Abstract
Metabolic adaptation of Saccharomyces cerevisiae cells from a nonfermentable carbon source to glucose induces selective, rapid breakdown of the gluconeogenetic key enzyme fructose-1,6-bisphosphatase (FBPase), a process called catabolite degradation. Herein, we identify eight novel GID genes required for proteasome-dependent catabolite degradation of FBPase. Four yeast proteins contain the CTLH domain of unknown function. All of them are Gid proteins. The site of catabolite degradation has been controversial until now. Two FBPase degradation pathways have been described, one dependent on the cytosolic ubiquitin-proteasome machinery, and the other dependent on vacuolar proteolysis. Interestingly, three of the novel Gid proteins involved in ubiquitin-proteasome–dependent degradation have also been reported by others to affect the vacuolar degradation pathway. As shown herein, additional genes suggested to be essential for vacuolar degradation are unnecessary for proteasome-dependent degradation. These data raise the question as to whether two FBPase degradation pathways exist that share components. Detailed characterization of Gid2p demonstrates that it is part of a soluble, cytosolic protein complex of at least 600 kDa. Gid2p is necessary for FBPase ubiquitination. Our studies have not revealed any involvement of vesicular intermediates in proteasome-dependent FBPase degradation. The influence of Ubp14p, a deubiquitinating enzyme, on proteasome-dependent catabolite degradation was further uncovered.
INTRODUCTION
Life depends on the ability of cells to adapt to environmental changes. The presence of glucose is vital to all cells, and as such it is the most important nutrient and signal trigger of cellular metabolism. When Saccharomyces cerevisiae cells are cultivated in media containing a nonfermentable carbon source, glucose is synthesized via the gluconeogenic pathway. Shifting these cells to glucose-containing media leads to a rapid switch from gluconeogenesis to glycolysis. During this metabolic adaptation, the key regulatory gluconeogenetic enzyme, fructose-1,6-bisphosphatase (FBPase), is rapidly inactivated and then degraded (half-life ∼20–30 min) in a process called catabolite inactivation (Gancedo, 1971; Holzer, 1976; Funayama et al., 1980). This inactivation process consists of two separate steps: 1) phosphorylation of the enzyme and 2) degradation of the protein. Degradation of the enzyme blocks gluconeogenesis; this prevents an otherwise ongoing futile cycle of ATP hydrolysis between the glycolytic phosphofructokinase and the gluconeogenic FBPase reactions.
The site of FBPase degradation is the subject of an ongoing debate (Schork et al., 1994a,b). The import of FBPase into Vid-vesicles with a diameter of 30–40 nm and its subsequent vacuolar degradation have been reported (Chiang and Schekman, 1991; Huang and Chiang, 1997). This vacuolar catabolite degradation process has been analyzed genetically by isolating and characterizing so-called vid-mutants (Hoffman and Chiang, 1996). Proteinase yscA-dependent catabolite degradation of FBPase has been shown to require, besides other factors, Vid22p, Cpr1p, and Vid24p (Chiang and Chiang, 1998; Brown et al., 2001, 2002). Vid24p has been reported to be peripherally attached to Vid-vesicles, and its absence leads to accumulation of FBPase trapped in these vesicles (Chiang and Chiang, 1998).
Our laboratory has found that catabolite degradation of FBPase occurs independent of vacuolar proteolysis (Wolf and Ehmann, 1979; Mechler and Wolf, 1981; Teichert et al., 1989). Instead, we find that the breakdown of FBPase, depends on polyubiquitination and the activity of the cytosolic 26S proteasome (Schork et al., 1994a,b, 1995). The linkage of polyubiquitin chains onto proteins via an enzyme cascade consisting of an ubiquitin activating enzyme (E1), ubiquitin conjugating enzymes (E2), and ubiquitin-protein ligases (E3) marks these proteins for degradation via the cytoplasmic and nuclear proteolytic nanocompartment, the proteasome (Hilt and Wolf, 1996, 2000; Baumeister et al., 1998). We furthermore showed that FBPase degradation is independent of the phosphorylation of the protein (Hämmerle et al., 1998). In a genetic approach isolating mutants defective in the glucose-induced degradation process of FBPase (gid-mutants) (Hämmerle et al., 1998; Schüle et al., 2000), we uncovered the essential function of the ubiquitin-conjugating enzyme Ubc8p (Gid3p) for glucose-induced proteasome-dependent degradation of FBPase (Schüle et al., 2000).
Herein, we report the identification of the GID1 and GID2 genes by complementation of the previously isolated point mutants with a yeast genomic library. We further characterize the localization and function of Gid2p. In addition, we present a reverse genomic approach identifying six additional novel GID genes involved in proteasome-dependent catabolite degradation of FBPase. We also report on overlapping functions of some GID gene products in vacuolar and proteasome-dependent FBPase degradation. Furthermore, we discuss new and reported findings in the context of the existence of two independent catabolite degradation pathways of FBPase.
MATERIALS AND METHODS
Construction and Growth Conditions of Strains
Previously described standard methods were used in media preparation and for genetic and molecular biological techniques (Guthrie and Fink, 1991; Ausubel et al., 1992). The S. cerevisiae strains used in this study are summarized in Table 1. Yeast strains were grown at 30°C.
Table 1.
S. cerevisiae strains used in this study
| Name | Genotype | Source |
|---|---|---|
| BY4743 | Mata/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0MET15/met15Δ0 lys2Δ0/LYS2 ura3Δ0/ura3Δ0 | EUROSCARF |
| Y34594 (gid1Δ) | BY4743 ygl227wΔ∷KANMX4/ygl227wΔ∷KANMX4 | EUROSCARF |
| Y33614 (gid2Δ) | BY4743 ydr255cΔ∷KANMX4/ydr255cΔ∷KANMX4 | EUROSCARF |
| Y36577 (gid3Δ) | BY4743 yel012wΔ∷KANMX4/yel012wΔ∷KANMX4 | EUROSCARF |
| Y33244 (gid4Δ) | BY4743 ybr105cΔ∷KANMX4/ybr105cΔ∷KANMX4 | EUROSCARF |
| Y31410 (gid5Δ) | BY4743 yil017cΔ∷KANMX4/yil017cΔ∷KANMX4 | EUROSCARF |
| Y33195 (gid6Δ) | BY4743 ybr058cΔ∷KANMX4/ybr058cΔ∷KANMX4 | EUROSCARF |
| Y33446 (gid7Δ) | BY4743 ycl039wΔ∷KANMX4/ycl039wΔ∷KANMX4 | EUROSCARF |
| Y36576 (gid8Δ) | BY4743 ymr135cΔ∷KANMX4/ymr135cΔ∷KANMX4 | EUROSCARF |
| Y31488 (gid9Δ) | BY4743 yil097wΔ∷KANMX4/yil097wΔ∷KANMX4 | EUROSCARF |
| Y35282 (vid22Δ) | BY4743 ylr373cΔ∷KANMX4/ylr373cΔ∷KANMX4 | EUROSCARF |
| Y32000 (vid27Δ) | BY4743 ynlL212wΔ∷KANMX4/ynl212wΔ∷KANMX4 | EUROSCARF |
| Y33513 (cpr1Δ) | BY4743 ydr155cΔ∷KANMX4/ydr155cΔ∷KANMX4 | EUROSCARF |
| Y33075 (ybl049wΔ) | BY4743 ybl049wΔ∷KANMX4/ybl049wΔ∷KANMX4 | EUROSCARF |
| Y35286 (fbp1Δ) | BY4743 ylr377cΔ∷KANMX4/ylr377cΔ∷KANMX4 | EUROSCARF |
| WAY.5-4A | Mata his3Δ1 ura3-52 | K.D. Entian |
| WAY.5-4A/D1 | WAY.5-4A gid1-3 | K.D. Entian |
| WAY.5-4A/B2 | WAY.5-4A gid2-1 | K.D. Entian |
| JK9-3da | Mata leu2-3/112 ura3-52 rme1 trp1 his4 GAL+ HMLa | M.N. Hall |
| AE7-6c | JK9-3da gid1Δ∷URA3-1 | M.N. Hall |
| W3031B | Matα ade2 leu2-3,112 his3 trp1 ura3 | H.L. Chiang |
| W3031BKO | W3031B fbp1Δ∷LEU2 | H.L. Chiang |
| YTS1 | W3031B gid2Δ∷KAN | This work |
| YTS2 | W3031B gid3Δ∷KAN | This work |
| YTS3 | W3031B GID2-HA3∷HIS5(s.p.) | This work |
| YTS6 | W3031B gid2Δ∷KAN gid3Δ∷KAN | This work |
| YFJ1 | W3031B gid4Δ∷KAN | This work |
Identification of GID Genes
gid1-3 and gid2-1 mutant cells were each transformed with a YCp50-based genomic library (Rose et al., 1987) and grown for 3 d at 30°C on selective plates containing 2% glucose. Transformants were replica plated and grown on selective plates with 2% ethanol as the sole carbon source. Thereafter, colonies were transferred onto nitrocellulose sheets and soaked with YPD for 2–2.5 h at 30°C. Cells were then lysed by incubating with 0.1% SDS, 0.2 M NaOH, 0.5% β-mercaptoethanol for 30 min and washed off the membrane. Nitrocellulose sheets were incubated with 10% milk in 20 mM Tris-HCl pH 7.6, 137 mM NaCl, 0.1% Tween 20 overnight at 4°C and then probed with FBPase antibody in 20 mM Tris-HCl pH 7.6, 137 mM NaCl, 0.1% Tween 20 for 1 h. After several washes, filters were incubated with peroxidase-coupled goat anti-rabbit antibody (Medac, Hamburg, Germany), and peroxidase activity was detected using the enhanced chemiluminescence system (Amersham Biosciences, Braunschweig, Germany).
