins to fully complement CPY sorting was determined by colony overlay assays (Conibear and Stevens, 2002).
Immunoprecipitation and Subcellular Fractionation
Immunoprecipitation of CPY, Vps10, and ALP was performed under denaturing conditions from radiolabeled extracts using appropriate polyclonal antisera as described previously (Conibear and Stevens, 2000, 2002). Nondenaturing immunoprecipitations and detection of copurifying proteins were also carried out as described (Conibear and Stevens, 2000). For immunoprecipitation of Tlg1p-associated proteins, spheroplasts were prepared from 20 OD600 units of cells, lysed in hypotonic buffer (25 mM KPO4, 2 M sorbitol, pH 7.5), and incubated for 30 min at 23°C with either 200 or 400 μg/ml DSP. After quenching the reaction by a 15-min incubation in the presence of 50 mM Tris, pH 7.5, the buffer was adjusted to 50 mM NaCl and 2% TX-100 to solubilize membranes, and nondenaturing immunoprecipitation with anti-Tgl1p antiserum (a gift from Hugh Pelham, MRC Laboratory of Molecular Biology, Cambridge, UK) was performed as described above.
Microscopy
Fluorescent microscopy of yeast cells expressing GFP-tagged proteins, stained with the vital dye CDCFDA, or fixed and labeled with antibodies to HA and/or myc epitopes was carried out as described (Conibear and Stevens, 2000, 2002). Cells were viewed using a 100× oil-immersion objective on a Zeiss Axioplan II fluorescence microscope, and images were captured with a Hamamatsu Orca CCD camera using OpenLab software and adjusted using Adobe Photoshop.
In Vitro Binding to GST Fusion Proteins
A 1-liter culture of E. coli strain BL21 (DE3) expressing either pLC158, pLC159, or pLC160 was induced with 0.5 mM IPTG for 3 h at 30°C, harvested, and lysed by sonication in PBS. Aliquots of the clarified lysates were stored at −80°C. GST fusion proteins were purified by incubating thawed lysates with glutathione-sepharose (Sigma, St. Louis, MO) for 2 h at 4°C washing with PBS and equilibrating with binding buffer (25 mM Tris, pH 8, 100 mM KCl, 5 mM MgCl2, 1 mM DTT, and protease inhibitors). Fifty microliters of a 50% slurry of sepharose-bound GST fusion protein was incubated for 2 h at 4°C with 10 OD600 units of yeast spheroplasts that had been lysed in binding buffer and clarified by centrifugation at 13,000 × g. After washing with binding buffer, bound proteins were eluted with sample buffer and analyzed by SDS-PAGE followed by Western blotting. The yield of GST-full length and GST-Nterm fusion proteins was approximately twofold lower than that of GST and GST-Cterm fusions.
RESULTS
Identification and Cloning of VPS51
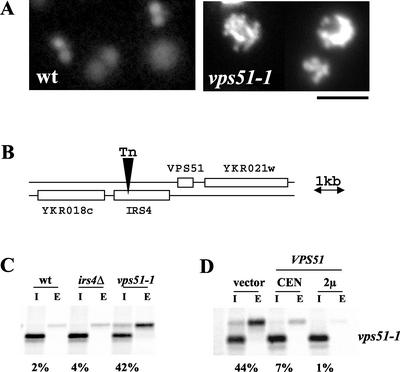
VPS52, VPS53, and VPS54 were isolated in a genetic screen designed to find new VPS genes that act at the Golgi and were found to share similar transport defects as well as a strikingly similar vacuolar morphology when stained with the vital dye CDCFDA (Conibear and Stevens, 2000). A single allele belonging to a fourth complementation group, vps51, was isolated in this same screen and found to exhibit the same aberrant vacuolar morphology (Figure 1A) even though its CPY missorting defect was less severe. Because mutants exhibiting similar vacuolar morphology often act at the same sorting step (Conibear and Stevens, 1998) the vps51-1 allele was chosen for further study.
Figure 1.
Identification and cloning of the vacuolar protein sorting gene VPS51. (A) The aberrant vacuolar morphology of vps51 mutants. Log phase cultures of wild-type (SEY6210) or vps51-1 (LCY131) cells were incubated with CDCFDA at 25°C for 45 min and examined under FITC optics. Vacuoles of vps51 mutants appear fragmented, tubular, and clustered. Scale bar, 4 μm. (B) Chromosomal features map showing the transposon insertion site in IRS4. (C) Deletion of IRS4 does not result in a CPY missorting defect. SEY6210 (wt), LCY159 (irs4Δ) and LCY131 (vps51-1) were pulse-labeled at 30°C for 10 min in the presence of [35S]methionine, and chased for 60 min after adding an excess of unlabeled cysteine and methionine. CPY was immunoprecipitated from the intracellular (I) and extracellular (E) fractions and visualized by SDS-PAGE followed by fluorography. (D) Expression of YKR020w rescues the CPY missorting defect of the vps51-1 mutant. YKR020w expressed from CEN (pLC31) or high copy (pLC32) vectors complemented the sorting defect of LCY131 cells by pulse-chase immunoprecipitation of CPY.
Backcrossing of the vps51-1 allele demonstrated that the transposon insertion was tightly linked to the CPY secretion phenotype (unpublished data). By rescue and sequencing of adjacent genomic DNA, the transposon insertion site in vps51-1 was identified in the C-terminal part of the IRS4 open reading frame (ORF; Figure 1B). Surprisingly, although vps51-1 mutants secrete ∼40% of newly synthesized CPY, precise deletion of the IRS4 ORF did not result in a CPY sorting defect (Figure 1C). Although a 2.7-kb fragment containing both IRS4 and YKR020w was able to rescue sorting, growth, and vacuolar morphology phenotypes, further analysis showed that YKR020w and not IRS4 complements the vps51-1 mutation (Figure 1D). Deletion of the YKR020w ORF gave rise to levels of CPY secretion and vacuolar morphology defects indistinguishable from those of the vps51-1 mutant (compare Figures 1C and 2A), consistent with the results of a recent genome-wide screen for vps mutants (Bonangelino et al., 2002). Taken together, these results indicate that VPS51 corresponds to YKR020w. VPS51 is predicted to encode a soluble 18.9-kDa protein with extensive coiled-coil regions but no other conserved functional domains.
