Abstract
Oxidative stress has been implicated in many diseases. The chief source of reactive oxygen species within the cell is the mitochondrion. We have characterized a variety of the biochemical and metabolic effects of inactivation of the mouse gene for the mitochondrial superoxide dismutase (CD1-Sod2tm1Cje). The Sod2 mutant mice exhibit a tissue-specific inhibition of the respiratory chain enzymes NADH-dehydrogenase (complex I) and succinate dehydrogenase (complex II), inactivation of the tricarboxylic acid cycle enzyme aconitase, development of a urine organic aciduria in conjunction with a partial defect in 3-hydroxy-3-methylglutaryl-CoA lyase, and accumulation of oxidative DNA damage. These results indicate that the increase in mitochondrial reactive oxygen species can result in biochemical aberrations with features reminiscent of mitochondrial myopathy, Friedreich ataxia, and 3-hydroxy-3-methylglutaryl-CoA lyase deficiency.
The pathophysiology of mitochondrial diseases has been attributed to decreased mitochondrial ATP production and toxicity resulting from increased mitochondrial reactive oxygen species (ROS) generation. Mammalian mitochondria generate most of the ATP for cells by the process of oxidative phosphorylation (OXPHOS). During OXPHOS, between 0.4 and 4% of the oxygen consumed is reduced to form superoxide anion (O2⨪) (1–5). Under normal circumstances, superoxide is reduced to H2O2 by the mitochondrial form of superoxide dismutase (SOD2). Within the mitochondria, the H2O2 then either is converted to water by mitochondrial glutathione peroxidase or can participate in Fenton type chemistry, giving rise to further ROS such as the hydroxyl radical (6).
ROS have been implicated in a wide variety of disorders, including Alzheimer disease, Parkinson disease, ischemic heart disease, and many mitochondrial diseases (7–12). Moreover, the pathogenic role of ROS has been strongly implicated in familial amyotrophic lateral sclerosis and Friedreich ataxia. Specifically, certain cases of familial amyotrophic lateral sclerosis have been shown to be caused by mutations in the cytosolic Cu/ZnSOD (13), and the mutant protein in Friedreich ataxia has been associated with high levels of mitochondrial iron and a reduction in mitochondrial complexes I, II, and III and aconitase (7, 9), presumably because of oxidative damage.
Mitochondrial diseases genetically can be classified into two types: disease that result from mutations of the mitochondrial genome (14) and those that are caused by mutations in nuclear genes encoding mitochondrial proteins (15). Recent examples of nuclear mitochondrial disease include Friedreich ataxia, Wilson disease, and spastic paraplegia (7, 16, 17). Nuclear mitochondrial defects can affect many different types of metabolic pathways. In addition to mitochondria being the main source of ATP within the cell, they are also responsible for fatty acid oxidation and are a major source of metabolic intermediates for cytosolic processes.
A number of inherited diseases disrupt metabolic pathways in the mitochondria. One such disease results in a 3-methylglutaconic aciduria (3-MGC) and can be caused by defects in 3-hydroxy-3-methylglutaryl (HMG)-CoA lyase. HMG-CoA lyase is a mitochondrial matrix protein that catalyzes cleavage of HMG-CoA into acetyl-CoA and acetoacetate. This enzyme is a critical enzyme in ketogenesis and is required in the last step of leucine catabolism, and mutations in the HMG-CoA gene have been associated with some forms of 3-MGC aciduria (18, 19). In urine, 3-MGC is detectable by gas chromatography/mass spectrometry (GC/MS), and patients with 3-MGC aciduria show a marked clinical heterogeneity that can encompass cardiomyopathy, bilateral optic atrophy, movement disorders, disorders of the respiratory chain, and hepatomegaly (20–24).
All three genes for superoxide dismutase (Sod1, Sod2, and Sod3) have been inactivated genetically in mice through homologous recombination (25–27). However, inactivation of the mitochondrial gene Sod2 has resulted in the most severe phenotype (25, 28). This indicates that the toxicity of the mitochondrial ROS is particularly deleterious to health. Mice lacking SOD2 have been generated in two genetic backgrounds, an outbred background, CD1 (25), and a hybrid background (28). In both cases, the phenotype is neonatal lethal, with the most severely affected tissues, such as the heart and brain, being postmitotic and having high-energy demands (25, 28, 29). In mice mutant for Sod2 on the CD1 background, dilated cardiomyopathy develops within the first week of life, accompanied by a profound accumulation of fat within the liver. A metabolic acidosis and a dramatic reduction in succinate dehydrogenase (complex II) histochemical staining have been reported (25). To characterize further the biochemical and physiological basis of the lethal effects of the Sod2 mutation, we have extensively analyzed the biochemistry of the mutant mouse mitochondria. We have confirmed that the mice have severe deficiencies in some iron-sulfur cluster-containing enzymes of the respiratory chain and tricarboxylic acid cycle, a partial defect in HMG-CoA lyase, and extensive ROS damage to the DNA. Hence, these animals have many of the biochemical features of mitochondrial disease associated with ROS toxicity.
MATERIALS AND METHODS
Mice Husbandry, Breeding, and Genotyping.
The Sod2 mutant allele was maintained on a CD1 background by backcrossing to CD1 male mice obtained from Charles River Breeding Laboratories. Heterozygous crosses were set up, and genotyping of the resultant pups was carried out at 2–3 days of age as described (29). After mating, pregnant dams were housed singly and were fed a diet of Rodent laboratory chow 5001 (PMI feeds, St. Louis) under standard conditions. All procedures with animals were carried out under Emory University Institutional Animal Use and Care Committee ethical guidelines (Institutional Animal Use and Care Committee 255-97).