The 5000 strains of the yeast deletion collection (EUROSCARF, Frankfurt, Germany) were grown in 96-well microtiter plates in YPD for 5.5 h to screen for strains deficient in the catabolite degradation of FBPase. Then, they were dropped onto YPEthanol plates and grown overnight. After transfer onto nitrocellulose membranes, strains were processed as described above.
Chromosomal Deletion of GID2
A DNA fragment was created by polymerase chain reaction (PCR) with oligonucleotides TS1 (TCTTCAAGAGAGATGCAGCACTGAGTAGGGAACCAAGAAACGCAGCTGAAGCTTCGTACGC) and TS2 (AAAAAAAAAAAAAAAAAACCTATGCAAAAATTTCAGAGCATAGGCCACTAGTGGATCTG) and plasmid pUG6 (Güldener et al., 1996). This PCR fragment was used to chromosomally replace chromosomal GID2 with a LoxP-KANR-LoxP cassette in the W303-1B strain background (Chiang and Schekman, 1991), yielding the strain YTS1 (Matα ade2 leu2-3, 112 his3 trp1 ura3 gid2Δ::KANR). Southern blotting confirmed correct gene replacement (our unpublished data).
Construction of Hemagglutinin (Ha)-tagged Gid2p (Gid2-Ha3)
Strain YTS3 (Matα ade2 leu2-3, 112 his3 trp1 ura3 GID2-HA3::HIS5) was constructed by chromosomal integration of a PCR fragment consisting of a triple Ha-tag and a Schizosaccharomyces pombe HIS5 marker (Cottarel, 1995) in W303-1B cells. The PCR fragment was generated using oligonucleotides TS9 (CCGTAAATACTTCAATGAGCAGTACAAAAAAGGTTCGTTTTGTTATGCTTGGAGCAGG-GGCGGGTGC) and TS10 (TTATCGCTTCCAATAAAAAAAAAAAAAAAAAACCTATGCAAAAATTTCAGGAGGTCGA-CGGTATCGATAAG) and plasmid p3xHA HIS5 (S. Munro, MRC Laboratory of Molecular Biology, Cambridge, UK). Correct integration was confirmed by Southern analysis (our unpublished data).
Sucrose Density Gradient Fractionation
Subcellular fractionation was done as described previously (Schüle et al., 2000). Cells were grown in YPD (2% glucose) medium to an optical density of 5–6, resuspended in YPEthanol (2% ethanol) at an OD600 of 0.2, and grown for 16 h at 30°C to derepress FBPase. Then, 2% glucose was added and 1000 OD600 cells were harvested at 0 and 30 min. Cells were then spheroplasted and proteinase inhibitors (Complete; Roche Diagnostics, Mannheim, Germany) were added. After homogenization, cell debris was removed by centrifugation (2000 × g, 10 min at 4°C), and the supernatant was loaded onto a 18–54% sucrose gradient (10 ml). The gradient was then centrifuged at 100,000 × g for 3 h at 4°C and 0.6-ml fractions were collected from the top to bottom and analyzed by immunoblotting. Guanosine diphosphatase activity was determined according to Abeijon et al. (1989).
Cell Fractionation and Proteinase K Digestion
This method was adopted from Schüle et al. (2000). Cells were grown in YPD containing 2% glucose to an optical density of 5–6, harvested, and resuspended at an OD600 of 0.2/ml in YPEthanol medium (2% ethanol). After 16 h of growth at 30°C, glucose was added to a final concentration of 2%, and aliquots (60 OD600) were withdrawn at 0 and 30 min after glucose addition. Cells were then converted to spheroplasts and lysed hypotonically. A 300 × g centrifugation step removed nonlysed cells. Centrifugation for 20 min at 6000 × g yielded an S6 supernatant and a P6 pellet fraction. The S6 fraction was further centrifuged at 100,000 × g for 3 h yielding an S1 supernatant and a P1 pellet fraction. After immunoblotting, samples were analyzed with antibodies against phosphoglycerate kinase (PGK), carboxypeptidase yscY (CPY) (Molecular Probes, Leiden, The Netherlands), Kar2p (R. Scheckman, Howard Hughes Medical Institute and Department of Molecular and Cell Biology, University of California, Berkeley, CA), and aminopeptidase I (API) (Barth and Thumm, 2001). Peroxidase-coupled secondary antibodies were from Medac (goat anti-rabbit) and Dianova (Hamburg, Germany) (goat anti-mouse), respectively. Peroxidase activity was monitored with the enhanced chemiluminescence system (Amersham Biosciences).
For proteinase K digestion, total cell lysates were generated as described above and incubated with 50 μg/ml proteinase K or 50 μg/ml proteinase K plus 0.2% Triton X-100, respectively, for 30 min on ice. Digestion was stopped by addition of 10% trichloroacetic acid, and samples were processed for immunoblotting.
Pulse-Chase Analysis and Western Blotting
Pulse-chase experiments were done as described in Schork et al. (1995). For quantification a PhosphorImager (Amersham Biosciences, Sunnyvale, CA) was used. Immunodetection of Ha-tagged proteins, and ubiquitin conjugates were described in Schüle et al. (2000). Antibodies directed against the Ha-tag were from Babco (Richmond, CA) (clone 16B12). Pulse-chase analysis of the N-end rule substrate Arg-β-gal was done according to Schüle et al. (2000).