Figure 2.
Aberrant trafficking of the CPY receptor, Vps10p, in vps51 mutants. (A) vps51 mutants have a less severe CPY missorting defect than vps52, vps53, or vps54 mutants. Wild-type (SNY36-B), vps52::kanr (LCY222), vps53:: Tn3URA (LCY221), vps54Δ:: TRP1 (LCY200), and vps51Δ:: TRP1 (LCY271) strains were radiolabeled for 10 min with [35S]methionine and chased for another 60 min in the presence of 50 μg/ml each of cysteine and methionine. CPY was immunoprecipitated from intracellular (I) and extracellular (E) fractions, separated by SDS-PAGE, and visualized by fluorography. The positions of the Golgi-modified (p2) and mature (m) forms of CPY are indicated. (B and C) Wild-type (SNY36-B), vps52::kanr (LCY222), and vps51Δ:: TRP1 (LCY271) strains containing a centromere-based plasmid expressing ALP (pSN92) were pulse-labeled for 10 min with [35S]methionine. After chasing for the indicated times, aliquots of cells were removed and subjected to immunoprecipitation with antibodies to Vps10p (B) or ALP (C) Samples were resolved by SDS-PAGE and visualized by fluorography. Vps10p turnover in vps51 mutants is faster than in wild-type cells but slower than in vps52 mutants (B). In contrast, conversion of proALP (pALP) to its mature, vacuolar form (mALP) was slowed only slightly in vps51 and vps52 cells (C). PEP4-dependent cleavage products (*) are indicated.
To understand how a transposon insertion 1700 base pairs upstream from the beginning of the VPS51 ORF can result in loss of function, spontaneous suppressors of the vps51-1 growth and CPY secretion phenotypes were isolated and cloned. Mutation of SIN3 or SET2, genes involved in heterochromatin formation and gene silencing, completely suppressed the CPY secretion defect of the vps51-1 strain but not that of ykr020wΔ, suggesting that the upstream transposon insertion leads to transcriptional repression of YKR020w (unpublished data).
Sorting Defects of vps51 Mutants
Defects in VPS52, VPS53, or VPS54 result in identical mutant phenotypes, which include fragmented tubular vacuoles, end4 synthetic lethality, defective recycling of Golgi membrane proteins, and missorting of ∼70% of newly synthesized CPY. Although vps51-1 mutant cells share many of these same phenotypes, their CPY sorting defect is much less severe. To get a more quantitative estimate of CPY secretion, a vps51 null mutant was constructed by deleting most of the YKR020w ORF and compared with congenic strains carrying null mutations in vps52, vps53, and vps54. After pulse-chase radiolabeling and immunoprecipitation of CPY from both the intracellular and extracellular fractions (Figure 2A), vps51Δ strains were found to secrete 40% of newly synthesized CPY into the medium as the p2 Golgi-modified form, whereas vps52, vps53, and vps54 strains had a more severe sorting defect, secreting 70% of their newly synthesized CPY, consistent with our previous findings (Conibear and Stevens, 2000).
In wild-type cells, newly synthesized CPY is recognized at the Golgi by its receptor Vps10p, and diverted away from the secretory pathway for delivery to the prevacuolar compartment (reviewed in Conibear and Stevens, 1998). Vps10p must continuously recycle back from the PVC to the Golgi to carry out further rounds of CPY sorting, and mutations that block this recycling step result in its mislocalization to the vacuole where it is subsequently degraded. Although Vps10p is quite stable over the course of the chase period in wild-type cells, it undergoes cleavage characteristic of vacuolar degradation in vps51 mutants with a half time of ∼3 h (Figure 2B). The greater half-life of Vps10p in vps51 strains compared with that in vps52 strains (2 h) is consistent with the milder CPY sorting defect of vps51 mutants. Resident Golgi membrane proteins maintain their localization by a process of continuous retrieval from the PVC, and we found that the stability and therefore the retention of the Golgi-resident protease Kex2p was also defective in the vps51 strain (unpublished data), indicating a general defect in the PVC-to-Golgi recycling pathway.
The vacuolar membrane protein alkaline phosphatase (ALP) follows an alternative pathway to the vacuole that bypasses the PVC and therefore does not depend on a functional PVC-to-Golgi recycling pathway. vps51 mutants, like vps52 mutants, show essentially wild-type ALP transport (Figure 2C). Although a slight delay in ALP maturation is evident both in vps51 and vps52 mutants at early time points, processing to its mature, vacuolar form is complete at 30 min. Taken together, these results indicate that loss of VPS51 results in sorting defects that are qualitatively similar to those seen in cells lacking VPS52, VPS53, and VPS54 even though they are somewhat less severe.
Vps51p Is a Subunit of the Tetrameric GARP Complex
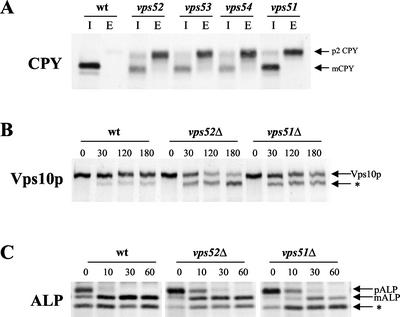
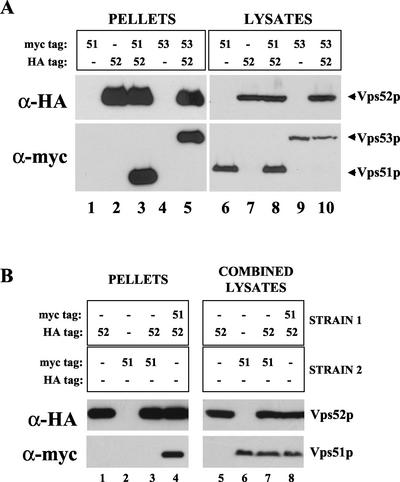
Because vps mutants that share a similar set of phenotypes generally act at the same sorting step, and often as components of a complex, we sought to discover by coimmunoprecipitation experiments if Vps51p is associated with the Vps52/53/54 complex. The chromosomal integration of either myc or HA epitope tags at the C terminus of the VPS51 ORF gave rise to proteins that were fully functional for CPY sorting. Different pair-wise combinations of myc- or HA-tagged Vps51p, Vps52p, and Vps53p were coexpressed in the same strains, as indicated in Figure 3A. Immunoprecipitation of HA-tagged Vps52p under nondenaturing conditions led to the copurification of either myc-tagged Vps51p (Figure 3A, lane 3) or myc-tagged Vps53p (Figure 3A, lane 5) when these were expressed in the same strain. However, neither Vps51-myc nor Vps53-myc was detected in immunoprecipitates from strains that did not also express Vps52-HA (Figure 3A, lanes 1 and 4). Most of the Vps51p present in the lysates could be coprecipitated with the rest of the complex, with little remaining in the supernatant (unpublished data), suggesting that Vps51p is the fourth subunit of the Vps52/53/54 complex. We have named this complex GARP, for Golgi-associated retrograde protein complex.