Mitochondrial Isolation and OXPHOS Analysis.
Pups of the indicated age and genotypes were killed by decapitation, and the tissues were rapidly removed and placed on ice in H-Buffer (210 mM mannitol/70 mM sucrose/1 mM EGTA/5 mM Hepes/0.5% BSA, pH 7.2) (30). Four to six hearts or hind-limb skeletal muscles from each genotype were pooled and homogenized on ice in 9 volumes of H-buffer by using a 1-ml sintered glass homogenizer (Wheaton Scientific). Before homogenization, the hind-limb muscles were “sliced” into smaller pieces by using a sterile razor blade. For brain, the cerebellum, cortex, striatum, and brainstem were dissected out by using a dissecting microscope, and the tissues were transferred to a cold Eppendorf tube. Homogenization was carried out in the Eppendorf with 9 volumes of H-buffer by using a pellet pestle homogenizer (Kontes). For all tissues, homogenization was carried out for 6–10 strokes or until a uniform color was obtained. The homogenate was transferred into a cold 1.5-ml Eppendorf tube, and the mitochondria were collected by centrifugation at 4°C, 600 × g for 5 min. The supernatant then was transferred to a new, cold 1.5-ml Eppendorf tube and was recentrifuged at 7,200 × g at 4°C for 10 min. The supernatant was aspirated, and the mitochondrial pellet was resuspended in 100 μl of cold H-buffer, was aliquoted, and was snap frozen in liquid nitrogen. Mitochondrial protein was measured by using the Lowry method, and OXPHOS enzyme analyses were carried out as described (30) except that 7.5 μg of mitochondrial protein was used per assay instead of 15 μg. For OXPHOS analysis, each data point represents enzyme results from 4–6 pooled animal tissues, with ≈45 Sod2 mutant and 55 control animals being assayed for mitochondrial OXPHOS.
Statistical Analysis.
All comparisons between groups were carried out by using a Mann–Whitney nonparametric t test in the program prism (GraphPad, San Diego). In addition, all comparisons between Sod2 mutant mice and controls were coded, and analysis was carried out without knowledge of genotype. (In the figures, ∗, 0.05 > P > 0.01; ∗∗, 0.01 > P > 0.001; and ∗∗∗, P < 0.001.) After analysis, the coding was broken, and the results were plotted by using prism.
Mitochondrial Aconitase and Fumarase Activity Assays.
Mitochondria were isolated as described for OXPHOS enzyme analysis from pups of the indicated age and genotype, and mitochondrial aconitase and fumarase measurements were measured spectrophotometrically. This was carried out by monitoring the formation of cis-aconitate from isocitrate at 240 nm in 50 mM Tris⋅HCl (pH 7.4) containing 0.6 mM MnCl2 and 20 mM isocitrate at 25°C (31). Fumarase activity in mitochondrial fractions was measured by monitoring the increase in absorbance at 240 nm at 25°C in a 1-ml reaction mixture containing 30 mM potassium phosphate (pH 7.4), 0.1 mM EDTA, and 5 mM l-malate (32). Protein concentrations were determined by using Coomassie Plus reagents (Pierce).
Organic Acids Analysis and Mitochondrial HMG-CoA Lyase Assessment.
Urine was collected daily from Sod2 mutant mice, as well as from wild-type or heterozygous littermates at 3 days of age. Because a minimum of ≈0.5 ml of urine is required for urine organic acids analysis, we pooled urine from each individual animal up until the day of sacrifice in a 1.5-ml Eppendorf tube. Urine was stored at −20°C between daily collections. Organic acids in urine were analyzed by GC/MS as described (33), except that dimethylmalonic acid was used as an internal standard instead of malonic, and chromatography was carried out on a 30-m column of DB-17 (J & W Scientific, Folsom, CA).
For HMG-CoA lyase activity, mitochondria were isolated from liver by homogenization and differential centrifugation as described above. A 15-μl sample of mitochondria suspended in H-buffer was diluted into 300-μl of cold 20 mM sodium phosphate (pH 7.2) containing 1 mM EDTA and 1 mM DTT. This sample was sonicated twice (15-second periods), and the resulting lysate was centrifuged at 12,000 × g for 30 min (4°C). The supernatant was used for spectrophotometric assays of HMG-CoA lyase, which is coupled to NADH production (340 nm). A 1-ml reaction mixture contained 0.2 M Tris⋅Cl (pH 8.2), 2.5 mM malate, 1.5 mM NAD+, 10 mM MgCl2, 5 mM DTT, 0.05 mM NADH, 2 units of malate dehydrogenase, 2 units of citrate synthase, and an aliquot of the soluble mitochondrial extract. The background rate of change in 340 nm absorbance was measured for 6 min before initiation of the reaction by addition of HMG-CoA (100 μM final concentration). A linear increase in 340-nm absorbance ensued because each cleavage of HMG-CoA resulted in production of one NADH. Activity at 340 nm (units per milliliter extract) was calculated by using a millimolar extinction coefficient for NADH = 6.2. Protein concentrations of the extracts were determined by using the Bradford method (Bio-Rad reagent).
Oxidative Damage Assessment.
Total DNA was extracted from pups of the indicated age and genotype by using the Puregene DNA extraction kit (Gentra Systems). The concentration of DNA was measured by either UV absorbance at 260 nM or by MS. Modified DNA bases, their stable isotope-labeled analogues, and other materials for GC/isotope-dilution MS (GC/IDMS) were obtained as described (34). Labeled 2′-deoxyguanosine (2′-deoxyguanosine-15N5) was purchased from Cambridge Isotope Laboratories (Cambridge, MA).
