Abstract
The yeast DEAD-box protein Dbp5p/Rat8p is an essential factor for mRNA export and shuttles between the nucleus and the cytoplasm. It is concentrated at the cytoplasmic fibrils of the nuclear pore complex where it interacts with several nucleoporins. On the basis of this localization, it has been suggested that it might participate in a terminal step of RNA export, the release from the mRNA of proteins that accompany the mRNA during translocation through nuclear pores. In this report, we present evidence linking Dbp5p to transcription. Two different screens identified genetic interactions between DBP5 and genes involved in early transcription events, initiation and promoter clearance. Mutations of transcription proteins expected to impair transcription act as suppressors of dbp5 mutants, whereas those that may act to increase transcription are synthetically lethal with dbp5 mutations. We also show that growth and mRNA export in dbp5 mutant strains are dependent on the carboxy-terminal domain of the RNA pol II largest subunit. Finally, we show that Dbp5p associates physically with components of transcription factor IIH. Because these interactions affect not only growth but also mRNA export, they are likely to reflect a functional relationship between Dbp5p and the transcription machinery. Together, our results suggest a nuclear role for Dbp5 during the early steps of transcription.
INTRODUCTION
In eukaryotic cells, transcription by RNA polymerase II yields pre-mRNAs that are converted into mRNAs through the RNA-processing events of 5′ capping, splicing, and 3′ cleavage/polyadenylation (for reviews, see Proudfoot et al., 2002; Reed and Hurt, 2002). The export of the mRNA to the cytoplasm for translation requires that all of these steps be completed correctly and that certain RNA-binding proteins associate with the mRNA to form heterogeneous ribonucleoprotein particles (hnRNPs) (Reed and Magni, 2001; for review, see Dreyfuss et al., 2002). Correct and efficient packaging and processing are facilitated by the cotranscriptional recruitment to the RNA of processing factors and RNA-binding proteins (Hirose and Manley, 2000; Lei et al., 2001; Strasser et al., 2002). Although the RNA processing steps are complex and involve many different proteins, it is probable that only a subset of the factors needed for each step associates with RNA polymerase during transcription. The others would likely be recruited once the polymerase-associated factors bound the pre-RNA at appropriate processing sites. Many of these factors are subsequently released as processing steps proceed and are completed, with only a subset remaining with the processed mRNA during the export process.
The complex interplay of the events in nuclear mRNA biogenesis is reflected in the observation that defects in export result in defects in processing and defects in processing result in a failure to export the mRNA (Damelin and Silver, 2000; Hilleren et al., 2001; Hillerin and Parker; 2001; Jensen et al., 2001b; Hammell et al., 2002). This means that many proteins must function properly for both pre-mRNA processing and mRNA export to occur and, in this sense, are both RNA-processing and RNA export factors. An example is the yeast splicing factor Sub2p and its mammalian ortholog UAP56, which have recently been connected with export (Gatfield et al., 2001; Jensen et al., 2001a; Luo et al., 2001; Strasser and Hurt 2001). Sub2p/UAP56 interacts directly with the conserved mRNA export factor Yra1p/Aly, suggesting that Sub2p/UAP56 plays a role in recruiting Yra1p/Aly to the mRNA (Luo et al., 2001; Strasser and Hurt, 2001). Several recent studies have also provided links between export factors and the transcription machinery. Lei et al., (2001) showed that the hnRNP protein Npl3p can be found in a complex with RNA pol II and that both Npl3p and Yra1p are recruited to genes in a transcription-dependent manner. Other studies (Jimeno et al., 2002; Strasser et al., 2002) indicate that Yra1p and Sub2p associate with THO, a four-protein complex involved in transcription elongation. The resulting complex, TREX, is recruited to activated genes during transcription and then travels with the RNA pol II along the entire length of the gene (Strasser et al., 2002). Yra1p and Aly also bind directly to Mex67p (in yeast) or TAP (in metazoan), respectively (Strasser and Hurt, 2000; Stutz et al., 2000). Mex67p/TAP is a major mRNA export factor that shuttles between the cytoplasm and the nucleus. At steady state, it is mainly localized at nuclear pore complexes (NPCs), where it interacts with FG repeat-containing nucleoporins (for review, see Weis, 2002). It is thought to function as a receptor for mRNA export by interacting simultaneously with both the hnRNPs and nucleoporins.
Another conserved factor essential for mRNA export is the DEAD-box protein Rat8p/Dbp5p (Snay-Hodge et al., 1998; Tseng et al., 1998). A large family of DEAD-box proteins is found in every organism, and one or more have been associated with each step in mRNA biogenesis and degradation (for review, see Linder, 2000; Tanner and Linder, 2001). Some DEAD-box proteins are known to be able to unwind short RNA:RNA duplexes, and one has been shown capable of removing stably-bound proteins from RNA in vitro (Jankowsky et al., 2001). However, the enzymatic activities of most DEAD-box proteins have not been determined. Dbp5p binds to NPCs where it interacts with Nup159p, Gle1p, and other proteins located on or near the cytoplasmic fibrils of the NPC (Snay-Hodge et al., 1998; Hodge et al., 1999; Strahm et al., 1999). In addition, Dbp5p shuttles between the nucleus and the cytoplasm. The precise function performed by Dbp5p is not known. We and others have suggested that Dbp5p's association with the cytoplasmic face of the NPC positions it to catalyze removal of proteins that accompany mRNAs through NPCs. Zhao et al. (2002) used immunoelectron microscopy to show that Chironomus tentans Dbp5p can be detected along the entire length of nuclear Balbiani ring hnRNPs, but there is little data about what Dbp5p actually does, the mechanism by which it becomes associated with hnRNPs, or whether it functions in one or multiple subcellular locations.