Glycerol Density Gradient Fractionation
Cells were grown for 16 h in YPEthanol and 50 OD600 cells were shifted to YPD medium for 25 min. The cells were then harvested, resuspended in 0.1 M KH2PO4, pH 7.0, in the presence of Complete (protease inhibitor cocktail tablets; Roche Diagnostics) and 10 mM phenylmethylsulfonyl fluoride, lysed with glass beads, and centrifuged at 10,000 × g for 15 min. Then, 200-μl aliquots of the resulting cell extract were layered on top of a glycerol step gradient (450 μl each of 50, 40, 30, and 20% of glycerol in 20 mM PIPES buffer, pH 6.8) and centrifuged for 4 h at 55,000 rpm and 15°C in a TLS-55 rotor (Beckman Coulter, Fullerton, CA). Thereafter, 200-μl fractions were collected and processed for immunoblotting.
RESULTS
Identification of GID1 and GID2 Genes
Using a fusion protein consisting of the amino-terminal part of FBPase fused to the marker β-galactosidase, we previously isolated three mutants defective in the GID1, GID2, and GID3 genes (gid1-3, gid2-1, and gid3-1) necessary for glucose-induced degradation of FBPase (Hämmerle et al., 1998). We uncovered GID3 as the gene encoding the ubiquitin-conjugating enzyme Ubc8p as a crucial component involved in catabolite degradation of FBPase (Schüle et al., 2000).
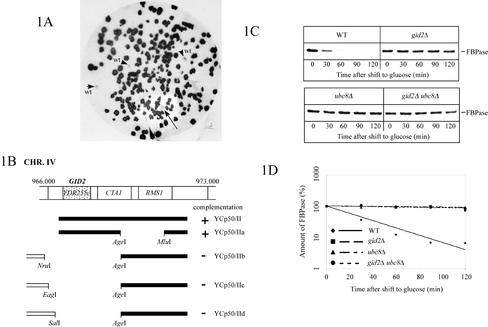
Applying the immunological colony screen with FBPase antibody and peroxidase-coupled goat anti-rabbit antibody (Schüle et al., 2000) to the gid1-3 and gid2-1 mutants, we now identified the respective GID1 and GID2 wild-type genes. As shown in Figures 1A and 2A, noncomplemented gid-mutant colonies look dark, due to a high FBPase level, whereas cells carrying a complementing plasmid can breakdown FBPase after a shift to glucose and therefore look white. As controls, wild-type colonies were included on each nitrocellulose sheet (Figures 1A and 2A, arrowheads). One positive clone was detected out of the 45,000 gid2-1 colonies screened (Figure 1A, arrow). The rescued plasmid contained an ∼5.7-kb genomic fragment from chromosome IV (Figure 1B). Subcloning identified YDR255c as the complementing open reading frame (ORF) (Figure 1B). To construct a YDR255c deleted strain, we integrated a PCR-generated kanamycin resistance cassette into the respective locus of wild-type cells (see MATERIALS AND METHODS for details). Correct gene replacement was confirmed by Southern blotting (our unpublished data). As expected, pulse-chase analysis of YDR255c-deleted cells showed a defect in degradation of FBPase after glucose addition (Figure 1, C and D). Analysis of gid2-1 ydr255cΔ diploid cells confirmed the identity of YDR255c as GID2 (our unpublished data).
Figure 1.
Identification of the GID2 gene. (A) After transformation with a YCp50-based genomic library, complemented gid2-1 colonies (WAY.5-4A/B2) are detected in a colony screen. Cells were transferred onto nitrocellulose sheets as outlined in MATERIALS AND METHODS and probed with antibodies against FBPase. Wild-type (WAY.5-4A) colonies (arrowheads) and gid2-1 colonies, carrying a complementing genomic fragment (arrow) are able to degrade FBPase and look white, due to their low level of FBPase. (B) Subcloning identifies YDR255c (GID2) as the complementing ORF. (C) Pulse-chase analysis of glucose-induced degradation of FBPase. (D) Quantification of C shows a significant stabilization of FBPase in gid2Δ (YTS1) cells. Control, wild-type (W303-1B) and UBC8 deleted (YTS2) cells.
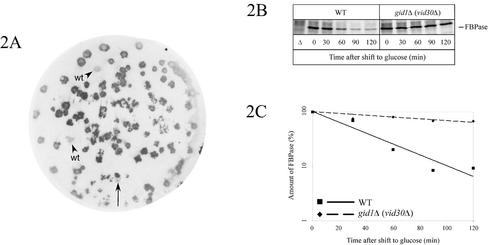
Figure 2.
Identification of the GID1 gene. (A) gid1-3 (WAY.5-4A/D1) cells were transformed with a YCp50-based genomic library, and after transfer of the colonies to nitrocellulose they were processed as described in MATERIALS AND METHODS. The level of FBPase was then detected with antibodies. Complemented colonies (arrow) and wild-type (WAY.5-4A) colonies (arrowheads) look white, due to degradation of FBPase. (B) Pulse-chase analysis of gid1Δ (vid30Δ) (AE7-6c) cells confirms the defect in glucose-induced degradation of FBPase. (C) Quantification of B. Control, wild-type cells (JK9-3da).
In a similar approach using gid1-3 mutant cells, a complementing plasmid carrying a fragment from chromosome VII was isolated (Figure 2A). YGL227w was identified as the complementing ORF encoding a protein of 108 kDa. Deletion of YGL227w also showed a significant defect in glucose-induced degradation of FBPase in pulse-chase measurements (Figure 2, B and C). The defect of gid1-3 ygl227wΔ diploid cells in breakdown of FBPase after glucose addition confirmed the identity of GID1 with YGL227w (our unpublished data).
Features of Gid1p and Gid2p
Based on unpublished results presented in the Saccharomyces genome database (SGD; http://genome-www.stanford.edu/saccharomyces/) and Incyte Genomics (http://www.incyte.com/proteome/YPDsearch-quick.html), we identified Gid1p as Vid30p, a protein that is proposed to be involved in glucose-induced degradation of FBPase in the vacuole. A function of Vid30p in the regulation of nitrogen metabolism has also been suggested (van der Merwe et al., 2001). A search for amino acid motifs identified a LisH and a CTLH motif within Gid1p/Vid30p. According to the SMART database, the LisH domain might be involved in regulation of microtubule dynamics, and the CTLH domain of unknown function is typically found C-terminal to the LisH motif. Interestingly, Gid2p also contains a CTLH motif. Furthermore, a putative transmembrane domain is found in Gid2p located between amino acids 301–323. The occurrence of these domains, however, does not suggest any function for Gid1p or Gid2p. Herein, we focus on the functional characterization of Gid2p.
Gid2p Is Essential for Glucose-induced Ubiquitination of FBPase
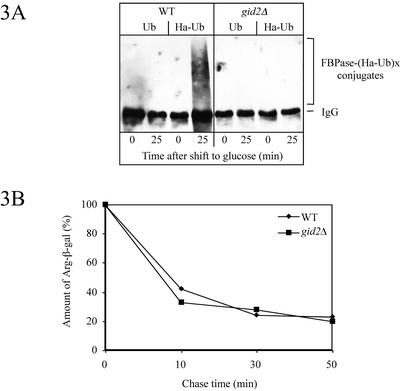
Our previous work demonstrated that glucose-induced degradation of FBPase depends on ubiquitination of the enzyme via the ubiquitin-conjugating enzymes Ubc1p, Ubc4p, Ubc5p, and Ubc8p followed by proteolysis dependent on the activity of the proteasome (Schork et al., 1995; Schüle et al., 2000). By using these standard conditions, FBPase degradation is impaired in mutants defective in subunits of the 20S core and the 19S cap of the proteasome, whereas wild-type-like proteolysis occurred in cells devoid of vacuolar proteinase yscA (Schork et al., 1994a,b, 1995). To learn more about the function of Gid2p in the proteasome-dependent degradation of FBPase, we measured glucose-induced ubiquitination of FBPase in gid2Δ cells under our standard conditions of catabolite degradation. In contrast to wild-type cells, no FBPase-ubiquitin conjugates were detectable in gid2Δ cells overexpressing Ha-tagged ubiquitin (Ellison and Hochstrasser, 1991) under conditions of catabolite degradation (Figure 3A) (Schork et al., 1995). This suggests that Gid2p has an essential function in glucose-induced ubiquitination of FBPase.