Aliquots of stable isotope-labeled analogues of DNA base products and 2′-deoxyguanosine-15N5 were added as internal standards to DNA samples containing ≈50 μg of DNA. The samples were dried under vacuum in a SpeedVac. The use of 2′-deoxyguanosine-15N5 permits the determination of the DNA amount by GC/IDMS. On hydrolysis, this compound yields guanine-15N5, which is used as an internal standard for the determination of the guanine amount in DNA and, consequently, the amount of DNA (35). DNA samples were hydrolyzed with 0.5 ml of 60% formic acid in evacuated and sealed tubes at 140°C for 30 min. The hydrolyzes were frozen in vials placed in liquid nitrogen and then were lyophilized for 18 h. DNA hydrolysates were derivatized and analyzed by GC/IDMS with selected-ion monitoring as described (36).
RESULTS
Respiratory Chain Deficiencies in Heart and Skeletal Muscle.
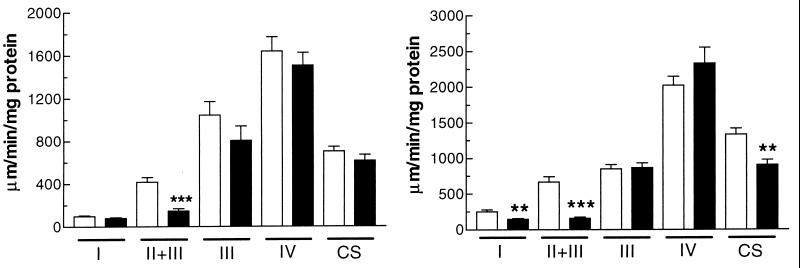
It was reported that Sod2 mutant mice on a CD1 background showed a marked deficiency in complex II, as detected by histochemical staining of frozen skeletal muscle and heart (25). Given this decrease in complex II activity, we wished to determine whether this and other components of the respiratory chain enzymes were catalytically impaired in heart and skeletal muscle. Using mitochondria isolated from 4- to 6-day-old mouse heart and hind-limb skeletal muscle from homozygous Sod2 mutant mice, heterozygotes, and wild-type mice, we observed a marked reduction in succinate dehydrogenase [succinate dehydrogenase (SDH) complex II+III] activity in both skeletal muscle (35% of control) and heart (24% of control) mitochondria, confirming our previous histochemical results (Fig. 1). In heart mitochondria, we also observed a complex I deficiency (59% of control) (Fig. 1b) and a citrate synthase deficiency (32% of control). These differences are all highly statistically significant (Fig. 1). We did not observe a significant complex I or citrate synthase deficiency in skeletal muscle nor significant differences in complexes III or IV for either tissue relative to controls.
Figure 1.
OXPHOS analysis of 4- to 6-day-old Sod2−/− mice vs. controls. Open bars, Sod2+/+ and -±; closed bars, Sod2−/−. Complexes I, II+III, III, and IV and citrate synthase (CS) are shown for mitochondria isolated from skeletal muscle and heart. SEMs are shown. n = 11 for controls; n = 9 for Sod2−/− mice. (a) OXPHOS enzymology from skeletal muscle. (b) OXPHOS enzymology from heart.
Mitochondrial Aconitase Deficiencies in Brain and Heart.
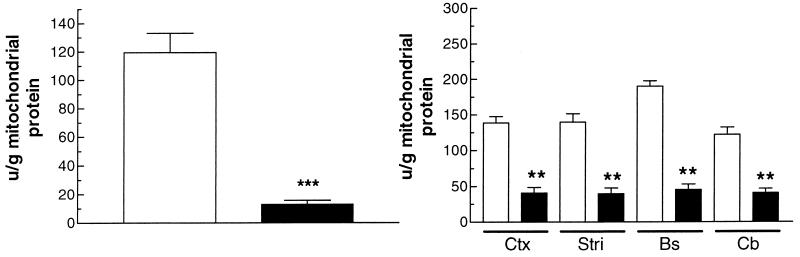
Aconitase, a mitochondrial enzyme, has been reported to be a highly sensitive to ROS inactivation (37–40). It also was reported that the total tissue aconitase activity of Sod2 mutant mice was reduced 22–43% (25). However, there are two aconitase isoforms, one mitochondrial and the other cytosolic, and previous studies did not distinguish between the two. We therefore assayed aconitase from mitochondria isolated from various tissues. To control for an overall reduction in tricarboxylic acid cycle enzymes by oxidative damage, we also assayed for fumarase activity. Mammalian fumarase is insensitive to O2⨪ (40, 41). In heart mitochondria isolated from 4- to 5-day-old Sod2−/− mice, we observed an 89% reduction in the aconitase activity, which is highly statistically significant (Fig. 2a). We also observed highly significant 67–76% reductions in aconitase activities in mitochondria isolated from the brainstem, cortex, striatum, and cerebellum of brains from 9- to 10-day-old Sod2−/− mice (Fig. 2b). By contrast, we found no significant reductions in the levels of mitochondrial fumarase activity in Sod2−/− mice (data not shown). Hence, the ROS resulting from SOD2 inactivation appears to have specifically inactivated the mitochondrial aconitase.
Figure 2.