Figure 3.
Vps51p coprecipitates with subunits of the Vps52/53/54 complex. (A) Strains containing chromosomal copies of one or more epitope-tagged proteins (as indicated) were subjected to immunoprecipitation under nondenaturing conditions with rabbit anti-HA antibodies. Copurifying proteins were detected by Western blotting with anti-HA or anti-myc mouse mAbs and species-specific secondary antibodies and visualized by chemiluminescence. Total cell lysates corresponding to 30% of the material used for immunoprecipitation were also analyzed by Western blotting with the same antibodies to confirm the presence of each of the epitope-tagged proteins. The following strains were used: lanes 1 and 6, LCY320; lanes 2 and 7, LCY226; lanes 3 and 8, LCY325; lanes 4 and 9, LCY228; and lanes 5 and 10, LCY227. (B) Lysates were prepared from two different strains, as indicated, and mixed together before immunoprecipitation with anti-HA antibodies as described for A. The following strains were used: lanes 1, 3, 5, and 7, LCY226; lanes 2, 3, 6, and 7, LCY320; lanes 4 and 8, LCY325; and lanes 1, 4, 5, and 8, SNY36–9B.
We have previously shown that the entire cellular pools of Vps52p, Vps53p, and Vps54p are tightly associated in a complex. To see if Vps51p undergoes cycles of association and dissociation from the rest of the GARP complex, Vps52p-HA and Vps51p-myc were expressed in two different strains, and cell lysates were mixed before carrying out the nondenaturing immunoprecipitation (Figure 3B). Vps51p and Vps52p could be coimmunoprecipitated only when coexpressed in the same strain, indicating that the two proteins do not appreciably dissociate and rebind during the course of the experiment. The same was true for Vps52p and Vps53p (unpublished data).
Vps51p Is Not Associated with Other Protein Complexes
Vps52p, Vps53p, and Vps54p have been shown to be interdependent for their stability, each being rapidly degraded in the absence of the others (Conibear and Stevens, 2000). The fact that Vps51p appears to dissociate more readily from the rest of the complex suggests that it might not be essential for the stability of the remaining subunits. Indeed, there was little difference in the half-life of Vps52p and Vps54p in vps51 mutants vs. wild-type cells, although both are rapidly turned over in vps53 mutants (unpublished data). Quantitative Western blotting confirmed that the levels of Vps52p, Vps53p, and Vps54p remain essentially unchanged in vps51Δ strains. Conversely, the abundance of Vps51p is unaffected by deletion of VPS52, VPS53, or VPS54 (see below). Therefore, the possibility exists that Vps51p is not simply a structural subunit of the complex, but instead has additional functions or associates with more than one type of protein complex.
To determine whether Vps51p is largely associated with the GARP complex or interacts with additional complexes, native immunoprecipitation was carried out on metabolically labeled strains expressing either myc or HA-tagged versions of Vps51p (Figure 4A). Although Vps51p-HA itself is not readily detected by 35S labeling, three major proteins were coimmunoprecipitated with Vps51p. Two of these three bands are of the expected size for Vps53p and Vps54p and comigrate with bands that coprecipitate with Vps52p-HA, whereas the third is the expected size for endogenous Vps52p. No new copurifying bands were identified, although low-affinity interactions may be disrupted during purification. To detect any additional pools of Vps51p, double-label immunofluorescence microscopy was carried out on strains coexpressing Vps51p-myc and Vps52p-HA (Figure 4B, top panels) or Vps51p-myc and Vps53p-HA (Figure 4B, bottom panels). The high degree of overlap between these markers suggests that Vps51p does not participate in multiple protein complexes, but instead is predominantly associated with the GARP complex in vivo.
Figure 4.
Vps51p copurifies and colocalizes with the major cellular pool of Vps52/53/54 complex. (A) Metabolically labeled strains expressing integrated, epitope-tagged forms of Vps52p (LCY226) or Vps51p (Vps51p-myc, LCY320; Vps51p-HA, LCY330) were converted to spheroplasts and lysed under nondenaturing conditions. The extracts were subjected to immunoprecipitation with rabbit anti-HA (lanes 1–3) or rabbit anti-myc (lane 4) antisera. The positions of wild-type and tagged forms of Vps52p, Vps53p, and Vps54p are indicated on the right. The triple-HA tag is predicted to add an additional 5 kDa to the molecular mass of Vps52p. Vps51p cannot be visualized by labeling with [35S]methionine due to a lack of sulfur-containing amino acids and therefore that region of the gel is not shown. (B) Vps51p colocalizes with Vps52p and Vps53p by indirect immunofluorescence microscopy. Strains coexpressing integrated, epitope-tagged forms of Vps51 and either Vps52p (LCY325, top panels) or Vps53p (LCY326, bottom panels) were fixed, spheroplasted, and double-labeled with an affinity-purified anti-HA polyclonal antiserum and anti-myc mAb 9E10. The distribution of Vps51p (left panels) overlaps closely with that of Vps52p or Vps53p (center panels). Scale bar, 4 μm.