Figure 3.
(A) Glucose-induced ubiquitination of FBPase is affected in gid2Δ (YTS1) cells. Crude extracts from wild-type (W303-1B) and gid2Δ (YTS1) cells overexpressing ubiquitin or Ha-tagged ubiquitin were immunoprecipitated with antibodies against FBPase. Proteins were separated by SDS-PAGE, blotted, and probed with antibodies against Ha. (B) In gid2Δ (YTS1) cells degradation of the N-end rule substrate Arg-β-gal is not affected. After pulse labeling, aliquots were withdrawn at the indicated times and the amount of Arg-β-gal was determined with a PhosphorImager after immunoprecipitation with antibodies against β-galactosidase.
N-End Rule Pathway Is Not Affected in gid2Δ Cells
Short-lived N-end rule substrates such as the Arg-β-galactosidase fusion protein (Arg-β-gal) are known to be rapidly degraded via the ubiquitin proteasome pathway (Bachmair et al., 1986; Richter-Ruoff et al., 1992; Seufert and Jentsch, 1992). Previous experiments suggested an increased level of Arg-β-gal in gid2-1 mutant cells compared with wild type (Hämmerle et al., 1998). Following the fate of Arg-β-gal in gid2Δ cells via pulse-chase measurements did not uncover an altered degradation of the substrate (Figure 3B). This finding rules out a function of Gid2p in the N-end rule pathway.
Gid2p Is Part of a Soluble Protein Complex
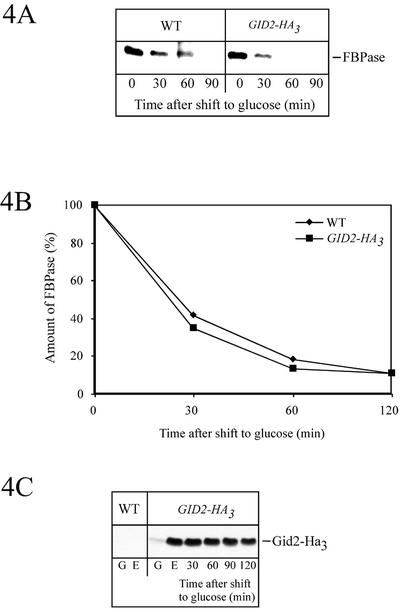
Using a PCR fragment consisting of a triple Ha-tag and a selectable marker (see MATERIAL AND METHODS), we generated a chromosomally integrated, carboxy-terminally tagged version of Gid2p expressed from its native promoter. Correct integration at the GID2 locus was confirmed by Southern analysis (our unpublished data). Wild-type-like degradation of FBPase, monitored by pulse-chase analysis (Figure 4, A and B), confirmed biological activity of the Ha-tagged Gid2p-version Gid2-Ha3. Consistent with its role in glucose-induced degradation of FBPase, only minor amounts of Gid2-Ha3 were detectable in cells grown in the presence of glucose (log-phase cells) (Figure 4C). Most interestingly, the level of Gid2-Ha3 significantly increased, when the cells were grown in ethanol medium (Figure 4C). After addition of glucose to ethanol-derepressed cells Gid2-Ha3 is relatively stable, consistent with its important function during the metabolic switch from gluconeogenesis to glycolysis.
Figure 4.
Ha-tagged Gid2p expressed from the chromosome with its native promoter is functional during catabolite inactivation of FBPase (strain YTS3). After pulse labeling, the cells were chased in glucose-containing medium. After immunoprecipitation with antibodies against FBPase and SDS-PAGE, the amount of FBPase was detected with a PhosphorImager (A) and quantified (B). (C) Steady-state levels of Ha-tagged Gid2p. Crude extracts of cells (YTS3) grown in glucose- (G) or ethanol (E)-containing media were analyzed on immunoblots with antibodies against Ha. Ethanol-grown cells were further shifted to glucose media and at the indicated times aliquots were analyzed. As a control, wild-type (W303-1B) cells lacking an Ha-tag were included.
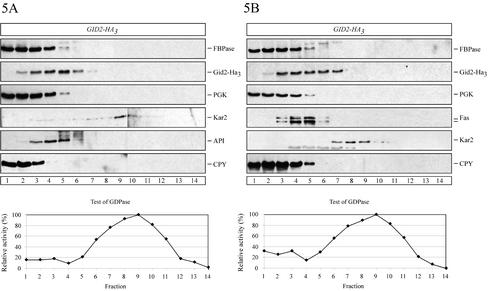
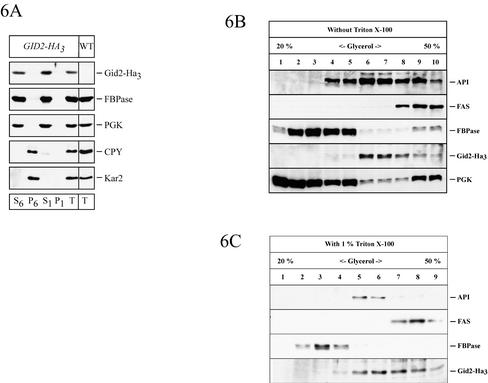
The localization of Gid2-Ha3 was analyzed on sucrose density gradients. We used cells grown on ethanol medium and cells grown on ethanol and thereafter incubated for 30 min in medium containing glucose, conditions that induce catabolite degradation. Under both conditions FBPase cosedimented with the cytosolic marker enzyme PGK and the soluble vacuolar carboxypeptidase yscY (CPY) (Figure 5, A and B). In the same gradient Gid2-Ha3 moves somewhat further into higher density fractions (Figure 5, A and B), in which large, soluble protein complexes such as fatty acid synthase (Fas, molecular mass ∼2.4 MDa; Egner et al., 1993) and API (molecular mass 600 kDa; Kim et al., 1997) are also located. Because the calculated molecular mass of Gid2p is 49 kDa, this suggests that Gid2-Ha3 may either be part of a large protein complex or may be associated with a low-density membrane fraction. To distinguish between these possibilities and to check whether Gid2-Ha3 is localized in the vacuole or in the cytosol, we performed an additional cell fractionation experiment (Schüle et al., 2000). Cells taken 30 min after shift to glucose medium were spheroplasted, hypotonically lysed (without affecting the integrity of the vacuole), and centrifuged at 6000 × g. The centrifugation yields a S6 supernatant and a P6 pellet fraction. The P6 fraction was enriched with the endoplasmic reticulum (ER) and the vacuole as shown by the appearance of Kar2p (BiP), a lumenal ER chaperone, and the soluble vacuolar CPY (Figure 6A). FBPase and Gid2-Ha3 were almost completely absent from the P6 fraction (Figure 6A). The 6000 × g supernatant (S6) was subjected to a high-speed centrifugation at 100,000 × g, yielding a P1 pellet and a S1 supernatant fraction. Both FBPase and Gid2-Ha3 were found almost exclusively in the S1 fraction together with the cytosolic marker enzyme PGK (Figure 6A), suggesting that they are soluble, cytosolic proteins. This finding is compatible with localization studies performed using indirect immunofluorescence where Gid2-Ha3 is seen in the cytosol (our unpublished data).