To gain further insight into the roles of Dbp5p, we performed multiple genetic screens. In a search for high-copy suppressors of the rat8–6 temperature-sensitive allele of DBP5, we identified an N-terminally truncated form of Ssl1p (called Ssl1p-t). Ssl1p is a component of transcription factor IIH (TFIIH), a multiprotein complex that participates in both transcription and excision repair (Yoon et al., 1992; Feaver et al., 1993; Wang et al., 1995). In transcription, TFIIH acts late during initiation and promoter clearance. One of its functions is the phosphorylation of the carboxy-terminal domain (CTD) of the largest subunit of RNA pol II (Feaver et al., 1994; Marshall et al., 1996), a modification that contributes to the transition from initiation to elongation (Dahmus, 1996; Komarnitsky et al., 2000). A synthetic lethal screen with dbp5-2 identified a mutant allele of BUR6. BUR6 encodes a histone fold-containing protein homologous to DRAP1/NC2α, a component of the mammalian repressor of basal transcription NC2. The NC2 heterodimer binds to the TATA-box binding protein (TBP) and thereby inhibits the binding of both TFIIA and TFIIB to DNA-bound TBP (Goppelt and Meisterernst, 1996; Goppelt et al., 1996).
Identifying genes involved in the initiation of transcription as genetically connected to DBP5 was unexpected and led us to investigate what effects mutations and conditions affecting various transcription processes would have on growth and mRNA export in dbp5 mutant cells. We report that mutations and conditions that are likely to decrease the rate of transcription initiation and promoter clearance partially suppress dbp5-2, whereas mutations that may increase the rates of these processes are synthetically lethal with dbp5-2. This indicates that some function or activity of Dbp5p is sensitive to modulation of very early events in mRNA synthesis. We also report that the ability of mutant forms of Dbp5p to function is affected negatively by truncation of the CTD of the largest RNA pol II subunit, Rpb1p. In addition, we demonstrate that Dbp5p interacts physically with multiple subunits of TFIIH. These findings are consistent with the idea that Dbp5p associates with the mRNA early during transcription and acts on the hnRNP during mRNA biogenesis.
MATERIALS AND METHODS
Yeast Strains and Genetic Methods
Yeast strains used in this study are listed in Table 1. Strains were grown using standard methods. 6-Azauracil (6-AU) was added at different concentrations to minimal complete media containing 1 g/l 5-fluoroorotic acid (5-FOA) and 20 mg/l uracil. For growth assays, yeast cells were diluted to an A600 of 0.1, and serial dilutions (1:10) were spotted onto YPD or selective plates and incubated at various temperatures. The GAL10::HA-BUR6 allele, the C-terminal hemagglutinin (HA)- and Myc-tagged strains and deletions of SSL1 and HRP1 were obtained as described previously (Longtine et al., 1998).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| FY23 | MATa leu2Δ1 ura3-52 trp1Δ63 | Winston et al. (1995) |
| FY86 | MATα leu2Δ1 ura3-52 his3Δ200 | Winston et al. (1995) |
| PDY1 | MATa leu2Δ1 ura3-52 trp1Δ63 rat8-6 | This study |
| CSY550 | MATa leu2Δ1 ura3-52 trp1Δ63 rat8-2 | Snay-Hodge et al. (1998) |
| PDY2 | MATa leu2Δ1 ura3-52 trp1Δ63 his3Δ200 ssl1Δ::KAN [pSSL1] | This study |
| PDY3 | MATa leu2Δ1 ura3-52 trp1Δ63 his3Δ200 ssl1Δ::KAN rat8-6 [pSSL1] | This study |
| JJ836 | MATa ura3-52 ino1-13 ssl1-1 | Yoon et al. (1991) |
| JJ837 | MATa ura3-52 ino1-13 ssl1-2 | Yoon et al. (1991) |
| PDY4 | MATa ura3-52 his3Δ200 ssl1-1 rat8-2 | This study |
| PDY5 | MATa ura3-52 ssl1-2 rat8-2 | This study |
| CSY564 | MATα leu2Δ1 ura3-52 his3Δ200 rat8-2 [pRAT8.31] | This study |
| GY215 | MATa ura3-52 trp1Δ63 his4-912δ lys2-128δ bur6-1 | Prelich (1997) |
| PDY6 | MATα leu2Δ1 ura3-52 lys2-128δ rat8-2 bur6-1 [pRAT8.31] | This study |
| PDY7 | MATα leu2Δ1 ura3-52 his3Δ200 GAL10::HA-BUR6::KAN | This study |
| PDY8 | MATα leu2Δ1 ura3-52 his3Δ200 GAL10::HA-BUR6::KAN rat8-2 | This study |
| PDY9 | MATα leu2Δ1 ura3-52 trp1Δ63 rat8-2 mot1-301 [pRAT8.31] | This study |
| CSY560 | MATa leu2Δ1 ura3-52 trp1Δ63 rat8-2 [pRAT8.31] | This study |
| GY238 | MATα ura3-52 leu2Δ1 his4-912δ lys2-128δ mot1-301 | Prelich (1997) |
| Z828 | MATa leu2-3,112 ura3-52 his3Δ200 srb4Δ2::HIS3 [pCT181 (srb4-138/LEU2/CEN)] | Gadbois et al. (1997) |
| PDY10 | MATa leu2-3, 112 srb4Δ2::HIS3 ura3-52 his3Δ200? rat8-2 [pCT181 (srb4-138/LEU2/CEN)] | This study |
| PDY12 | MATα ura3-52 leu2-3, 112 (and/or leu2Δ1)rat8-2 rpb1Δ187::HIS3 [pRP112 (RPB1/CEN/URA3)] | This study |
| PDY13 | MATa ura3-52 leu2-3, 112 (and/or leu2Δ1) rpb1Δ187::HIS3 [pRP112 (RPB1/CEN/URA3)] | This study |
| Z26 | MATα ura3-52 his3Δ200 leu2-3,112 rpb1Δ187::HIS3 [pRP112 (RPB1/CEN/URA3)] | Nonet et al. (1987) |
| CS41-4.3 | MAT? ura3-52 leu2-3 his3-11 trp1-1 ade2-1 bur6::HIS3 (pbur6-ts/CEN/LEU2) | D, Reinberg |
| YMH202 | MAT? ura3-52 leu2-3 his3-11 trp1-1 ade2-1 bur6::HIS3 (pBUR6/CEN/URA3) | D. Reinberg |
| PDY 14 | MATα leu2Δ1 ura3-52 his3Δ200 DBP5-HA::HIS3 | This study |
| PDY15 | MAT? leu2Δ1 ura3-52, DBP5-HA::HIS3 TFB2-Myc:: KAN | This study |
| PDY16 | MAT? leu2Δ1 ura3-52, DBP5-HA::HIS3 RAD3-Myc:: KAN | This study |
| mex67-5 | MATa ade2 leu2 ura3 his3 trp1 mex67::HIS3 [pmex67-5 (TRP1/CEN)] | Segref et al. (1997) |
| yra1-1 | MATa ade2 leu2 ura3 his3 trp1 yra1::HIS3 [pyra1-1 (TRP1/CEN)] | Strasser and Hurt (2000) |
| sub2-85 | MATa ade2 leu2 ura3 his3 trp1 sub2::kanMX4 [psub2-85 (TRP1/CEN)] | Strasser and Hurt (2001) |
High-Copy Suppression Screen
The rat8-6 allele of DBP5 was generated by a polymerase chain reaction (PCR)-based mutagenesis procedure as described previously (Snay-Hodge et al., 1998). It contains a single amino acid change, of Leu to Pro at amino acid 220 (Hodge and Cole, unpublished data). A plasmid containing this mutant allele, pCS616, was used to replace the DBP5 wild-type gene in the strain FY23 with rat8-6, as described previously (Snay-Hodge et al., 1998), yielding the PDY1 strain. This strain was used for the high-copy suppression screen by transforming it with two different yeast genomic libraries cloned in the vectors YEp13 (LEU2/2 μm; Nasmyth and Tatchell, 1980) or YEp24 (URA3/2 μm; Carlson and Botstein, 1982). Transformants (40,000 for the YEp24 library and 5,000 for the YEp13 library) were grown overnight on selective medium at room temperature and then transferred to 34 or 37°C. Plasmids able to suppress the mutation were isolated and used to transform PDY1 (rat8-6) and CSY550 (rat8-2). The yeast insert in the suppressor plasmids was sequenced from both sides.