Mitochondrial aconitase activity in isolated mitochondria from heart and different regions of the brain. Open bars, Sod2+/+ and -±; closed bars, Sod2−/−. (a) Mitochondrial aconitase activity in hearts of 4- to 5-day-old mice. n = 13 for control hearts; n = 9 for Sod2−/− hearts. (b) Mitochondrial aconitase activity of brain in 9- to 10-day-old mice. Ctx, Cortex; Str, striatum; Bs, brainstem; Cb, cerebellum. SEMs are shown. n = 5 for controls; n = 6 for Sod2−/−.
Organic Acid Abnormalities in Urine and HMG-CoA Lyase Deficiency in Liver.
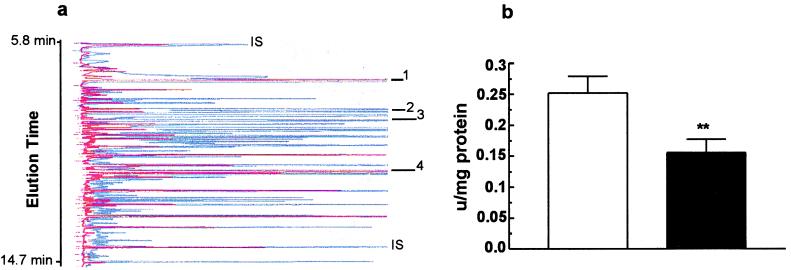
To characterize further the metabolic defects of the Sod2−/− mice, we analyzed the urine organic acids by GC/MS of four individual Sod2−/− mice aged 9, 10, 14, and 15 days old and compared them to age matched controls. The oldest two animals were from the 12% of animals surviving to this age (29). Large amounts of 3-methylglutaconic, 2-hydroxyglutaric, 3-hydroxy-3 methylglutaric, and 3-hydroxyisovaleric acids were found in the urine of the Sod2−/− mice and not in controls. The internal standard of dimethylmalonic acid added to each sample based on urine creatinine makes these analyses semiquantitative. Normalization to the internal standard revealed a relative increase of each metabolite in the two older animals (14 and 15 days old) over the younger animals (9 and 10 days old), indicating a probable worsening of the metabolic defect with age. The urine organic acid profile in the mutant mice suggested a defect in HMG-CoA lyase (18, 42). Consistent with this deduction, the assay of this enzyme in liver mitochondria revealed a statistically significant 36% decrease in HMG-CoA lyase activity relative to controls (Fig. 3b).
Figure 3.
Organic aciduria and HMG-CoA lyase deficiency in Sod2−/− mice. (a) Representative gas chromatogram of a 9-day-old Sod2−/− mouse urine (blue) superimposed on a chromatogram from a 9-day-old control (red). Two of the abnormal metabolites associated with HMG-CoA lyase deficiency are indicated (peaks 2 and 3), and the increased levels of succinic and citric acids are shown (peaks 1 and 4). IS, internal standards; peak 1, succinic acid; peak 2, 3-OH-3-methyl glutaric acid; peak 3, 3-methyl glutaconic acid; peak 4, citric acid. Peak identities were confirmed by MS. (b) HMG-CoA lyase activity in isolated mitochondria from livers of 9- to 10-day-old Sod2−/− mice. n = 5 for Sod2−/− animals (closed bars); n = 6 for wild-type animals (open bars).
Oxidative Damage in Total Genomic DNA.
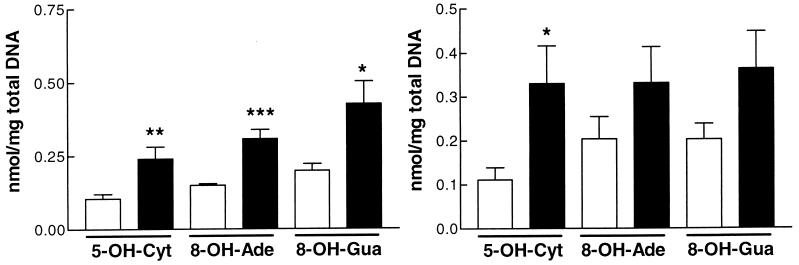
The high levels of mitochondrial ROS caused by the SOD2 deficiency would be expected to damage DNA. To detect oxidative damage to DNA, we used GC/IDMS, which can detect >20 different types of oxidative DNA lesions (34). Total DNA was extracted from heart, liver, and brain of 4- to 6-day-old Sod2−/− mice and controls, was hydrolyzed, and was analyzed by GC/IDMS. This revealed 215–300% increases of 8-OH-guanine, 8-OH-adenine, and 5-OH-cytosine in heart, which were highly significant over age matched controls (Fig. 4a). Similar increases were seen in brain DNA, although only 5-OH-cytosine levels were statistically increased (Fig. 4b). By contrast, no difference was observed in the liver DNA between Sod2−/− mice and controls (data not shown).
Figure 4.
Levels of modified DNA bases in total DNA from brain and hearts of Sod2−/− mice. Open bars, Sod2+/+ and ±; closed bars, Sod2−/−. (a) Modified bases in heart of Sod2−/− mice. n = 6 for control and mutant brains. (b) Modified bases in brain of Sod2−/− mice. n = 6 for controls; n = 7 for mutants.
DISCUSSION
Oxidative stress often has been hypothesized to be the chief etiological factor in the pathogenesis of a variety of diseases as well as in aging (12, 43–49). Because the mitochondria are a major source of ROS and the mitochondria play a central role in energy metabolism, intermediary metabolism, and apoptosis (50), it has been hypothesized that mitochondrially generated ROS are a major cause of degenerative diseases (51). This hypothesis is strongly supported by our results on mice lacking the mitochondrial SOD, which have shown severe effects on the heart and liver, the inactivation of key respiratory chain and tricarboxylic acid cycle enzymes, inhibition of HMG-CoA lyase, and the accumulation of oxidative DNA damage.