Figure 5.
Sucrose density gradient fractionation of cells (YTS3) expressing Ha-tagged Gid2p from the chromosome. After 16 h of growth on ethanol medium, the cells were shifted to glucose medium. The cells were analyzed before (A) and 30 min after shift to glucose medium (B). From the sucrose gradient, fractions were collected from the top of the gradient (fraction 1) and analyzed by immunoblotting with antibodies against Ha (Ha-tagged Gid2p), cytosolic PGK, Fas (cytosolic fatty acid synthase), CPY (vacuolar carboxypeptidase yscY); vacuolar API, and ER lumenal Kar2p. The fractions were further tested for their Guanosine diphosphatase (Golgi) activity. Further details are given in the text.
Figure 6.
(A) FBPase and Ha-tagged Gid2p are found in the soluble fraction 30 min after shift to glucose medium. Cells expressing Ha-tagged Gid2p from the chromosome (YTS3) were converted to spheroplasts. After mild hypotonic lysis, the cell lysate was fractionated by centrifugation. S6 and P6 refer to the 6000 × g supernatant and pellet, respectively; S1, 100,000 × g supernatant; P1, 100,000 × g pellet. (B and C) Glycerol gradient fractionation of cell lysates from cells expressing Ha-tagged Gid2p from the chromosome (YTS3). Fractions were immunoblotted with antibodies against API, Fas (fatty acid synthase), FBPase, Gid2-Ha3, and PGK. Glycerol gradient fractionation was done without (B) and with 1% Triton X-100 (C).
To further analyze whether Gid2-Ha3 is part of a protein complex, we examined the sedimentation behavior of the protein in a glycerol density gradient with extracts from cells taken 30 min after onset of glucose-triggered inactivation (Figure 6B). Gid2-Ha3 sedimented mainly in fractions 6 and 7, where the maximum of aminopeptidase I with an approximate mass of 600 kDa was also found (Figure 6B). Fatty acid synthase was detected in fractions 8–10. To check whether the sedimentation of Gid2-Ha3 is due to membrane association, we repeated the glycerol gradient fractionation in the presence of 0.2% (our unpublished data) and 1% Triton X-100 (Figure 6C). A similar sedimentation pattern of Gid2-Ha3 in the presence of detergent argues against membrane association as the reason for the high Mr sedimentation pattern of Gid2-Ha3. Together, these results suggest that Gid2-Ha3 is part of a cytosolic protein complex with a molecular mass of at least 600 kDa.
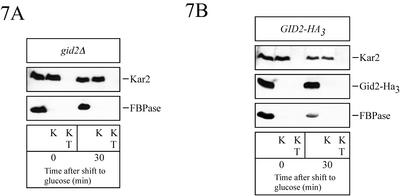
The site of catabolite degradation of FBPase is a matter of an ongoing debate (Schork et al., 1994b). Although we find a cytosolic, ubiquitin-proteasome–dependent catabolite degradation of FBPase (Schork et al., 1994a,b, 1995; Hämmerle et al., 1998; Schüle et al., 2000), vacuolar degradation of FBPase (Chiang and Schekman, 1991) and the involvement of vesicular intermediates in this process has also been suggested (Huang and Chiang, 1997; Chiang and Chiang, 1998). We therefore were interested in whether some FBPase became membrane protected during the catabolite degradation process in gid2Δ cells. To test this, ethanol grown cells were taken before and after a 30-min shift to glucose medium, spheroplasted, and hypotonically lysed. Aliquots of the lysed spheroplasts were incubated with proteinase K and, as a control, with proteinase K and Triton X-100. As shown in Figure 7A, no proteinase-protected FBPase protein was detectable under the conditions used. In a similar experiment, we tested for membrane trapped Gid2p in wild-type cells expressing Gid2-Ha3 from the chromosome. No proteinase-protected Gid2-Ha3 was detectable before or after shifting the cells for 30 min to glucose medium (Figure 7B). Together, our data suggest that under our testing conditions, Gid2-Ha3 is part of a cytosolic, soluble protein complex and that FBPase is not engulfed into vesicles in gid2Δ cells during glucose-induced degradation.
Figure 7.
FBPase is not engulfed in vesicles under the inactivation conditions used (see MATERIALS AND METHODS). At the indicated times after shift to glucose medium, cells were spheroplasted, hypotonically lysed, and incubated with proteinase K (K) and proteinase K with Triton X-100 (KT). Samples were then immunoblotted and analyzed with antibodies against ER lumenal Kar2p, Ha, and FBPase. In A, gid2Δ (YTS1) cells and in B cells (YTS3) chromosomally expressing Ha-tagged Gid2p were used.
Identification of Six Additional GID Genes, Essential for Proteasome-dependent FBPase Catabolite Degradation
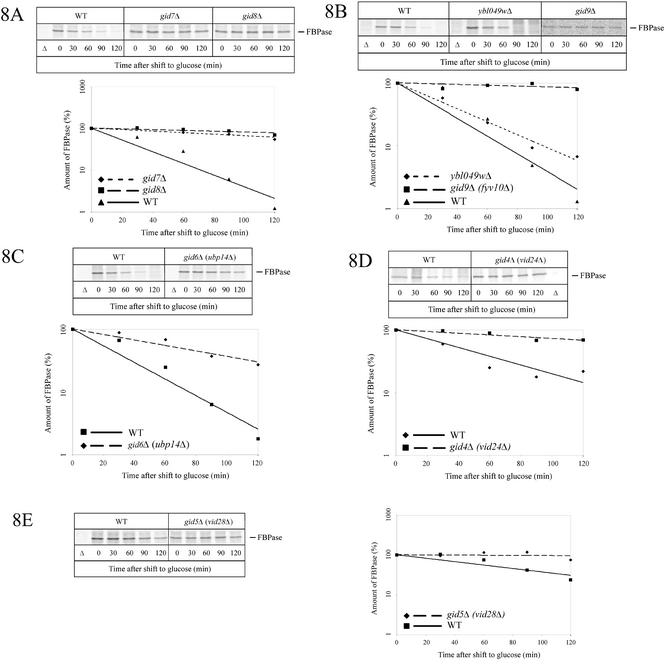
Because Gid2-Ha3 is part of a protein complex with a molecular mass of at least 600 kDa, it is highly probable that the complex contained additional gene products involved in the catabolite degradation of FBPase. Because our initial screen identified only three complementation groups (Hämmerle et al., 1998), we developed an alternative genomic approach to identify further GID genes. The “yeast gene deletion project” created about 5000 yeast strains, each deleted for an individual gene (EUROSCARF). This yeast deletion strain collection represents all nonessential genes and covers ∼85% of the yeast genome. The deletion strains, supplied in 96-well microtiter plates, were grown in rich medium containing glucose, and then each strain was dropped onto YPEthanol plates and grown for 1 d to derepress FBPase. Subsequently, cells were transferred onto nitrocellulose sheets, incubated with glucose medium to induce degradation of FBPase, and the remaining amount of FBPase was detected using specific antibodies (for details, see MATERIALS AND METHODS). Putative gid-mutants were identified on the basis of their darker immunostaining compared with wild-type cells. Candidate colonies were picked and catabolite degradation of FBPase was examined by immunoblotting (our unpublished data) and pulse-chase analysis (Figure 8, A–E). In this way, besides GID1, GID2, and GID3, we identified six novel GID genes (Table 2). All cells deleted in GID genes grow normally on rich media containing either glucose or ethanol as carbon source (our unpublished data).