The GARP Complex Is Required for Retrograde Transport from Early Endosomes as well as Late Endosomes
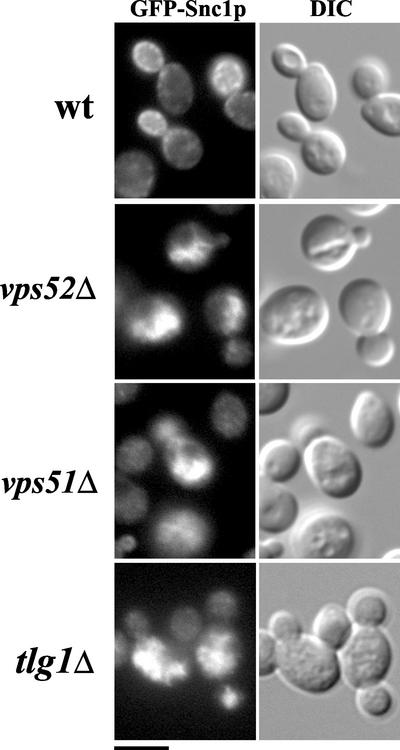
The exocytic v-SNARE Snc1p follows a recycling itinerary in which it is transported through an early endosomal compartment before recycling back to the TGN to be incorporated into newly forming secretory vesicles that must be targeted to fuse with the plasma membrane (Lewis et al., 2000). Recycling Snc1p does not reach the PVC/late endosome, because mutations that disrupt transport to and from the PVC have no effect on Snc1p recycling. To test the role of GARP in transport from the early endosome back to the TGN, a GFP-tagged version of Snc1p was integrated into vps52 and vps51 mutants (Figure 5). In wild-type cells, Snc1-GFP is localized to the plasma membrane of the growing bud. This localization is unchanged in vps5 mutants (Lewis et al., 2000, and unpublished data), which are defective in the formation of recycling vesicles at the PVC. However, Snc1p is clearly mislocalized in both vps51 and vps52 mutants (Figure 5). Instead of being found at the cell surface, Snc1-GFP was largely found intracellularly, where it colocalized with the vacuole as seen by DIC. The hazy cytosolic fluorescence of these mutants suggests that a portion of Snc1p might be trapped in transport vesicles. This mislocalization, which is also seen in vps53 and vps54 mutants, is strikingly similar to that seen in the absence of the TGN t-SNARE Tlg1p (Lewis et al., 2000 and Figure 5). Therefore, all four subunits of the GARP complex are required for the recycling of two different vesicle populations back to the late Golgi: those derived from early endosomes (carrying Snc1p) as well as those carrying retrograde cargo (such as Vps10p) from the late endosomal/PVC compartment.
Figure 5.
The v-SNARE Snc1p is mislocalized in vps51 and vps52 mutants. Wild-type (SNY36-B), vps52Δ::kanr (LCY222), vps51Δ:: TRP1 (LCY271) and tlg1Δ:: TRP1 (LCY300) strains containing integrated copies of the GFP-Snc1p were grown to log phase in synthetic medium and examined by fluorescence microscopy under FITC optics (left panels) to visualize Snc1p or DIC optics (right panels) to visualize vacuoles. Snc1p is localized to the plasma membrane of the growing bud tip in wild-type cells, whereas in vps51, vps52, and tlg1 mutants it is found in intracellular structures that coincide with the vacuole as seen in DIC. Scale bar, 4 μm.
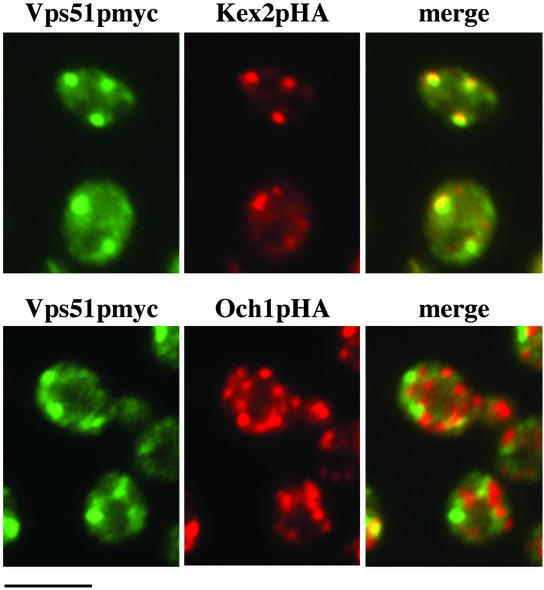
Previously identified genes required for the recycling of both Vps10p and Snc1p include other components of the TGN fusion machinery: the t-SNARE Tlg2p, the Rab Ypt6p, and the Ric1p/Rgp1p heterodimer, which acts as a GTP exchange factor for Ypt6p (Lewis et al., 2000; Panek et al., 2000; Siniossoglou et al., 2000). Although Ypt6p and Tlg1p are thought to work together with GARP at the TGN, a role for these Rab and SNARE proteins at the cis-Golgi has also been proposed. Therefore, localization experiments were performed to determine if the GARP complex might also act at the cis-Golgi in addition to the TGN. By double-label immunofluorescence microscopy, little colocalization was seen between Vps51p and a chromosomally integrated, HA-tagged version of the cis-Golgi marker Och1p (Figure 6). However, the distribution of Vps51p showed extensive overlap with the TGN protein Kex2p, consistent with previous findings that Vps52p colocalizes with A-ALP, another well-characterized marker of the late Golgi (Conibear and Stevens, 2000). Although we cannot rule out the presence of low levels of GARP at the cis-Golgi, it seems unlikely that the GARP complex plays a significant role in mediating fusion with early Golgi compartments.
Figure 6.
Vps51p colocalizes with the late Golgi marker Kex2p, but not with the cis-Golgi marker Och1p. Strains containing integrated copies of myc-tagged Vps51p and either Kex2p-HA (LCY329, top) or Och1p-HA (LCY328, bottom) were fixed, spheroplasted, and double-labeled with an anti-myc mAb and an affinity-purified HA rabbit antiserum. The merged images (right panels) show that the punctate staining pattern of Vps51p (green) coincides well with that of Kex2p (red, top panels) but shows little or no overlap with that of Och1p (red, bottom panels). Scale bar, 4 μm.