Yeast Plasmids Construction
Plasmids pGFD1 and pGFD2 were obtained from the YEp24-based genomic library. Plasmid pPD1 was obtained from the YEp13-based yeast genomic library. Plasmid pPD2 was obtained by subcloning a DNA fragment extending from the BamHI site in the SSKI open reading frame (ORF) to the SalI site located in the YEp13 vector into YEp24. Plasmid pPD3 was obtained by deleting the SphI fragment from pPD1, which extends from the SphI site located in the SSL1 ORF to a SphI site located in the YEp13 vector. Plasmid pPD5 was obtained by subcloning an XhoI-SphI PCR fragment containing the 5′ region of the SSL1 gene into pPD2 digested with SalI and SphI. Plasmid pSSL1 was obtained by subcloning a BamHI-EagI fragment from pPD5 into pRS316 (Sikorski and Hieter, 1989). Plasmids pC1, pC23, pC3, and pV5 encode truncated forms of Rbp1p lacking different numbers of the repeats found in the CTD of Rpb1p and have been described previously (Nonet et al., 1987).
In Situ Hybridization
To localize poly(A)+ RNA within cells, we used in situ hybridization with an oligo(dT)50 probe coupled to digoxigenin, which was performed as described previously (Cole et al., 2002).
Immunoprecipitation and Western Blotting
Immunoprecipitation was performed as described previously (Fan et al., 2001) by using extracts expressing Dbp5p-HA (strain PDY14), Dbp5p-HA, and Tfb2p-Myc (strain PDY15) or Dbp5p-HA and Rad3p-Myc (strain PDY16). Extracts were incubated with 3 μl of mouse monoclonal anti-HA antibody (Sigma-Aldrich, St. Louis, MO) at 4°C for 4 h. Then 30 μl of EZview red protein G affinity gel (Sigma-Aldrich) was added to the extracts and incubated for an additional hour at 4°C. Washes and elutions were performed according to the manufacturer's instructions. Western blot analyses were performed using peroxidase-conjugated mouse monoclonal anti-c-myc (Roche Diagnostics, Indianapolis, IN) to detect Tfb2p-Myc and Rad3p-Myc, rabbit polyclonal anti-Tfb1p to detect Tfb1p or mouse monoclonal anti-HA to detect Dbp5p-HA.
Determination of Poly(A)+ Levels and Poly(A) Tail Length Measurements
Total RNA was isolated as described previously (Martinez-Pastor et al., 1996). RNA transfer and hybridization with the poly(dT) probe were performed as described previously (Kuldell and Buratowski, 1997). Poly(A) tail length measurements were performed as described previously (Hammell et al., 2002).
RESULTS
Identification of a New Extragenic High-Copy Suppressor of the rat8-6 Mutant
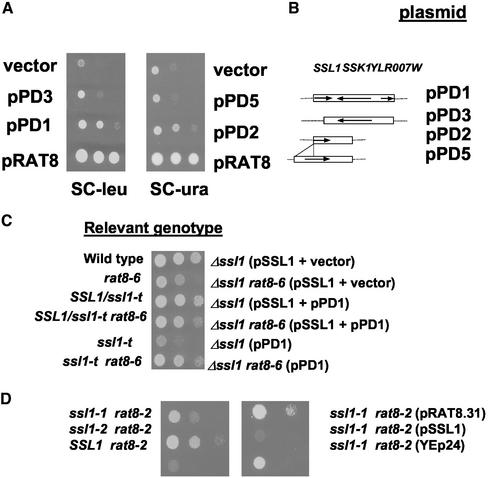
To gain further insight into the functions of the DEAD-box protein and mRNA export factor Dbp5p/Rat8p, we performed a high-copy suppressor screen at 34 and 37°C in cells carrying the integrated rat8-6 allele of DBP5 (see MATERIALS AND METHODS). At 37°C, all plasmid clones isolated encoded Dbp5p, suggesting that Rat8-6p loses all function at 37°C and that there are no bypass suppressors of a complete loss of function allele of DBP5/RAT8. At 34°C, besides DBP5 and the previously identified suppressor GFD1 (Hodge et al., 1999), we isolated another previously identified but unpublished high-copy suppressor, GFD2 (Colot and Cole, unpublished data) and also obtained suppression of the growth defect by a plasmid (pPD1) whose yeast genomic DNA fragment from chromosome XII includes all or parts of the SSL1, SSK1, and YLR007w genes. The level of suppression provided by plasmid pPD1 is similar to that found by overexpression GFD2 but less than by overexpressing GFD1 (our unpublished data). Extragenic suppression was mapped to the SSL1 locus (Figure 1, A and B), because deletion of SSL1 sequences from pPD1 (plasmid pPD3) eliminated suppression, and subcloning a fragment containing only the SSL1 sequences of pPD1 into the multicopy vector YEp24 (plasmid pPD2) partially suppressed the growth defect at 34°C. No suppression was observed at 37°C (our unpublished data). Ssl1p is a component of TFIIH, which is believed to function in both transcription and excision repair (Wang et al., 1995).