The heart has a high requirement for ATP, generated by the oxidation of organic and fatty acids. The extraordinary rate of development of the dilated cardiomyopathy in the Sod2−/− mutant mice may be attributable to the substantial increase in work load that occurs in the neonatal heart at ≈3 days of age (52). This increase in oxidative energy to do work implies a dramatic increase in the flux of electrons through the respiratory chain, thereby causing an increase in mitochondrial ROS. A transgenic mouse model of ATP deficiency in the heart has been shown to cause hypertrophic cardiomyopathy (53). In humans, both dilated and hypertrophic cardiomyopathies have been associated with defects of the respiratory chain, aconitase inactivation, and 3-MGC aciduria (21, 22, 54–58). The respiratory chain defects in the Sod2−/− mice, in conjunction with the dilated cardiomyopathy, aconitase deficiency, and organic aciduria argue that mitochondrial ROS may give rise to a number of human diseases of diverse presentation.
The mitochondrial complexes I and II and aconitase defects observed in the Sod2−/− mice are consistent with ROS inactivation of iron–sulfur cluster-containing enzymes. These enzymological abnormalities are consistent with the biochemical defects observed in Friedreich ataxia, which also have been attributed to ROS mediated inactivation of iron–sulfur clusters (7).
Complex I consists of at least 43 polypeptides encompassing an FMN and at least six iron-sulfur clusters (59, 60). These iron–sulfur clusters also may be subject to superoxide-mediated inactivation, consistent with the decrease in complex I activity in mitochondria from hearts of the Sod2−/− mice. The lack of reduction of complex I in skeletal muscle may reflect tissue-specific differences in enzyme subunit structure or function or, alternatively, metabolic rate and, hence, levels of superoxide and the subsequent oxidative damage.
Complex II (succinate dehydrogenase) consists of four main polypeptides encompassing FAD, three iron-sulfur centers, and a cytochrome b (61). The two “anchor” polypeptides encompass a cytochrome b and interact with ubiquinone whereas the large peripheral membrane proteins are the FAD-associated flavoprotein and the iron–sulfur cluster-containing protein (Ip). Ip contains three iron–sulfur clusters, only one of which is a tetranuclear (4Fe-4S) cluster (61). The decrease in SDH activity in Sod2−/− mice may be caused by oxidation of the 4Fe-4S cluster, although, because the structure of complex II is not yet known, it is uncertain whether the cluster is accessible to oxidation within the membrane (62). Alternatively, SDH expression might be inhibited at the transcriptional or post-translational level because of oxidative damage.
The inactivation of aconitase in the SOD2-deficient mice is likely to be caused by oxidation of its 4Fe-4S cluster by superoxide, which liberates the iron as Fe2+ (62). This iron then can participate in the Fenton reaction, generating hydroxyl radical and leading to further oxidative damage. A decrease in aconitase activity also is seen in Friedreich ataxia (7); therefore, the inactivation of aconitase in the heart and brain of SOD2-deficient mice is consistent with ROS-mediated inactivation of iron–sulfur centers.
The mRNAs for the Ip subunit of SDH in Drosophila melanogaster and the mitochondrial aconitases contain iron-regulatory elements (IRE) in the 5′ untranslated regions (63, 64). Translational control of messages containing an IRE can occur when iron-responsive proteins bind with high affinity to the IRE (65). Iron-responsive protein 1 is a dual function protein: when lacking its 4Fe-4S cluster, it functions as an iron-responsive protein, and, when iron is replete in the cell, it functions as cytoplasmic aconitase. One mechanism for removal of the tetranuclear Fe-S cluster from iron-responsive protein 1 is via oxidative stress (66–68). Although the message for the Ip of SDH in mammals lacks an IRE in its 5′ UTR (69), there may be other elements within the message that function in a similar manner to the IRE in mitochondrial aconitase. Such elements could negatively regulate the expression of the Ip subunit in response to oxidative stress. This may cause a decrease in the translation of the Ip, thereby resulting in a reduction in activity of SDH in the Sod2−/− mice.
The elevated levels of citrate and succinate in the urine of the Sod2−/− mice are consistent with the preferential inactivation of aconitase and succinate dehydrogenase. The reason for the reduction of heart citrate synthase, which condenses acetyl-CoA and oxaloacetate into citrate in the tricarboxylic acid cycle, is unclear. The metabolic profile is consistent with the organic aciduria in human patients with a near complete deficiency of HMG-CoA lyase. The mechanism by which the HMG-CoA lyase would be inhibited by ROS is unclear because this enzyme lacks iron–sulfur clusters found in complex II and aconitase. However, HMG-CoA lyase exists as a homodimer of 31-kDa subunits and requires that a C-terminal cysteine (Cys-323) be in a reduced state for optimal activity in vitro (42, 70). Hence, in vivo oxidative stress may partially inactivate HMG-CoA lyase by oxidizing Cys-323, thus contributing to the organic aciduria seen in the Sod2−/− mice.
The high levels of oxidative damage to DNA are also consistent with elevated ROS production. DNA is subject to a wide variety of base modifications resulting from ROS damage (71). Because of difficulty in isolating mtDNA from animals of this age, we were unable to determine whether the marked increases in modified bases were caused by nuclear DNA damage, mtDNA damage, or both. However, it seems likely that the mtDNA is a major target for oxidative damage because of its close physical proximity to the site of production of superoxide. It is unlikely that the nuclear DNA is a primary target because superoxide formed inside the mitochondria will not pass through the mitochondrial membranes and, hence, ROS damage may be contained largely within the mitochondria. These inferences are supported by the finding that there is an accumulation of 8-OH-guanine in the liver of mtDNA of Sod2± mice on a C57BL/6 background at 2–4 months of age (72).