Figure 8.
Pulse-chase analysis of FBPase degradation in novel gid mutant cells. gid7Δ (Y33446), gid8Δ (Y36576) (A), gid9Δ (fyv10Δ) (Y31488), ybl049wΔ (Y33075) (B), gid6Δ (ubp14Δ) (Y33195) (C), gid4Δ (vid24Δ) (YFJ1) (D), and gid5Δ (vid28Δ) (Y31410) (E) cells. Details are given in the text. In the experiment shown for gid5Δ, labeling was extended to 16 h to enhance the amount of labeled protein.
Table 2.
GID genes
| ORF | ORF name | |
|---|---|---|
| GID1 | YGL227w | VID30 |
| GID2 | YDR255c | |
| GID3 | YEL012w | UBC8 |
| GID4 | YBR105c | VID24 |
| GID5 | YIL017c | VID28 |
| GID6 | YBR058c | UBP14 |
| GID7 | YCL039w | |
| GID8 | YMR135c | |
| GID9 | YIL097w | FYV10 |
Among these six new proteins, Gid7p (Figure 8A), Gid8p (Figure 8A), and Gid9p (Figure 8B) are of unknown function. Ubp14p/Gid6p (Figure 8C), a deubiquitinating enzyme was also found to affect proteasome-dependent catabolite degradation of FBPase. Ubp14p is proposed to prevent inhibition of proteasomal degradation by removing ubiquitin chains, which may otherwise compete with substrate molecules binding to the proteasome (Amerik et al., 1997). Pulse-chase analysis showed a threefold decreased rate in catabolite degradation of FBPase in cells lacking Ubp14p/Gid6p (Figure 8C). Pulse-chase experiments using cells lacking GID9 and also GID5/VID28 showed a somewhat smaller amount of labeled FBPase. This could be overcome by extending the labeling time from 3.5 h to 16 h. These conditions lead to a slightly delayed degradation kinetics of FBPase in wild-type cells (half-life ∼60 min).
Most interestingly, our genomic screen also identified Vid24p/Gid4p (Figure 8D) and Vid28p/Gid5p (Figure 8E) as components essential for proteasome-dependent catabolite degradation of FBPase. This was surprising because Vid24p had been previously described as a peripheral membrane protein located at Vid-vesicles implicated in transport of FBPase to the vacuole during glucose-induced vacuolar degradation (Chiang and Chiang, 1998). Because these researchers reported that in the absence of Vid24p FBPase accumulated trapped inside Vid-vesicles, Vid24p had been proposed to function in vacuolar targeting of FBPase-containing vesicles (Chiang and Chiang, 1998).
Only a Subset of Vid-Proteins Affect Proteasome-dependent Catabolite Degradation of FBPase
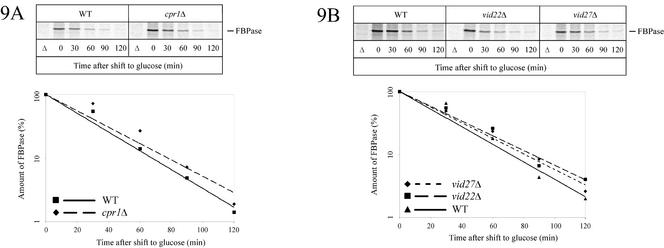
Based on database entries, Vid30p/Gid1p and Vid28p/Gid5p also should be implicated in glucose-induced vacuolar degradation of FBPase, but so far the respective data have not been published. The finding that three gene products, Vid30p/Gid1p, Vid24p/Gid4p, Vid28p/Gid5p, which were reported to affect vacuolar FBPase degradation, also act in proteasome-dependent FBPase degradation raised the question whether there may be a general overlap between these two sets of genes, GID and VID, in FBPase degradation. We therefore additionally analyzed Vid22p and Cpr1p, cyclophilin A, a mediator of Vid22p function, which were both shown to affect vacuolar FBPase degradation (Brown et al., 2001, 2002), for their involvement in proteasome-dependent degradation of FBPase. We also included cells devoid of Vid27p in our analysis. Involvement of Vid27p in vacuolar catabolite inactivation of FBPase is not published but is mentioned in the SGD and the Incyte Genomics YPD database. Under our standard conditions, pulse-chase analysis demonstrates that in cells lacking either Vid22p, Cpr1p, or Vid27p proteasome-dependent degradation of FBPase is not affected (Figure 9, A and B). Together, our data suggest the existence of two glucose-induced degradation pathways for FBPase exhibiting a partial, but not complete overlap.
Figure 9.
Vid27p, Vid22p, and Cpr1p are not essential for proteasome-dependent catabolite degradation of FBPase. Pulse-chase analysis of FBPase degradation was measured in cpr1Δ (Y33513) (A), vid22Δ (Y35282), and vid27Δ (Y32000) (B) cells.
GID Genes Are Not Involved in Catabolite Degradation of Galactose Permease (Gal2p)
Not only cytoplasmic FBPase undergoes glucose-induced catabolite degradation. The plasma membrane bound sugar transporter galactose permease (Gal2p) is also a target of this mechanism. In contrast to ubiquitin-proteasome–mediated degradation of FBPase, Gal2p undergoes ubiquitin-linked endocytosis followed by degradation in the vacuole (Horak and Wolf, 1997, 2001). Interestingly, both proteolytic pathways are induced by the same components of glucose signaling (Horak et al., 2002). We therefore tested whether the Gid-proteins identified herein to be involved in FBPase degradation, play any role in the catabolite degradation of Gal2p. We did not find an alteration of Gal2p degradation (our unpublished data) in any of the nine mutants defective in a single GID gene (gid1Δ to gid9Δ), indicating the specificity of these gene products in ubiquitin-proteasome–dependent degradation of FBPase. Also, deletions in VID22 and VID27, suggested to be involved in vacuolar FBPase degradation (Brown et al., 2001, 2002; database entry) did not affect the glucose-induced Gal2p breakdown (our unpublished data).
DISCUSSION
As a result of two independent screens, we identified eight novel GID genes involved in glucose-induced proteasome-dependent catabolite degradation of FBPase. In an initial screen using chemical mutagenesis, we isolated gid-mutant cells defective in the degradation of a FBPase-β-galactosidase fusion protein. By complementation of the FBPase degradation phenotype of gid1-3 and gid2-1 mutant cells with a yeast genomic library, we identified the respective genes as YGL227w/GID1 and YDR255c/GID2. In addition, we started a novel reverse genomic approach by using a yeast deletion collection consisting of ∼5000 individual clones, each lacking a nonessential gene. The collection covers ∼85% of the total genome. This approach was possible because a defective degradation of FBPase is not lethal. When using this mutant collection, identification of a deletion strain is synonymous with the identification of the respective gene. The screen detected the known genes GID1/VID30, GID2, and the previously identified UBC8/GID3 gene (Schüle et al., 2000) as expected. In addition, six novel genes involved in proteasome-dependent catabolite inactivation of FBPase were discovered: GID4/VID24, GID5/VID28, GID6/UBP14, GID7, GID8, and GID9.