Figure 1.
Suppression of rat8 growth defect by ssl1 mutations. (A) Suppression of rat8-6 ts growth defect by overexpression of ssl1-t. Strain rat8-6 (PDY1) was transformed with the high-copy plasmid obtained from the library (pPD1), plasmids containing different fragments obtained from the DNA genomic insert of pPD1 (pPD2 and pPD3, see B) or a plasmid containing the full-length SSL1 gene (pPD5). The restriction fragments from the DNA genomic insert isolated from the library were subcloned in the multicopy vectors YEp13 (left) or YEp24 (right). Full-length SSL1 was obtained in plasmid pPD5 by adding to the truncated allele (ssl1-t) of plasmid pPD2 a PCR fragment containing the promoter and 5′ sequences of SSL1 ORF. As controls, the same strain was transformed with the empty vectors (YEp181 for leucine selection, or YEp24 for uracil selection) or plasmids containing the wild-type RAT8 gene (pCS8.31 in SC-leu plate, or pRAT8.31 in SC-ura plate). Cells were diluted to an OD600 of 0.1, and 1:10 serial dilutions were spotted onto selective plate, which were incubated for 3 d at 34°C. (C) Complementation of Δssl1 by ssl1-t. The Δssl1 allele was introduced into the diploid strain FY86 × PDY1. This strain was transformed with pSSL1 (wild-type SSL1/URA3/CEN). Haploids carrying a genomic deletion of SSL1 were selected by their inability to grow on plates containing 5-FOA. Δssl1 haploids were transformed with plasmids pPD1 (ssl1-t) or YEp1ac181 (vector). To select for cells that had lost pSSL1, cells were plated on 5′-FOA plates. Growth of the different strains was checked by spotting serial dilutions (1:10) on selective media (SC-leu) and incubated at 30°C for 2 d. (D) Suppression of rat8-2 ts growth defect by ssl1 mutant alleles. Left, double mutant strains were segregants from the cross CSY550 (rat8-2) × JJ636 (ssl1-1) and CSY550 × JJ637 (ssl1-2). Serial dilutions (1:10) were spotted onto a YPD plate and incubated for 3 d at 34°C. Right, double mutant strain rat8-2 ssl1-1 was transformed with pRAT8.31 (RAT8/CEN/URA3), pSSL1 (SSL1/CEN/URA3), or YEp24 (2 μm/URA3 vector), and 1:10 serial dilutions were spotted onto selective plate (SC-ura), which were incubated for 3 d at 34°C.
Interestingly, the yeast genomic fragment with suppressing activity did not contain the full-length SSL1 gene. The suppressing plasmid lacks the SSL1 promoter and the part of the gene encoding the first 100 amino acids, and seems to be expressed from a cryptic promoter present in the bacterial portion of the plasmid. Truncation of Ss1p seems critical for suppression because a multicopy plasmid containing the full-length SSL1 gene (pPD5) under control of its own promoter was unable to suppress the rat8-6 mutation. Similar partial suppression of the rat8-2/dbp5-2 allele was also observed (our unpublished data). Therefore, suppression is specific to the truncated form of SSL1, which we designate ssl1-t.
The ssl1-t allele encodes a protein that is partially functional, as shown by the ability of plasmid pPD1 to rescue the lethality of a Δssl1 mutation (Figure 1C). However, growth of a strain carrying ssl1-t as the only SSL1 allele was reduced relative to wild type. Interestingly, the rat8-6 strain carrying the ssl1-t allele grows better at 30°C than the RAT8 ssl1-t strain, indicating that the presence of the rat8-6 allele in place of RAT8 also partially suppresses the growth defect of ssl1-t cells.
ts Mutations in the SSL1 Gene Suppress the rat8 Growth Defect at 34°C
The fact that a N-terminally truncated version of Ssl1p, but not the full-length protein, is able to partially suppress the growth defect at 34°C of rat8-2 and rat8-6 mutants led us to investigate how other SSL1 mutant alleles would affect growth of rat8 mutant strains. We tested the ssl1-1 and ssl1-2 ts alleles (Yoon et al., 1992). Double mutants ssl1-1/rat8-2 and ssl1-2/rat8-2 were constructed and growth at different temperatures was compared with growth of the parental strains. The ssl1-1 and ssl1-2 mutations partially suppressed the growth defect of rat8-2 at 34°C, but, like the ssl1-t allele, neither ts allele conferred growth at 37°C (Figure 1D; our unpublished data). The ability of ts ssl1 alleles to suppress the growth defect of rat8-2 at 34°C was completely ablated by the introduction into the ssl1-1 rat8-2 double mutant strain of a plasmid containing the wild-type SSL1 gene (Figure 1D).
To determine the specificity of the genetic interaction between to DBP5 and SSL1 we analyzed the effect of ssl1-t on mutants of other export factors. Figure 2 shows that overexpression of ssl1-t suppresses the growth defect at 37°C of a sub2-85 mutant but not the ts phenotype of either the mex67-5 or yra1-1 mutants at this temperature.
Figure 2.
Suppression of sub2-85 ts growth defect by overexpression of ssl1-t. yra1-1, mex67-5, and sub2-85 mutant strains were transformed with high-copy plasmids containing the SSL1 wild-type gene (pPD5), the ssl1-t allele (pPD2), or the empty vector YEp24 (vector). Serial dilutions (1:10) were spotted onto a YPD plate and incubated for 3 d at different temperatures.