Vps51 Links the GARP Complex to the t-SNARE Tlg1p
A role for tethering factors in binding to SNAREs and activating them for fusion has been proposed previously, though as yet there is little evidence for this in vivo. We initially used the yeast two-hybrid system to see if any of the GARP subunits were able to interact with TGN SNAREs (Figure 7A). Using this system, Vps51p, but not Vps52p, Vps53p, or Vps54p, was found to interact with the Golgi t-SNARE Tlg1p (Figure 7A, and unpublished data). Tlg1p and Tlg2p form a complex in vivo (Nichols et al., 1998), but Vps51p did not interact with Tlg2p in the two-hybrid system, although Tlg1p-Tlg2p two-hybrid interactions were observed.
Figure 7.
Vps51p is required for the association of Vps52p with Tlg1p in vivo. (A) Haploid strains expressing GALAD fusion proteins were mated in duplicate with strains expressing GALBD fusion proteins, as indicated. Interaction between fusion proteins activates the HIS3 reporter and permits the growth of diploids on selective media lacking histidine. (B) Strains expressing the indicated epitope-tagged proteins were lysed, treated with the cross-linker DSP, and subjected to immunoprecipitation under nondenaturing conditions using Tlg1p antiserum. Coprecipitating proteins were detected by immunoblotting with anti-HA and anti-myc mouse mAbs and species-specific secondary antibodies. Cell lysates were TCA precipitated, solubilized, and analyzed in parallel to confirm expression of the tagged proteins. Vps52p does not copurify with Tlg1p from strains lacking Vps51p, whereas Vps51p maintains its association with Tlg1p in the absence of Vps52p. The following strains were used: lane 1, LCY325; lane 2, LCY321; lane 3, LCY421; and lane 4, LCY423.
These results led us to investigate whether Vps51p binds directly to Tgl1p in vivo. Strains containing epitope-tagged versions of Vps51p and Vps52p were lysed in the presence of different concentrations of cross-linkers and subjected to immunoprecipitation under nondenaturing conditions using anti-Tlg1p antiserum (Figure 7B). Under these conditions, both Vps52p and Vps51p copurified with Tlg1p. At the highest cross-linker concentration used, immunoprecipitation of Tlg1p resulted in the copurification of a relatively small fraction (∼1%) of the intracellular pool of Vps51p and Vps52p. This is likely to underestimate the amount of Tlg1p-bound GARP complex present in the cell, because detection of these complexes was dependent on the cross-linking efficiency. No interactions were detected in the absence of cross-linker, suggesting that the interactions between GARP and Tlg1p may be of low affinity or may be detergent sensitive. Binding of Tlg1p to ProtA-tagged Vps54p in the presence of cross-linkers has previously been reported (Siniossoglou and Pelham, 2001). Interestingly, no Vps52p was recovered in the pellet when the same immunoprecipitation protocol was carried out on a strain lacking Vps51p (Figure 7B, lane 2), even though Vps52p is present at normal levels in vps51Δ cells. However, Vps51p remained associated with Tlg1p even in the absence of Vps52p (Figure 7B, lane 3). These data strongly suggest that Vps51p mediates the interaction of the GARP complex with Tgl1p.
Tlg1p Is Not Required for the Golgi Association of Vps51p and Vps52p
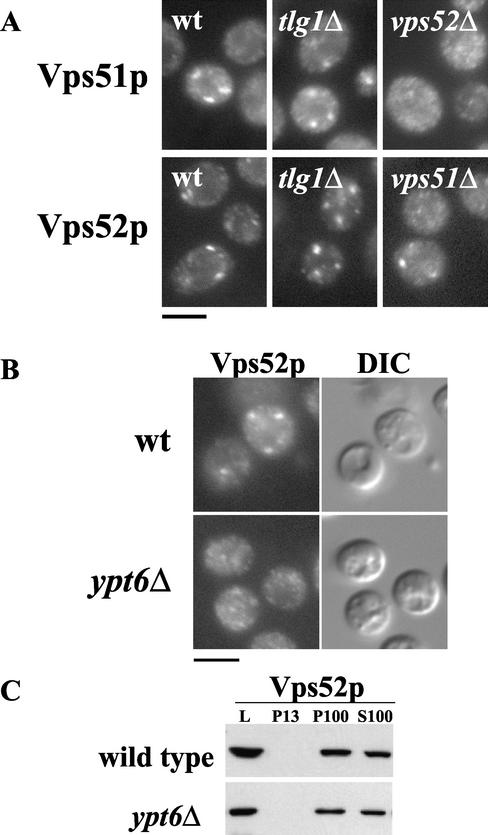
Both Vps51p and Tlg1p localize to the late Golgi, and thus binding of Vps51p to Tlg1p could provide a mechanism for its association with Golgi membranes. However, both Vps51p and Vps52p still show a characteristic Golgi-like staining pattern in strains lacking Tlg1p (Figure 8A). The localization of both Vps51p (Figure 8A) and Tgl1p (unpublished data) is more dispersed in vps52 mutants. Despite the lack of obvious Golgi staining pattern, the fact that these proteins still coprecipitate (Figure 7) suggests that they are able to maintain their interaction to some extent, perhaps instead associating at the cytosolic face of transport vesicles. Loss of the late Golgi Rab protein Ypt6p has no effect on the localization of the late Golgi marker Sec7p (Siniossoglou and Pelham, 2001), yet causes the redistribution of Vps52p to a dispersed, finely punctate staining pattern (Figure 8B). Because the proportion of Vps52p associated with membranes is unaffected by the absence of Ypt6p (Figure 8C), this dispersed staining pattern does not represent mislocalization of the GARP complex to the cytosol but may instead reflect binding to transport vesicles. Therefore, membrane association of the GARP complex must be mediated by an unidentified mechanism that is independent of both the Rab protein Ypt6p and the t-SNARE Tlg1p.
Figure 8.
Loss of Tlg1p does not prevent the Golgi association of Vps51p or Vps52p. (A) The localization of Vps51p-myc by indirect immunofluorescence (labeled with the anti-myc mAb 9E10; top panels) is unaffected by loss of TLG1 but becomes diffuse in the absence of VPS52. Loss of either VPS51 or TLG1 does not abolish the punctate staining of Vps52p-HA labeled with anti-HA mAb HA.11 (bottom panels). (B) Although the localization of Vps52p-HA becomes diffuse in cells lacking ypt6Δ (LCY392); (C) subcellular fractionation indicates that the proportion of Vps52p sedimenting with high-speed membranes is unchanged. L, lysate; P13, 13,000 × g pellet; P100, 100,000 × g pellet; S100, 100,000 × g supernatant.