Lack of Ubp14p/Gid6p resulted in a threefold decrease of FBPase breakdown (Figure 8C). Ubp14p is thought to act as a “helper” enzyme during proteasomal breakdown of ubiquitinated proteins (Amerik et al., 1997). It disassembles free ubiquitin chains, which would otherwise compete with binding of ubiquitinated proteins to the proteasome. Ubp14p thus accelerates proteasomal breakdown of ubiquitinated proteins. Under our conditions used previously for measuring catabolite degradation (Schork et al., 1994a,b; Hämmerle et al., 1998; Schüle et al., 2000; this article), breakdown of FBPase was only dependent on the activity of the cytosolic ubiquitin-proteasome system. Consistent with this finding, we demonstrated an essential function of ubiquitin conjugation (Schork et al., 1995) and of the ubiquitin-conjugating enzyme Ubc8p (Schüle et al., 2000). Retardation of FBPase degradation in ubp14Δ cells further supports our view that proteasomal degradation rather than ubiquitination alone is an essential step of catabolite inactivation.
Most interestingly, with Gid4p/Vid24p, Gid1p/Vid30p, and Gid5p/Vid28p we also detected some VID gene products in our genomic screen. A functional study is only reported for Vid24p (Chiang and Chiang, 1998). The involvement of Vid30p and Vid28p in vacuolar catabolite degradation is solely based on database entries (SGD and Incyte Genomics YPD database). Identification of Vid-proteins necessary for proteasomal degradation of FBPase was unexpected because the vacuolar catabolite degradation of the enzyme is supposed to start with the sequestration of FBPase into so-called Vid-vesicles in response to glucose addition to cells. These vesicles are thought to deliver FBPase to the vacuolar lumen, where proteinase yscA-dependent degradation should occur. Vid24p has been localized as a peripheral membrane protein to the cytosolic surface of the Vid-vesicles. Because lack of Vid24p leads to accumulation of FBPase in these vesicles, a targeting function had been attributed to Vid24p (Chiang and Chiang, 1998). In this context, the function of some Vid proteins and especially of Vid24p for proteasome-dependent FBPase degradation is an enigma.
We therefore examined whether some other VID genes might be essential for proteasome-dependent catabolite inactivation. We analyzed the involvement of Vid22p and Cpr1p in this process, which have been shown to affect vacuolar FBPase degradation (Brown et al., 2001, 2002) and Vid27p. The participation of Vid27p in vacuolar FBPase degradation is based on a database entry. Our pulse-chase analysis of FBPase degradation under conditions allowing proteasome-dependent inactivation showed wild-type-like kinetics of FBPase breakdown in vid22Δ, vid27Δ, and cpr1Δ cells (Figure 9). Also a deletion of Ssa2p, which was shown to affect vacuolar FBPase degradation (Brown et al., 2000), did not alter proteasome-dependent degradation of FBPase (our unpublished data). This suggests, that catabolite inactivation of FBPase might be mediated by two distinct pathways, which share some components such as Gid1p/Vid30p, Gid4p/Vid24p, and Gid5p/Vid28p. In contrast to Chiang et al. we were unable to find glucose triggered vacuolar degradation of FBPase (Wolf and Ehmann, 1979; Mechler and Wolf, 1981; Teichert et al., 1989; Schork et al., 1994a;b; Schüle et al., 2000). Because we discovered the participation of some VID-genes in proteasome-linked degradation of FBPase, we repeated catabolite degradation studies of the enzyme in mutants defective in VID22 under our conditions and under conditions used by the laboratories of Chiang (Hoffman and Chiang, 1996) and Broach (Jiang et al., 1998). No stabilization of FBPase was observed under both conditions in the vid22 mutant (our unpublished data; Figure 9B). These negative results prevented us from determining the influence of Gid-proteins on vacuolar-dependent FBPase degradation. It is therefore still unclear under which conditions the cells activate the vacuolar-dependent FBPase degradation pathway. The glucose signaling mechanisms for the vacuolar and the proteasomal catabolite degradation pathways of FBPase must be different. In fact, although vacuolar degradation of FBPase was reported to be dependent on the known cAMP-triggered protein kinase A cascade and FBPase phosphorylation (Jiang et al., 1998), the proteasome-dependent degradation pathway of FBPase is not (Hämmerle et al., 1998; Horak et al., 2002). Instead, other signal transduction components such as Grr1p and Reg1p are necessary for proteasomal FBPase degradation (Horak et al., 2002). These same signaling components are also necessary for endocytotic internalization and vacuolar catabolite degradation of the plasma membrane localized galactose transporter Gal2p (Horak et al., 2002). We therefore tested whether the GID genes are also involved in Gal2p proteolysis. Following the fate of Gal2p via immunoblotting in all gid-mutants, we observed wild-type–like degradation (our unpublished data). These data suggest a specific involvement of Gid-proteins in the catabolite inactivation of FBPase.
Our experiments (Figures 5–7) suggest that biologically active Gid2-Ha3 expressed from the chromosome with its native promoter is part of a soluble, cytosolic protein complex with a molecular mass of at least 600 kDa. We did not find any indication for membrane association of Gid2-Ha3, even though the sequence of Gid2p suggests the presence of a putative transmembrane domain. Gid2p is a necessary component for ubiquitination of FBPase to occur. Under the conditions used to induce proteasome-dependent catabolite inactivation of FBPase, we did not find any hint for the involvement of vesicular intermediates in the process so far.
Our genetic analysis for proteasome-dependent catabolite degradation of FBPase identified eight novel proteins required for this process. These novel Gid proteins are good candidates for being components of the high-molecular-mass complex in which Gid2p resides (Figures 5 and 6). Indeed, during the preparation of this article, a systematic mass spectrometry search for protein complexes by using Gid7p and Ybl049wp as bait proteins revealed that Gid2p may be part of a larger protein complex (Ho et al., 2002). Proteins interacting with both Gid7p and Ybl049wp were Gid1p/Vid30p, Gid2p, Gid5p/Vid28p, Gid7p, Gid8p, and Gid9p (Ho et al., 2002); furthermore, Vid24p/Gid4p was detected as an interactor of Gid7p. No function for most of these proteins has yet been suggested. The findings could imply the existence of a protein complex consisting of eight individual proteins, seven of which we have identified to be essential for proteasome-dependent catabolite degradation of FBPase. Because the study of Ho et al. (2002) also identified Ybl049wp as a putative member of this protein complex, we tested the fate of FBPase in YBL049w-deleted cells by pulse-chase analysis (Figure 8B). Surprisingly, under our conditions wild-type-like degradation of FBPase occurred. This might indicate that, with interchanging components, the “Gid-protein complex” may also be involved in functions distinct from proteasome-dependent catabolite degradation of FBPase.
A motif search identified the presence of six WD40 repeats in Gid7p (between amino acids 316–351, 363–399, 448–487, 608–643, 651–687, and 693–741). No common function is attributed to WD40 repeat proteins. Rather, it is thought that their common β-propeller fold formed by the WD40 repeats acts as a platform, allowing sequential and simultaneous interactions with several proteins (Smith et al., 1999). One might therefore speculate that Gid7p plays such a role in the proposed Gid–protein complex.
Interestingly, the previously mentioned LisH domain is present in Gid1p and in Gid8p. The occurrence of this motif is thought to be linked to regulation of microtubule dynamics (Smith et al., 2000), but no clear function can actually be deduced from its presence. C-terminal to the LisH domain the α-helical CTLH domain of unknown function is also present in some proteins. Most interestingly, in the yeast genome the CTLH domain is present only in four proteins: Gid1p, Gid2p, Gid8p, and Gid9p. Our study attributes function of these CTLH domain-containing proteins in proteasome-dependent glucose-induced catabolite degradation of FBPase. This is especially interesting because >90 proteins from different organisms ranging from Arabidopsis to human are known to also contain this domain. None of our newly discovered Gid proteins contain a signature sequence found in ubiquitin-protein ligases (E3s). Whether one of the Gid proteins or part of the complex represents a new type of E3 is unknown.