Genetic Interactions between RAT8 and the Global Transcription Regulators BUR6, MOT1, and SRB4
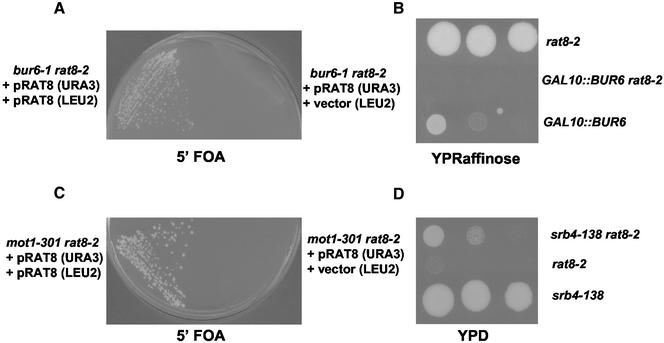
In a screen to identify mutations lethal in combination with the rat8-2 mutant allele, we isolated a mutant allele of the BUR6 gene (Hammell and Cole, unpublished data). BUR6 encodes a histone fold-containing protein homologous to one of the subunits (DRAP1/NC2α) of the heterodimeric mammalian transcription repressor negative cofactor 2 (NC2). NC2 binds to TBP and inhibits its function (Goppelt and Meisterernst, 1996; Goppelt et al., 1996). Synthetic lethality was also observed between rat8-2 and the previously published bur6-1 allele (Prelich, 1997) (Figure 3A). Furthermore, the slight growth in raffinose at 30°C shown by a strain containing the BUR6 gene under the control of the GAL10 promoter was not observed when this BUR6 allele was combined with the rat8-2 mutation (Figure 3B).
Figure 3.
Genetic interactions between rat8-2 and bur6-1, mot1-301 and srb4-138. (A) Double mutant rat8-2 bur6-1 [pRAT8.31] (pRAT8 [URA3] in the figure) was a segregant from cross CSY564 (rat8-2 [pRAT8.31]) × GY215 (bur6-1). This strain was transformed with pCS8.31 (pRAT8 [LEU2] in the figure) or YCplac181 (vector [LEU2] in the figure), and synthetic lethality was analyzed by streaking the transformants on a 5-FOA plate and incubation at 30°C for 4 d. (B) The GAL10::BUR6 allele was obtained by chromosomal replacement of the BUR6 promoter with a PCR fragment containing the GAL10 promoter and the HA epitope (Longtine et al., 1998) in frame with the BUR6 ORF. Replacement was performed in strain FY86 to obtain GAL10::BUR6 and in strain CSY550 to obtain rat8-2 GAL10::BUR6. Strains were spotted on a YPRaffinose plate and incubated 3 d at 30°C. Strain CSY550 (rat8-2) was used as control. (C) Double mutant rat8-2 mot1-301 [pRAT8.31] was a segregant from cross CSY560 (rat8-2 [pRAT8.31]) × GY236 (mot1-301). This strain was transformed with pCS8.31 or YCplac181 and synthetic lethality was analyzed by streaking the transformants on a 5-FOA plate and incubation at 30°C for 4 d. (D) Strains were segregants from cross CSY564 (rat8-2 pRAT8.31) × Z628 (srb4-138). Growth of the different strains was checked by spotting 1:10 serial dilutions onto a YPD plate, which was incubated for 3 d at 34°C.
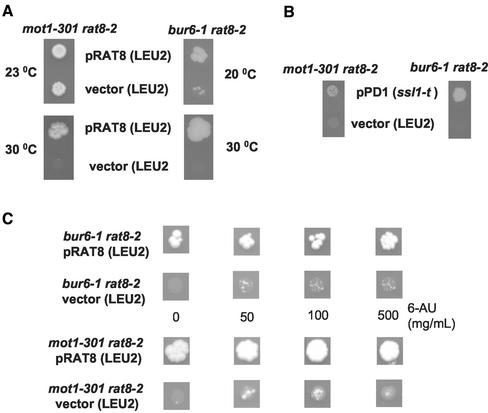
The original bur mutants were isolated in a genetic selection designed to identify mutations that affect basal transcription in Saccharomyces cerevisiae (Prelich and Winston, 1993). Based on their distinct sets of phenotypes and genetic interactions, it was proposed that the bur mutations comprised two groups. One group consists of BUR6 and BUR3/MOT1 (Prelich and Winston, 1993). Moreover, it has been reported that the activity of NC2 and Mot1p are regulated similarly under different physiological conditions (Lemaire et al., 2000) and both seem to repress transcription by acting through DNA-bound TBP. Interestingly, both Mot1p and Bur6p also have positive roles at some promoters in vivo (Prelich, 1997; Lemaire et al., 2000). These properties prompted us to investigate genetic interactions between mutations in MOT1 and RAT8. Figure 3C shows that rat8-2 mot1-301 double-mutant cells are unable to grow at 30°C, although they can grow at 23°C (our unpublished data; Figure 8A).
Figure 8.
Suppression of synthetic lethality of rat8-2 bur6-1 and rat8-2 mot1-301 by reducing the transcription rate. (A) Double mutants rat8-2 bur6-1 and rat8-2 mot1-301 containing pRAT8.31 were transformed with pCS8.31 (RAT8/CEN/LEU2) or YCplac181 (CEN/LEU2). Synthetic lethality was analyzed by spotting the transformants on a 5-FOA plate and incubating at 20, 23, or 30°C for 4 d. (B) Double mutants rat8-2 bur6-1 and rat8-2 mot1-301 containing pRAT8.31 were transformed with plasmid pPD1 (ssl1-t/2 μm/LEU2) or YEplac181 (2 μm/LEU2) and spotted on 5-FOA plates at 30°C for 2 d. (C) Double mutants rat8-2 bur6-1 and rat8-2 mot1-301 containing pRAT8.31 were transformed with pCS8.31 (RAT8/CEN/LEU2) or YCplac181 (CEN/LEU2) and spotted on 5-FOA plates containing the indicated amount of 6-azaurcil, and incubated at 30°C for 4 d.