Vps51p Is Required for the Binding of Vps52/53/54 to the N-Terminal Domain of Tlg1p In Vitro
The binding of regulatory factors to the N-terminal domain of syntaxin-like t-SNAREs promotes SNARE assembly and membrane fusion. Recent studies have demonstrated that many nonsyntaxin SNAREs, including the Tlg1p homolog syntaxin 6, have a conserved, independently folded N-terminal domain that is structurally similar to that of syntaxin (Dulubova et al., 2001; Tochio et al., 2001; Misura et al., 2002). To identify which domain of Tlg1p is responsible for binding Vps51p, we expressed the entire soluble (cytosolic) portion of Tlg1p, as well as C- and N-terminal domains, as GST fusion proteins in E. coli. When clarified detergent extracts of strains expressing epitope-tagged GARP subunits were incubated with immobilized GST fusion proteins, both Vps51p and Vps52p showed significant binding to full-length Tlg1p-GST compared with GST alone (Figure 9). Furthermore, both proteins bound efficiently to the N-terminal domain of Tgl1p but not to the GST fusion protein containing the Tlg1p C-terminal domain. This is consistent with observations that Vps54p binds the Tlg1p N-terminal domain in vitro (Siniossoglou and Pelham, 2001).
Figure 9.
VPS51 is required for the binding of Vps52p to the Tlg1p N-terminal domain in vitro. Binding to the N-terminal (N), C-terminal (C), or full-length cytosolic portion (C+N) of Tlg1p was assessed by incubating lysates from cells expressing Vps52p-HA (1 and 2) or Vps51p-myc (3) with GST-Tlg1 fusion proteins purified from E. coli and bound to sepharose beads. The binding of Vps52p-HA to the full-length and N-terminal domains of Tlg1p was abolished by loss of VPS51. The following strains were used: 1, LCY325; 2, LCY321; and 3, LCY327.
When cell lysates from strains lacking VPS51 were incubated with GST-Tlg1p fusion proteins, Vps52p no longer bound to full-length or N-terminal forms of Tlg1p, even though the levels of Vps52p protein as well as its TGN localization remain unchanged in vps51Δ cells. Thus Vps51p is required for the binding of the remaining subunits to the N-terminal domain of the t-SNARE Tlg1p both in vivo and in vitro. Conversely, in the absence of VPS52, VPS53, or VPS54, Vps51p is still able to bind specifically to the full-length and N-terminal domain GST-Tlg1p fusion proteins although the binding is drastically reduced (unpublished data), suggesting that the presence of the remaining GARP subunits is not essential for binding but does increase the affinity of Vps51p for Tlg1p. Efforts to express a functional form of Vps51p in E. coli have not yet been successful, so we cannot rule out the possibility that Vps51p requires additional factors in order to bind Tlg1p. However, these in vivo and in vitro binding studies clearly show a requirement for the small coil-coil protein Vps51p in mediating the interaction of the GARP complex with the N-terminal domain of the TGN t-SNARE Tlg1p.
DISCUSSION
Here we report the identification and characterization of Vps51p, a novel protein that is required for the association of the Vps52/53/54, or GARP, complex with the SNARE fusion machinery. A single allele of vps51 was isolated in the same genetic screen used to identify vps52, vps53, and vps54 (Conibear and Stevens, 2000) and found to have similar morphological defects but less severe missorting phenotypes. Nevertheless, Vps51p appears to be the fourth subunit of the GARP complex. As a tetramer, the GARP complex has half the number of subunits of the related exocyst and COG complexes. Interestingly, it has been suggested that the COG complex is composed of two subcomplexes of four subunits each, based on phenotypic analysis and deep-etch electron microscopy studies (Whyte and Munro, 2001, 2002; Ungar et al., 2002). Further work will be needed to see if the proposed fourfold nature of the exocyst, COG, and GARP complexes (Whyte and Munro, 2002) has functional significance. Loss of VPS51 did not alter the abundance of Vps52p, Vps53p, or Vps54p, even though these last three proteins are dependent on each other for their stability. Conversely, Vps51p levels were not appreciably changed by mutation of any other subunit. Therefore, Vps51p is unlikely to play a significant structural role in the complex but may instead have a regulatory function, or act in only a subset of late Golgi trafficking events.
GARP Is Required for the Fusion of Two Types of Vesicles with the TGN
Tethering complexes may mediate the fusion of more than one type of vesicle with the target membrane and therefore participate in more than one trafficking pathway. The cargo proteins Snc1p and Vps10p follow distinct recycling pathways to the TGN involving two different types of endosomes. We have found that mutation of any of the four GARP subunits causes defects in the retrograde transport of both Vps10p and Snc1p to the TGN. Therefore, the GARP complex appears to mediate the fusion of two different classes of vesicle, derived from distinct early and late endosomal populations.
Defects in both retrograde pathways to the late Golgi are known to result from loss of the TGN fusion machinery, include the SNAREs Tlg1p and Tgl2p, the Rab Ypt6p, and the GTP exchange factor Rgp1p/Ric1p (Lewis et al., 2000; Panek et al., 2000; Siniossoglou et al., 2000). Loss of Snc1p recycling in GARP mutants is therefore consistent with its proposed role in fusion at this step. However, failure to retrieve exocytic v-SNAREs to the Golgi would be expected to block secretion and result in severe growth defects if not death. It is possible that continued synthesis of Snc1/2p could maintain sufficient levels of secretion to support life. Alternatively, in the absence of GARP and other components of the TGN fusion machinery, Snc1/2p may follow alternative recycling pathways to the Golgi. The suppression of ypt6Δ and vps52Δ mutants by high levels of the cis-Golgi Rab protein Ypt1p (unpublished data) may be explained by the upregulation of such a salvage pathway to the cis-Golgi. Because little if any GARP complex colocalized with cis-Golgi markers by immunofluorescence microscopy, and its loss does not affect the distribution of cis-Golgi markers (Conibear and Stevens, 2000), it is unlikely that GARP itself has a role in tethering at this compartment.