Further studies will be necessary to determine the precise molecular composition, the topology and the dynamics of the putative “Gid–protein complex.” Finally, the function(s) of the entire complex will have to be elucidated.
ACKNOWLEDGMENTS
We thank Michael N. Hall for providing yeast strains. The expert help of E. Tosta with preparation of this article is acknowledged. This work was supported by the “Deutsche Forschungsgemeinschaft”, SFB 495, and the “Fonds der Chemischen Industrie”. J.H. was supported by grants 204/01/0272 and 204/02/1240 from the grant agency of the Czech Republic and by Institutional Research Concept No. AVOZ 5011922.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0456. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0456.
REFERENCES
- Abeijon C, Orlean P, Robbins PW, Hirschberg CB. Topography of glycosylation in yeast: characterization of GDPmannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc Natl Acad Sci USA. 1989;86:6935–6939. doi: 10.1073/pnas.86.18.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amerik A, Swaminathan S, Krantz BA, Wilkinson KD, Hochstrasser M. In vivo disassembly of free polyubiquitin chains by yeast Ubp14 modulates rates of protein degradation by the proteasome. EMBO J. 1997;16:4826–4838. doi: 10.1093/emboj/16.16.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Kingston RE, Seidman FG, Struhl K, Moore DD, Brent R, Smith FA. Current Protocols in Molecular Biology. New York: John Wiley & Sons; 1992. [Google Scholar]
- Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234:179–86. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- Barth H, Thumm M. A genomic screen identifies AUT8 as a novel gene essential for autophagy in the yeast Saccharomyces cerevisiae. Gene. 2001;274:151–156. doi: 10.1016/s0378-1119(01)00614-x. [DOI] [PubMed] [Google Scholar]
- Baumeister W, Walz J, Zuhl F, Seemüller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose-1,6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Cui DY, Hung GG, Chiang HL. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J Biol Chem. 2001;276:48017–48026. doi: 10.1074/jbc.M109222200. [DOI] [PubMed] [Google Scholar]
- Brown CR, McCann JA, Hung GG, Elco CP, Chiang HL. Vid22p, a novel plasma membrane protein, is required for the fructose-1,6-bisphosphatase degradation pathway. J Cell Sci. 2002;115:655–666. doi: 10.1242/jcs.115.3.655. [DOI] [PubMed] [Google Scholar]
- Chiang HL, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Chiang HL. Vid24p, a novel protein localized to the fructose-1, 6-bisphosphatase-containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998;140:1347–1356. doi: 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottarel G. The Saccharomyces cerevisiae HIS3 and LYS2 genes complement the Schizosaccharomyces pombe his5-303 and lys1-131 mutations, respectively: new selectable markers and new multi-purpose multicopy shuttle vectors, pSP3 and pSP4. Curr Genet. 1995;28:380–383. doi: 10.1007/BF00326437. [DOI] [PubMed] [Google Scholar]
- Egner R, Thumm M, Straub M, Simeon A, Schüller HJ, Wolf DH. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J Biol Chem. 1993;268:27269–27276. [PubMed] [Google Scholar]
- Ellison MJ, Hochstrasser M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- Funayama S, Gancedo JM, Gancedo C. Turnover of yeast fructose-bisphosphatase in different metabolic conditions. Eur J Biochem. 1980;109:61–66. doi: 10.1111/j.1432-1033.1980.tb04767.x. [DOI] [PubMed] [Google Scholar]
- Gancedo C. Inactivation of fructose-1,6-diphosphatase by glucose in yeast. J Bacteriol. 1971;107:401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. Vol. 194. New York: Academic Press; 1991. [Google Scholar]
- Hämmerle M, Bauer J, Rose M, Szallies A, Thumm M, Dusterhus S, Mecke D, Entian KD, Wolf DH. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome-catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:25000–25005. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Proteasomes: destruction as a program. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- Hilt W, Wolf DH. Proteasomes. The World of Regulatory Proteolysis. Georgetown, VA: Landes Bioscience; 2000. pp. 1–387. [Google Scholar]
- Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hoffman M, Chiang HL. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–1566. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzer H. Catabolite inactivation in yeast. Trends Biochem Sci. 1976;1:178–181. [Google Scholar]
- Horak J, Regelmann J, Wolf DH. Two distinct proteolytic systems responsible for glucose-induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem. 2002;277:8248–8254. doi: 10.1074/jbc.M107255200. [DOI] [PubMed] [Google Scholar]
- Horak J, Wolf DH. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak J, Wolf DH. Glucose-induced monoubiquitination of the Saccharomyces cerevisiae galactose transporter is sufficient to signal its internalization. J Bacteriol. 2001;183:3083–3088. doi: 10.1128/JB.183.10.3083-3088.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PH, Chiang HL. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Davis C, Broach JR. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 1998;17:6942–6951. doi: 10.1093/emboj/17.23.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Scott SV, Oda MN, Klionsky DJ. Transport of a large oligomeric protein by the cytoplasm to vacuole protein targeting pathway. J Cell Biol. 1997;137:609–618. doi: 10.1083/jcb.137.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechler B, Wolf DH. Analysis of proteinase A function in yeast. Eur J Biochem. 1981;121:47–52. doi: 10.1111/j.1432-1033.1981.tb06427.x. [DOI] [PubMed] [Google Scholar]
- Richter-Ruoff B, Heinemeyer W, Wolf DH. The proteasome/multicatalytic-multifunctional proteinase. In vivo function in the ubiquitin-dependent N-end rule pathway of protein degradation in eukaryotes. FEBS Lett. 1992;302:192–196. doi: 10.1016/0014-5793(92)80438-m. [DOI] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Schork S, Bee G, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 1994a;349:270–274. doi: 10.1016/0014-5793(94)00668-7. [DOI] [PubMed] [Google Scholar]
- Schork S, Bee G, Thumm M, Wolf DH. Site of catabolite inactivation. Nature. 1994b;369:283–284. doi: 10.1038/369283a0. [DOI] [PubMed] [Google Scholar]
- Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation occurs via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- Schüle T, Rose M, Entian KD, Thumm M, Wolf DH. Ubc8p functions in catabolite degradation of fructose-1,6-bisphosphatase in yeast. EMBO J. 2000;19:2161–2167. doi: 10.1093/emboj/19.10.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W, Jentsch S. In vivo function of the proteasome in the ubiquitin pathway. EMBO J. 1992;11:3077–3080. doi: 10.1002/j.1460-2075.1992.tb05379.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DS, Niethammer M, Ayala R, Zhou Y, Gambello MJ, Wynshaw-Boris A, Tsai LH. Regulation of cytoplasmic dynein behavior and microtubule organization by mammalian Lis1. Nat Cell Biol. 2000;2:767–75. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Teichert U, Mechler B, Müller H, Wolf DH. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J Biol Chem. 1989;264:16037–16045. [PubMed] [Google Scholar]
- van der Merwe GK, Cooper TG, van Vuuren HJ. Ammonia regulates VID30 expression and Vid30p function shifts nitrogen metabolism toward glutamate formation especially when Saccharomyces cerevisiae is grown in low concentrations of ammonia. J Biol Chem. 2001;276:28659–28666. doi: 10.1074/jbc.M102280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Ehmann C. Studies on a proteinase B mutant of yeast. Eur J Biochem. 1979;98:375–384. doi: 10.1111/j.1432-1033.1979.tb13197.x. [DOI] [PubMed] [Google Scholar]