The significant increase in 5-OH-cytosine in both heart and brain is likely to be important. This lesion has been reported to be extremely mutagenic, with predominantly C-T transitions resulting at a frequency of 2.5% (73). Of interest, despite substantial pathology (25), the liver displayed no statistically significant increases or trends in any DNA lesions detectable by GC/IDMS. This could be because of the level of damage being below our level of detection or, alternatively, the metabolic state of the tissue and, hence, level of mitochondrial ROS being produced.
The Sod2−/− mutant mice are the first model of mitochondrial disease based on increased mitochondrial ROS generation. This model shows that primary effects are the inactivation of some enzymes containing Fe-S centers, such as complexes I and II and mitochondrial aconitase. However, mitochondrial ROS may inactivate other enzymes with redox sensitive subunits such as HMG-CoA lyase. Whatever the final array of enzymatic targets, it is clear that the final common pathway involves a presumed inhibition of the mitochondrial capacity to make ATP. Hence, the hypertrophic cardiomyopathy of the adenine nucleotide translocater 1-deficient mouse (53) and the dilated cardiomyopathy of the Sod2−/− mice (25) appear to be the product of chronic and acute mitochondrial energetic failure, respectively.
Sod2−/− mice also have been shown to be a powerful tool for demonstrating the efficacy of antioxidants in treating endogenous mitochondrial ROS (29). Both the cardiomyopathy and the accumulation of lipid in the liver of Sod2−/− mice are rescued effectively by treatment of these animals with the synthetic antioxidant MnTBAP (29). By combining antioxidant therapeutics along with biochemical and genetic approaches, Sod2−/− mutant mice may uncover many new insights into the role of free radicals in mitochondrial disease.
Acknowledgments
We thank Dr. Brian Ackrell for helpful comments on the manuscript. This work was supported by National Institutes of Health Grants AG13154, HL45572, and NS21328 (to D.C.W.); DK21491 (to H.M.M.); HD04024 (to S.I.G.); and AG08938 and AG16998 (to C.J.E.), the Alzheimer’s Investigator Award for M.P., and the Johnson & Johnson Focused Giving Grant for D.C.W. Certain commercial equipment or materials are identified in this paper to specify adequately the experimental procedure. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
ABBREVIATIONS
- ROS
reactive oxygen species
- OXPHOS
oxidative phosphorylation
- SOD
superoxide dismutase
- 3-MGC
3-methylglutaconic aciduria
- HMG
3-hydroxy-3-methylglutaryl
- GC
gas chromatography
- MS
mass spectrometry
- IDMS
isotope-dilution MS
- Ip
iron–sulfur cluster-containing protein
- SDH
succinate dehydrogenase
- IRE
iron-regulatory elements
References
- 1.Boveris A. Methods Enzymol. 1984;105:429–435. doi: 10.1016/s0076-6879(84)05060-6. [DOI] [PubMed] [Google Scholar]
- 2.Chance B, Sies H, Boveris A. Physiol Rev. 1979;59:527–605. doi: 10.1152/physrev.1979.59.3.527. [DOI] [PubMed] [Google Scholar]
- 3.Turrens J F, Boveris A. Biochem J. 1980;191:421–427. doi: 10.1042/bj1910421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turrens J F, Alexandre A, Lehninger A L. Arch Biochem Biophys. 1985;237:408–414. doi: 10.1016/0003-9861(85)90293-0. [DOI] [PubMed] [Google Scholar]
- 5.Hansford R G, Hogue B A, Mildaziene V. J Bioenerg Biomembr. 1997;29:89–95. doi: 10.1023/a:1022420007908. [DOI] [PubMed] [Google Scholar]
- 6.Giulivi C, Boveris A, Cadenas E. Arch Biochem Biophys. 1995;316:909–916. doi: 10.1006/abbi.1995.1122. [DOI] [PubMed] [Google Scholar]
- 7.Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, Sidi D, Munnich A, Rustin P. Nat Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 8.Schapira A H V. Biochim Biophys Acta. 1998;1366:225–233. doi: 10.1016/s0005-2728(98)00115-7. [DOI] [PubMed] [Google Scholar]
- 9.Pandolfo M. Neuromuscul Disord. 1998;8:409–415. doi: 10.1016/s0960-8966(98)00039-x. [DOI] [PubMed] [Google Scholar]
- 10.Beal M F. Biochim Biophys Acta. 1998;1366:211–223. doi: 10.1016/s0005-2728(98)00114-5. [DOI] [PubMed] [Google Scholar]
- 11.Robinson B H. Biochim Biophys Acta. 1998;1364:271–286. doi: 10.1016/s0005-2728(98)00033-4. [DOI] [PubMed] [Google Scholar]
- 12.Chandra M, Chandra N, Agrawal R, Kumar A, Ghatak A, Pandey V C. Int J Cardiol. 1994;43:121–125. doi: 10.1016/0167-5273(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 13.Rosen D R, Siddique T, Patterson D, Figlewicz D A, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan J P, Deng H X, et al. Nature (London) 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 14.Wallace D C. J Bioenerg Biomembr. 1994;26:241–250. doi: 10.1007/BF00763096. [DOI] [PubMed] [Google Scholar]