VPS51 Is Required for the Binding of GARP to the TGN t-SNARE Tlg1p
By binding activated Rab proteins and SNARE regulatory regions, tethering proteins may form a critical link between components of the fusion machinery. The association of Vps52/53/54 with the Tgl1p N-terminal domain does not depend on the Rab YPT6 (Siniossoglou and Pelham, 2001). Instead, we found that this interaction requires Vps51p. In cells lacking VPS51, the remaining GARP subunits are stable, but no binding of GARP to Tlg1p could be observed either in vivo or in vitro. However, the binding of Vps51p to Tlg1p was independent of the other GARP subunits. These results are consistent with recently reported in vitro binding studies that map the Tlg1p-binding domain to an N-terminal region of Vps51p (Siniossoglou and Pelham, 2002).
In cells lacking Vps52p the amount of Vps51p that coimmunoprecipitated with Tlg1p was only slightly reduced, yet its localization is dramatically altered, becoming much more dispersed. A similar redistribution of the GARP complex is seen in the absence of YPT6, even though it remains membrane-bound. Taken together, this suggests that the association of GARP with Tlg1p is not restricted to the late Golgi, but must also occur at other compartments, presumably on transport vesicles. Other members of the “quatrefoil” family of tethering complexes (Whyte and Munro, 2002) are not only localized to donor compartments but also have additional functions there. The mammalian Sec6/8 (exocyst) complex functions at the TGN as well as at the basolateral membrane in polarized epithelial cells (Yeaman et al., 2001). Similarly, the COG (sec34/35) complex is required both for the sorting of cargo at the ER and for the tethering of these ER-derived vesicles at the cis-Golgi (Morsomme and Riezman, 2002). This role for tethering factors may ensure that newly formed vesicles carry the factors required for their subsequent targeting and fusion. Further research will be needed to see if the GARP has a role in sorting and vesicle formation at endosomal membranes.
In vitro, Vps51p still showed weak but specific binding to the N-terminus of Tlg1p expressed as a GST fusion protein, when each of the other GARP subunits was deleted in turn. Binding to t-SNARE regulatory domains may be a convenient way for tethering factors to recognize vesicle and/or target membranes and bring them together. Tethering complexes may do more than simply restrain vesicles in the proximity of the target membrane, but may instead interact with SNAREs to promote fusion. Despite having no sequence similarity to the N-terminal regions of syntaxin-like t-SNAREs, other SNAREs have independently folded N-terminal inhibitory domains and appear to exist in a closed conformation (Tochio et al., 2001; Antonin et al., 2002). This suggests that inhibition by N-terminal domains may be a general mechanism for regulating SNARE assembly at other transport steps and that a number of regulatory proteins must exist to remove the inhibition.
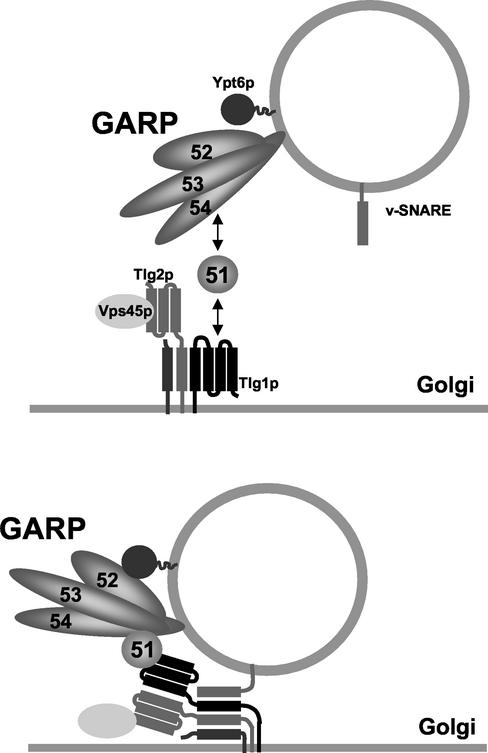
An attractive model is that the binding of the small coiled-coil protein Vps51p to the N-terminal domain of Tlg1p stabilizes it in the open conformation, thereby activating SNARE assembly and directly linking tethering to the fusion event (Figure 10). In a liposome fusion assay, the late Golgi t-SNAREs Tlg2p, Tgl1p, and Vti1p must be activated in order to bind their cognate v-SNARE Snc2p (Paumet et al., 2001). In vitro, preincubation of the t-SNAREs with a C-terminal peptide corresponding to the SNARE motif of Snc2p is sufficient to activate the t-SNAREs. In vivo, a cytosolic factor is likely to replace the Snc2p peptide in t-SNARE activation.
Figure 10.
Vps51p links Vps52/53/54 to the t-SNARE Tlg1p. In this model, Vps51p interacts with the N-terminal domain of Tlg1p and causes a conformational change that promotes the assembly of v- and t-SNAREs into a fusion complex. The Ypt6-dependent, Vps51/Tlg1-independent interactions that link the GARP complex to the Golgi are not well understood and are not shown here. See text for details.
The N-terminal domains of Tlg2p, Vti1p, and the Tlg1p homolog syntaxin 6 have recently been shown to all possess a strong structural similarity to that of syntaxin, but none appears to form a closed conformation in vitro (Antonin et al., 2002; Dulubova et al., 2002; Misura et al., 2002). It may be that intramolecular SNARE interactions are not always readily detected in vitro, because the results of the liposome fusion work are most consistent with the model that an unidentified factor catalyzes SNARE assembly by displacing an inhibitory domain. Although it is usually assumed that this domain corresponds to the N-terminus of the syntaxin-like Tlg2p, the recent identification of structurally related regions in Tlg1p and Vti1p argue that the N-terminal regions of these “nonsyntaxin” t-SNAREs are equally likely to mediate inhibitory intramolecular interactions that regulate fusion. We hypothesize that Vps51p may well be the cytosolic factor that carries out the activating function of the Snc2p peptide. A more detailed characterization of the interactions that link GARP subunits to the SNARE machinery should lead to a better understanding of how tethering factors connect vesicle and target membranes to promote docking and fusion.
ACKNOWLEDGMENTS
We are grateful to Lucy Robinson, Mike Lewis, and Hugh Pelham for their generous gifts of plasmids and reagents. We thank Ken Prehoda for technical advice and members of the Stevens lab for helpful discussions. This work was supported by grant GM32448 from the National Institutes of Health (T.H.S.).