BUR6 was also identified as a gene whose mutations can suppress a temperature-sensitive mutation in the subunit of the RNA polymerase II holoenzyme encoded by SRB4 (Gadbois et al., 1997). Srb4p has an essential and positive role in transcription at the majority of class II promoters in yeast (Thompson and Young, 1995). To check the possibility of a genetic interaction between SRB4 and RAT8, we constructed a double mutant strain srb4-138 rat8-2 and analyzed growth at different temperatures. The data shown in Figure 3D indicate that the srb4-138 mutation improves the growth of the rat8-2 strain at 34°C.
Defects in Transcription Elongation Functions Do Not Suppress dbp5 Mutations
Both Ssl1p and Srb4p are involved specifically in positive control of transcription initiation. Because recent studies tie mRNA export factors (Yra1p and Sub2p) to transcription elongation factors, we wondered whether the rat8-2 growth defect would be suppressed by mutants with defects in elongation. We analyzed the growth phenotype of double mutant strains containing the rat8-2 allele and a mutant allele of SPT6, or a disruption of ELP3 or HPR1. Elp3p, Spt6p, and Hpr1p have been reported to be involved in transcriptional elongation (Chavez and Aguilera, 1997; Hartzog et al., 1998; Wittschieben et al., 1999). In all cases, the double mutants grew no better at 34°C than the single rat8-2 mutant strain, which grows very poorly at this temperature (our unpublished data). This indicates that that there is specificity to suppression of the rat8 growth defect by mutants affecting initiation of transcription.
Interaction of DBP5 with rpb1 CTD-truncated Mutations
The CTD of the largest subunit of yeast RNA polymerase II contains 26 near-perfect heptapeptide repeats at its carboxy terminus, and this domain plays a central role in coupling transcription and pre-mRNA processing (for a recent review, see Maniatis and Reed, 2002). Yeast cells with a partially truncated CTD are unable to fully activate a variety of genes in vivo (Meisels et al., 1995, and references therein). Moreover, TFIIH includes a CTD-kinase activity (Cismowski et al., 1995; Valay et al., 1995). To determine whether the function of RAT8 depends on the CTD, we constructed a rat8-2 strain carrying a disruption of RPB1, covered by a wild-type RPB1 gene carried on a URA3/CEN plasmid. This strain was transformed with a series of LEU2 plasmids carrying rpb1 alleles truncated to remove different numbers of heptapeptide repeats. The effect on rat8-2 of these mutant alleles was analyzed by growth on plates containing 5-FOA media (Figure 4). Although all the double-mutant strains were viable, a strong growth defect was observed when the Rpb1p CTD was truncated to 10 repeats. A more modest growth defect was seen when 11 repeats were retained, and little or no growth defect when there were 12 or more repeats remaining. We conclude that the function of Dbp5p is sensitive to the length of the Rpb1p CTD.
Figure 4.
Analysis of rat8-2 rpb1-CTD truncation double mutants. Strains rat8-2 rpb1Δ-187::HIS3 pRPB1 (RPB1/CEN/URA3) and RAT8 rpb1Δ-187::HIS3 pRPB1 were segregants from cross Z26 (rpb1Δ-187::HIS3 pRPB1) × CSY550 (rat8-2). These strains were transformed with plasmids containing RPB1 genes encoding Rpb1p retaining different numbers of CTD repeats: pC3 (10 repeats), pC1 (11 repeats), pC23 (12 repeats), and pV5 (13 repeats). The synthetic interaction was analyzed by spotting serial 1:10 dilution of the transformants on a 5-FOA plate and incubation at 30°C for 3 d.
Dbp5p Physically Interacts with Components of the TFIIH
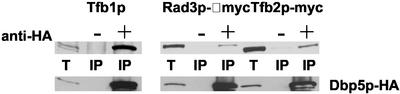
The genetic interaction of DBP5 with SSL1 and the rpb1 CTD-truncation mutants prompted us to investigate the possibility of a physical interaction between Dbp5p and components of the transcription machinery. Although we were unable to detect Ssl1p in a complex with Dbp5p (our unpublished data), we found that a fraction of Tfb1p coimmunoprecipitates with Dbp5p-HA (Figure 5). Using Myc-tagged proteins, we also found a physical interaction between Dbp5p-HA and two additional TFIIH components, Tfb2p and Rad3p (Figure 5). These results suggest that Dbp5p associates with TFIIH and could interact with mRNA beginning at the initial stages of transcription.
Figure 5.
Physical interaction between Dbp5p and TFIIH components. Rat8-HA (bottom) from cell extracts was immunoprecipitated with α-HA antibodies (+). Samples with no added primary antibody (−) were used as controls. Total cell extracts (T) contain a 1/200 of the amount of extract used for immunoprecipitation. Samples were analyzed on a 7.5% SDS-PAGE gel. The entire immunoprecipitate was analyzed on the gel. Tfb1p in the immunoprecipitates (IP) was detected with rabbit polyclonal α-Tfb1p and anti-rabbit immunoglobulin G-horseradish peroxidase as the secondary antibody. Genomically myc-tagged Tfb2p and Rad3p were detected with α-myc (mouse) horseradish peroxidase-conjugated antibodies.
Mutations in SSL1 Reduce the mRNA Export Defect of rat8-2 Mutant
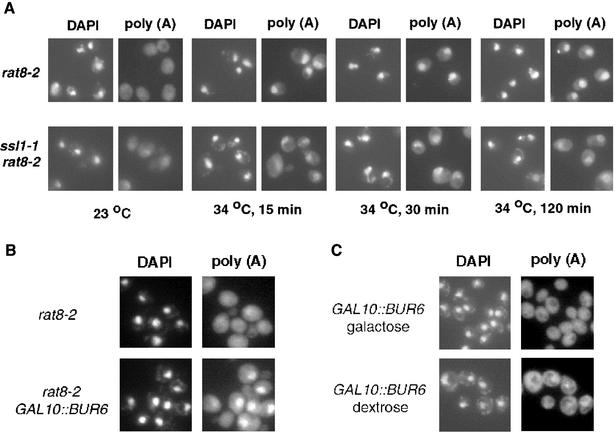
Strains carrying the rat8-2 allele show a partial nuclear accumulation of poly(A)+ mRNA at 23°C and a complete defect in mRNA export at 34 and 37°C (Figure 6A; Snay-Hodge et al., 1998). Because the ssl1-1 mutation partially suppresses the growth defect of rat8-2 mutants at 34°C, we analyzed its effect on export of poly(A)+ mRNA in rat8-2 cells. After a shift to 34°C, the mRNA signal is almost exclusively nuclear in rat8-2 mutant cells (Figure 6). Interestingly, in rat8-2 ssl1-1 double mutant cells, significantly less poly(A)+ mRNA accumulated in nuclei and there was a substantial increase in cytoplasmic signal for poly(A)+ mRNA. No mRNA export defect was observed in ssl1-1 cells at either at 34 or 37°C (our unpublished data). The data indicate that the ssl1-1 mutation not only suppresses the growth defect of rat8-2 cells but also increases mRNA export.