- 15.DiMauro S, Schon E A. Nat Genet. 1998;19:214–215. doi: 10.1038/883. [DOI] [PubMed] [Google Scholar]
- 16.Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, De Michele G, Filla A, Cocozza S, Marconi R, et al. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- 17.Lutsenko S, Cooper M J. Proc Natl Acad Sci USA. 1998;95:6004–6009. doi: 10.1073/pnas.95.11.6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell G A, Ozand P T, Robert M F, Ashmarina L, Roberts J, Gibson K M, Wanders R J, Wang S, Chevalier I, Plochl E, et al. Am J Hum Genet. 1998;62:295–300. doi: 10.1086/301730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scriver C R, Beaudet A L, Sly W S, Valle D. The Metabolic and Molecular Bases of Inherited Disease. New York: McGraw–Hill; 1995. [Google Scholar]
- 20.Gibson K M, Elpeleg O N, Jakobs C, Costeff H, Kelley R I. Pediatr Neurol. 1993;9:120–123. doi: 10.1016/0887-8994(93)90046-f. [DOI] [PubMed] [Google Scholar]
- 21.Figarella-Branger D, Pellissier J F, Scheiner C, Wernert F, Desnuelle C. J Neurol Sci. 1992;108:105–113. doi: 10.1016/0022-510x(92)90195-q. [DOI] [PubMed] [Google Scholar]
- 22.Draaisma J M, van Kesteren I C, Daniels O, Sengers R C. Pediatr Cardiol. 1994;15:89–90. doi: 10.1007/BF00817615. [DOI] [PubMed] [Google Scholar]
- 23.Gibson K M, Bennett M J, Mize C E, Jakobs C, Rotig A, Munnich A, Lichter-Konecki U, Trefz F K. J Pediatr. 1992;121:940–942. doi: 10.1016/s0022-3476(05)80348-8. [DOI] [PubMed] [Google Scholar]
- 24.Ibel H, Endres W, Hadorn H B, Deufel T, Paetzke I, Duran M, Kennaway N G, Gibson K M. Eur J Pediatr. 1993;152:665–670. doi: 10.1007/BF01955244. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Huang T T, Carlson E J, Melov S, Ursell P C, Olson J L, Noble L J, Yoshimura M P, Berger C, Chan P H, et al. Nat Genet. 1995;11:376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 26.Carlsson L M, Jonsson J, Edlund T, Marklund S L. Proc Natl Acad Sci USA. 1995;92:6264–6268. doi: 10.1073/pnas.92.14.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reaume A G, Elliott J L, Hoffman E K, Kowall N W, Ferrante R J, Siwek D F, Wilcox H M, Flood D G, Beal M F, Brown R H, Jr, et al. Nat Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- 28.Lebovitz R M, Zhang H, Vogel H, Cartwright J, Jr, Dionne L, Lu N, Huang S, Matzuk M M. Proc Natl Acad Sci USA. 1996;93:9782–9787. doi: 10.1073/pnas.93.18.9782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melov S, Schneider J A, Day B J, Hinerfeld D, Coskun P, Mirra S S, Crapo J D, Wallace D C. Nat Genet. 1998;18:159–163. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 30.Trounce I A, Kim Y L, Jun A S, Wallace D C. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 31.Krebs H A, Holzach O. Biochem J. 1952;52:527–528. doi: 10.1042/bj0520527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Racker E. Biochim Biophys Acta. 1950;4:211–214. doi: 10.1016/0006-3002(50)90026-6. [DOI] [PubMed] [Google Scholar]
- 33.Goodman S I, Markey S P. Lab Res Methods Biol Med. 1981;6:1–158. [PubMed] [Google Scholar]
- 34.Dizdaroglu M. Methods Enzymol. 1994;234:3–16. doi: 10.1016/0076-6879(94)34072-2. [DOI] [PubMed] [Google Scholar]
- 35.Senturker S, Karahalil B, Inal M, Yilmaz H, Muslumanoglu H, Gedikoglu G, Dizdaroglu M. FEBS Lett. 1997;416:286–290. doi: 10.1016/s0014-5793(97)01226-x. [DOI] [PubMed] [Google Scholar]
- 36.Doetsch P W, Zasatawny T H, Martin A M, Dizdaroglu M. Biochemistry. 1995;34:737–742. doi: 10.1021/bi00003a005. [DOI] [PubMed] [Google Scholar]
- 37.Gardner P R, Fridovich I. J Biol Chem. 1992;267:8757–8763. [PubMed] [Google Scholar]
- 38.Gardner P R, Nguyen D D, White C W. Proc Natl Acad Sci USA. 1994;91:12248–12252. doi: 10.1073/pnas.91.25.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gardner P R, Raineri I, Epstein L B, White C W. J Biol Chem. 1995;270:13399–133405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- 40.Patel M, Day B J, Crapo J D, Fridovich I, McNamara J O. Neuron. 1996;16:345–355. doi: 10.1016/s0896-6273(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 41.Flint D H, Tuminello J F, Emptage M H. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 42.Roberts J R, Narasimhan C, Hruz P W, Mitchell G A, Miziorko H M. J Biol Chem. 1994;269:17841–17846. [PubMed] [Google Scholar]
- 43.Beal M F. Ann Neurol. 1995;38:357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- 44.Ebadi M, Srinivasan S K, Baxi M D. Prog Neurobiol. 1996;48:1–19. doi: 10.1016/0301-0082(95)00029-1. [DOI] [PubMed] [Google Scholar]