Abbreviations used:
- ALP
alkaline phosphatase
- COG
conserved oligomeric Golgi complex
- CPY
carboxypeptidase Y
- GARP
Golgi-associated retrograde protein complex
- PVC
prevacuolar compartment
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–10–0654. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–10–0654.
REFERENCES
- Antonin W, Dulubova I, Arac D, Pabst S, Plitzner J, Rizo J, Jahn R. The N-terminal domains of syntaxin 7 and vti1b form three-helix bundles that differ in their ability to regulate SNARE complex assembly. J Biol Chem. 2002;11:11. doi: 10.1074/jbc.M204369200. [DOI] [PubMed] [Google Scholar]
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2486–2501. doi: 10.1091/mbc.02-01-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Scheller RH. SNARE-mediated membrane fusion. Nat Rev Mol Cell Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Multiple sorting pathways between the late Golgi and the vacuole in yeast. Biochim Biophys Acta. 1998;1404:211–230. doi: 10.1016/s0167-4889(98)00058-5. [DOI] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Studying yeast vacuoles. Methods Enzymol. 2002;351:408–432. doi: 10.1016/s0076-6879(02)51861-9. [DOI] [PubMed] [Google Scholar]
- Dulubova I, Sugita S, Hill S, Hosaka M, Fernandez I, Sudhof TC, Rizo J. A conformational switch in syntaxin during exocytosis: role of munc18. EMBO J. 1999;18:4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, Rizo J. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J. 2002;21:3620–3631. doi: 10.1093/emboj/cdf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I, Yamaguchi T, Wang Y, Sudhof TC, Rizo J. Vam3p structure reveals conserved and divergent properties of syntaxins. Nat Struct Biol. 2001;8:258–264. doi: 10.1038/85012. [DOI] [PubMed] [Google Scholar]
- Fernandez I, Ubach J, Dulubova I, Zhang X, Sudhof TC, Rizo J. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A. Cell. 1998;94:841–849. doi: 10.1016/s0092-8674(00)81742-0. [DOI] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis JC, Nichols BJ, Dhruvakumar S, Pelham HR. Two syntaxin homologues in the TGN/endosomal system of yeast. EMBO J. 1998;17:113–126. doi: 10.1093/emboj/17.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis MJ, Nichols BJ, Prescianotto-Baschong C, Riezman H, Pelham HR. Specific retrieval of the exocytic SNARE Snc1p from early yeast endosomes. Mol Biol Cell. 2000;11:23–38. doi: 10.1091/mbc.11.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Misura KM, Bock JB, Gonzalez LC, Jr, Scheller RH, Weis WI. Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog. Proc Natl Acad Sci USA. 2002;99:9184–9189. doi: 10.1073/pnas.132274599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morsomme P, Riezman H. The Rab GTPase Ypt1p and tethering factors couple protein sorting at the ER to vesicle targeting to the Golgi apparatus. Dev Cell. 2002;2:307–317. doi: 10.1016/s1534-5807(02)00133-8. [DOI] [PubMed] [Google Scholar]
- Nichols BJ, Holthuis JC, Pelham HR. The Sec1p homologue Vps45p binds to the syntaxin Tlg2p. Eur J Cell Biol. 1998;77:263–268. doi: 10.1016/s0171-9335(98)80084-8. [DOI] [PubMed] [Google Scholar]
- Panek HR, Conibear E, Bryan JD, Colvin RT, Goshorn CD, Robinson LC. Identification of Rgp1p, a novel Golgi recycling factor, as a protein required for efficient localization of yeast casein kinase 1 to the plasma membrane. J Cell Sci. 2000;113:4545–4555. doi: 10.1242/jcs.113.24.4545. [DOI] [PubMed] [Google Scholar]
- Parlati F, Weber T, McNew JA, Westermann B, Sollner TH, Rothman JE. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci USA. 1999;96:12565–12570. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumet F, Brugger B, Parlati F, McNew JA, Sollner TH, Rothman JE. A t-SNARE of the endocytic pathway must be activated for fusion. J Cell Biol. 2001;155:961–968. doi: 10.1083/jcb.200104092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R, Gallwitz D. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes. J Cell Biol. 2002;157:645–655. doi: 10.1083/jcb.200202006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyzes nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J. 2001;20:5991–5998. doi: 10.1093/emboj/20.21.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR. Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem. 2002;277:48318–48324. doi: 10.1074/jbc.M209428200. [DOI] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV. The Sec34/Sec35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio H, Tsui MM, Banfield DK, Zhang M. An autoinhibitory mechanism for nonsyntaxin SNARE proteins revealed by the structure of Ykt6p. Science. 2001;293:698–702. doi: 10.1126/science.1062950. [DOI] [PubMed] [Google Scholar]
- Ungar D, Oka T, Brittle EE, Vasile E, Lupashin VV, Chatterton JE, Heuser JE, Krieger M, Waters MG. Characterization of a mammalian Golgi-localized protein complex, COG, that is required for normal Golgi morphology and function. J Cell Biol. 2002;157:405–415. doi: 10.1083/jcb.200202016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte JR, Munro S. The Sec34/35 Golgi transport complex is related to the exocyst, defining a family of complexes involved in multiple steps of membrane traffic. Dev Cell. 2001;1:527–537. doi: 10.1016/s1534-5807(01)00063-6. [DOI] [PubMed] [Google Scholar]
- Whyte JR, Munro S. Vesicle tethering complexes in membrane traffic. J Cell Sci. 2002;115:2627–2637. doi: 10.1242/jcs.115.13.2627. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Dulubova I, Min SW, Chen X, Rizo J, Sudhof TC. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif. Dev Cell. 2002;2:295–305. doi: 10.1016/s1534-5807(02)00125-9. [DOI] [PubMed] [Google Scholar]
- Yeaman C, Grindstaff KK, Wright JR, Nelson WJ. Sec6/8 complexes on trans-Golgi network and plasma membrane regulate late stages of exocytosis in mammalian cells. J Cell Biol. 2001;155:593–604. doi: 10.1083/jcb.200107088. [DOI] [PMC free article] [PubMed] [Google Scholar]