Figure 6.
(A) Partial suppression of nuclear accumulation of poly(A)+ RNA at 34°C in cells carrying rat8-2 by ssl1-1. rat8-2 and rat8-2 ssl1-1 cells were grown to midlog phase at 23°C and incubated for different times at 34°C. (B) Exacerbation of the poly(A)+ transport defect of rat8-2 by depletion of Bur6p. (A) rat8-2 and rat8-2 GAL10::BUR6 cells were incubated for 3 h at 23°C in YPD. (C) Partial block to mRNA export in wild-type cells depleted of Bur6p. GAL10::BUR6 cells were grown in YPGalactose to midlog phase at 23°C and were incubated overnight in YPD at the same temperature. In all the cases, cells were fixed and in situ hybridization was performed using a digoxigenin-conjugated oligo(dT) probe, followed by incubation with a fluorescein isothiocyanate-conjugated anti-digoxigenin antibody.
dbp5 and other mRNA export mutants produce mRNAs with abnormally long poly(A) tails (Jensen et al., 2001b; Hammell et al., 2002). This defect of dbp5 mutants was not suppressed by ssl1, because the poly(A) length distribution in a rat8-2 ss1-1 double mutant was similar than in a rat8-2 single mutant strain (our unpublished data).
Depletion of Bur6p Exacerbates mRNA Export Defect of rat8-2 Mutants
The synthetic lethality between BUR6 and RAT8 mutant alleles prompted us to determine how depletion of Bur6p would affect the distribution of poly(A)+ RNA. We placed the BUR6 coding region under control of the inducible GAL10 promoter. Production of Bur6p can be terminated by transfer of cells growing on galactose to glucose-containing medium. Western blotting to monitor the levels of Bur6p after a shift to glucose indicated that Bur6p had almost completely disappeared after 3 h in glucose medium (our unpublished data).
In contrast to suppression of the mRNA export defect in rat8-2 ssl1-1 cells, a substantial increase in accumulation of poly(A)+ RNA in nuclei was seen when rat8-2 mutant cells containing the regulatable GAL10::BUR6 allele were transferred to dextrose for 4 h. In this condition, rat8-2 single mutants show only a slight mRNA export defect (Figure 6B). A slight nuclear accumulation of poly(A)+ mRNA can even be observed in the RAT8 GAL10::BUR6 mutant strain after overnight incubation in dextrose containing medium (Figure 6C), suggesting that optimal mRNA export depends on some function of Bur6p.
Poly(A)+ Levels in bur6 and mot1 Mutants
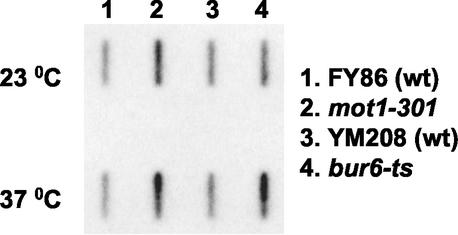
Both ssl1 and srb4 mutants are defective in RNA pol II transcription (Kim et al., 1994; Wang et al., 1995; Lee and Kim, 1998). However, Bur6p and Mot1p have been shown to be involved in both transcriptional repression and activation (Prelich, 1997; Lemaire et al., 2000; Geisberg et al., 2001). To measure the effect of the bur6 and mot1 mutations on total mRNA levels, we determined the amount of poly(A)+ RNA in cells by slot blot analysis, by using poly(dT) as a probe. The results shown in Figure 7 indicate that overall mRNA levels are higher in mot1 and bur6 mutants that in the respective isogenic wild-type strains, suggesting that Mot1p and Bur6p repression has a greater effect on overall mRNA levels than does Mot1p and Bur6p activation at other loci.
Figure 7.
bur6-1 and mot1-301 mutations increased total mRNA levels. Slot blot hybridization to detect total poly(A)+ RNA of mot1-301 (lane 2) and its isogenic wild-type strain (lane 1, FY86), and the bur6-ts mutant (lane 4) and its isogenic wild-type strain (YM208, lane 3). Exponentially growing cells in YPD (samples 23°C) were shifted to 37°C and incubated for 90 min. Total RNA was analyzed with a 32P-labeled poly(dT) probe. rRNA was used to verify the application of equal amounts of sample (our unpublished data).
Synthetic Lethality between RAT8 and the General Repressors BUR6 and MOT1 Can Be Suppressed by Reducing the Rate of Transcription
The increased levels of poly(A)+ RNA present in bur6 and mot1 mutant cells raised the possibility that the synthetic lethality observed between rat8-2 and either bur6 or mot1 mutants could be a consequence of production of more mRNA than can be properly handled by cells containing the defective mRNA export factor encoded by rat8-2. If this were the case, it might be possible to suppress the growth defects of rat8-2 bur6-1 and rat8-2 mot1-301 double mutants by reducing the level of transcription. To test this possibility, we used three strategies to reduce transcription rates in double-mutant cells.
First, we examined the effect of reducing the temperature of growth. Figure 8A shows that neither double-mutant strain can grow at 30°C but rat8-2 mot1-301 cells can grow at 23°C and rat8-2 bur6-1 cells are able to grow when temperature is reduced to 20°C. The second strategy to reduce transcription initiation in the rat8-2 bur6-1 and rat8-2 mot1-301 double mutants was to transform these strains with plasmid pPD1, encoding the truncated allele of SSL1 (ssl1-t). Figure 8B shows that double mutants rat8-2 bur6-1 and rat8-2 mot1-301 transformed with pPD1, but not with the vector YEplac181, were able to grow at 30°C. Finally, we reduced transcription by including 6-AU in the growth media. 6-AU impairs transcription by lowering of GTP and/or UTP levels (Exinger and Lacroute, 1992). Figure 8C shows that addition of 6-AU allows limited growth of the double mutants bur6-1 rat8-2 and mot1-301 rat8-2 at 30°C, whereas normally these strains show no growth at 30°C.