- 45.Lyras L, Cairns N J, Jenner A, Jenner P, Halliwell B. J Neurochem. 1997;68:2061–2069. doi: 10.1046/j.1471-4159.1997.68052061.x. [DOI] [PubMed] [Google Scholar]
- 46.Parkes T L, Elia A J, Dickinson D, Hilliker A J, Phillips J P, Boulianne G L. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 47.Ohkoshi N, Mizusawa H, Shiraiwa N, Shoji S, Harada K, Yoshizawa K. Muscle Nerve. 1995;18:1265–1271. doi: 10.1002/mus.880181108. [DOI] [PubMed] [Google Scholar]
- 48.Wallace D C, Melov S. Nat Genet. 1998;19:105–106. doi: 10.1038/448. [DOI] [PubMed] [Google Scholar]
- 49.Sohal R J, Weindruch R. Science. 1996;273:59–63. doi: 10.1126/science.273.5271.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marzo I, Brenner C, Zamzami N, Jurgensmeier J M, Susin S A, Vieira H L, Prevost M C, Xie Z, Matsuyama S, Reed J C, et al. Science. 1998;281:2027–2031. doi: 10.1126/science.281.5385.2027. [DOI] [PubMed] [Google Scholar]
- 51.Wallace D C. Science. 1992;256:628–632. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 52.Arber S, Hunter J J, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard J C, Chien K R, Caroni P. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 53.Graham B H, Waymire K G, Cottrell B, Trounce I A, MacGregor G R, Wallace D C. Nat Genet. 1997;16:226–234. doi: 10.1038/ng0797-226. [DOI] [PubMed] [Google Scholar]
- 54.Angelini C, Melacini P, Valente M L, Reichmann H, Carrozzo R, Fanin M, Vergani L, Boffa G M, Martinuzzi A, Fasoli G. Jpn Heart J. 1993;34:63–77. doi: 10.1536/ihj.34.63. [DOI] [PubMed] [Google Scholar]
- 55.Besley G T, Lendon M, Broadhead D M, Till J, Heptinstall L E, Phillips B. J Inherit Metab Dis. 1995;18:221–223. doi: 10.1007/BF00711772. [DOI] [PubMed] [Google Scholar]
- 56.Marin-Garcia J, Goldenthal M J, Ananthakrishnan R, Pierpont M E, Fricker F J, Lipshultz S E, Perez-Atayde A. J Inherit Metab Dis. 1996;19:309–312. doi: 10.1007/BF01799259. [DOI] [PubMed] [Google Scholar]
- 57.Muller-Hocker J, Ibel H, Paetzke I, Deufel T, Endres W, Kadenbach B, Gokel J M, Hubner G. Virchows Arch A Pathol Anat Histopathol. 1991;419:355–362. doi: 10.1007/BF01606527. [DOI] [PubMed] [Google Scholar]
- 58.Hall R E, Henriksson K G, Lewis S F, Haller R G, Kennaway N G. J Clin Invest. 1993;92:2660–2666. doi: 10.1172/JCI116882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walker J E. Q Rev Biophys. 1992;25:253–324. doi: 10.1017/s003358350000425x. [DOI] [PubMed] [Google Scholar]
- 60.Ohnishi T. Biochim Biophys Acta. 1998;1364:186–206. doi: 10.1016/s0005-2728(98)00027-9. [DOI] [PubMed] [Google Scholar]
- 61.Ackrell B C, Johnson M K, Gunsalus R P, Cecchini G. In: Chemistry and Biochemistry of Flavoenzymes. Muller F, editor. III. Boca Raton, FL: CRC Press; 1992. pp. 230–297. [Google Scholar]
- 62.Gardner P R. Biosci Rep. 1997;17:33–42. doi: 10.1023/a:1027383100936. [DOI] [PubMed] [Google Scholar]
- 63.Kim H Y, LaVaute T, Iwai K, Klausner R D, Rouault T A. J Biol Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- 64.Gray N K, Pantopoulous K, Dandekar T, Ackrell B A, Hentze M W. Proc Natl Acad Sci USA. 1996;93:4925–4930. doi: 10.1073/pnas.93.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schalinske K L, Chen O S, Eisenstein R S. J Biol Chem. 1998;273:3740–3746. doi: 10.1074/jbc.273.6.3740. [DOI] [PubMed] [Google Scholar]
- 66.Weiss G, Goossen B, Doppler W, Fuchs D, Pantopoulos K, Werner-Felmayer G, Wachter H, Hentze M W. EMBO J. 1993;12:3651–3657. doi: 10.1002/j.1460-2075.1993.tb06039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martins E A, Robalinho R L, Meneghini R. Arch Biochem Biophys. 1995;316:128–134. doi: 10.1006/abbi.1995.1019. [DOI] [PubMed] [Google Scholar]
- 68.Pantopoulos K, Hentze M W. EMBO J. 1995;14:2917–2924. doi: 10.1002/j.1460-2075.1995.tb07291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scheffler I E. Prog Nucleic Acid Res Mol Biol. 1998;60:267–315. doi: 10.1016/s0079-6603(08)60895-8. [DOI] [PubMed] [Google Scholar]
- 70.Hruz P W, Miziorko H M. Protein Sci. 1992;1:1144–1153. doi: 10.1002/pro.5560010908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dizdaroglu M. Mutat Res. 1992;275:331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- 72.Williams M D, Van Remmen H, Conrad C C, Huang T T, Epstein C J, Richardson A. J Biol Chem. 1998;273:28510–28515. doi: 10.1074/jbc.273.43.28510. [DOI] [PubMed] [Google Scholar]
- 73.Feig D I, Sowers L C, Loeb L A. Proc Natl Acad Sci USA. 1994;91:6609–6613. doi: 10.1073/pnas.91.14.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]