DISCUSSION
The gene encoding the DEAD-box protein Dbp5p was identified, together with several nucleoporins, in a screen for yeast ts mutant strains that accumulated poly(A)+ RNA in their nuclei at the nonpermissive temperature (Amberg et al., 1992). The implied role for a DEAD-box protein in mRNA export was not surprising, because these proteins play important roles in almost every step in mRNA metabolism (de la Cruz et al., 1999). The enzymatic activities of most DEAD-box proteins have not been defined, but the presence of several conserved motifs suggests that these proteins are able to bind and hydrolyze ATP and that some or all are able to harness the energy from ATP hydrolysis to unwind double-stranded RNA (Tanner and Linder, 2001). This has led to their being called DEAD-box protein RNA helicases, but the fact that they unwind only short oligoribonucleotides makes unwindase a more accurate term (Linder and Daugeron, 2000). It has been suggested that an RNA/protein complex might also be a substrate for DEAD-box proteins (de la Cruz et al., 1999), and one DEAD-box protein has been shown capable of removing a stably bound protein from single-stranded RNA (Jankowsky et al., 2001).
Dbp5p has ATP-binding and ATPase activity and, although purified Dbp5p is not an RNA unwindase, it may have this activity in extracts (Tseng et al., 1998). At steady state, Dbp5p is concentrated at the nuclear rim and a minor fraction can be observed in the cytoplasm (Snay-Hodge et al., 1998). To assign a role for Dbp5 in mRNA export, we must take into account that mRNA is exported as an RNA/protein complex (hnRNP) and most of the hnRNP proteins have to be removed during or soon after export, allowing them to shuttle back to the nucleus. This has led us and others to suggest that Dbp5p might facilitate mRNA export by removing these proteins from mRNA. The fact that Dbp5p associates with NPCs on the cytoplasmic face of the pore, where it binds directly to proteins in the Rat7p/Nup159p subcomplex (Hodge et al., 1999), positions it to perform such a role. Most nucleoporins do not seem to play an essential role in mRNA export, whereas those in the Rat7p/Nup159p subcomplex are those most important for mRNA export. In fact, it is possible that the primary function of the Rat7p/Nup159p subcomplex during export is to provide a binding site for Dbp5p. Previously, we showed that Dbp5p is a shuttling protein and accumulates rapidly in nuclei when the function of the exportin, Xpo1p, is blocked (Hodge et al., 1999). This suggested that Dbp5p might also function in the nucleus.
The studies presented herein support the idea that Dbp5p plays a function in the nucleus which is linked to transcription. First, we identified an allele of SSL1 as a high-copy suppressor of the rat8-6 and rat8-2 alleles (Figure 1). The suppressing ssl1-t allele encodes an N-terminally truncated form of Ssl1p and no suppression was observed by overexpression of the wild-type SSL1 gene. In addition, we found that the ts ssl1-1 and ssl1-2 alleles could also partially suppress rat8-2 mutant, suggesting that the suppression by Ssl1p-t is due to reduced Ssl1p function. Suppression in this case operates in both directions: the rat8-6 ssl1-t strain grows better than either single mutant. Second, the suppressive activity of ssl1-t is specific for some export factors. In addition to rat8-2 and rat8-6, we found that overexpression of ssl1-t is also able to suppress the thermosensitivity of sub2-85 but not mex67-5 or yra1-1 mutants (Figure 2). Interestingly, it has been proposed that Sub2p is recruited cotranscriptionally to the mRNA and performs a key function early in export by recruiting Yra1p (Strasser and Hurt, 2001; Strasser et al., 2002). Third, in a different genetic screen we identified an allele of BUR6 as synthetic lethal with rat8-2 (Figure 3; Hammell and Cole, unpublished data). Four, we observed strong effects on the growth of rat8-2 cells in strains where Rpb1p has been replaced by deletion alleles of rpb1 lacking different numbers of the heptapeptide repeats of the Rpb1p-CTD (Figure 4). Finally, we have detected physical interactions between Dbp5p and several components of the TFIIH complex (Figure 5), suggesting that Dbp5p may interact with TFIIH.
Both Ssl1p and Bur6p are involved in transcription initiation. Ssl1p is a part of TFIIH (Feaver et al., 1993), which is essential for initiation of pol II transcription and for promoter clearance, as well as for nucleotide excision repair (Wang et al., 1995). The function of Bur6p is less clear. It is homologous to one (NC2α) of the two subunits of the mammalian transcription repressor DRAP1/NC2. In yeast, NC2 interacts directly with the TATA-box binding protein (Goppelt and Meisterernst, 1996) and is able to repress transcription by RNA polymerase II in vitro (Gadbois et al., 1997). However, other studies indicate that the NC2 complex activates transcription at some promoters (Prelich, 1997; Geisberg et al., 2001). Overall, it probably affects more promoters negatively than positively because we found an increased amount of poly(A)+ RNA in bur6-ts cells after a shift to the nonpermissive temperature (Figure 7). The RNA pol II CTD plays multiple roles during mRNA synthesis, and dramatic changes in the phosphorylation pattern of the CTD occur as transcription moves from initiation to promoter clearance to elongation. At promoters, the CTD is phosphorylated on Ser 5 by TFIIH, which includes polypeptides that interact physically (Rad3p, Tfb1p, and Tfb2p) or genetically (Ssl1p) with Dbp5p. The CTD also interacts directly with factors involved in every step of pre-mRNA processing, helping to bring these factors to the growing mRNA (for review, see Lewis and Tollervey, 2000). We think that these genetic and physical interactions reflect truly functional interactions at the level of mRNA export because the mRNA export defect of rat8-2 cells at 34°C is alleviated in the presence of the ssl1-1 allele (Figure 6A) and depletion of Bur6p exacerbates the normally modest mRNA defect of rat8-2 cells at 23°C (Figure 6B).
Many of our studies with ssl1-t were performed in cells that also carried a wild-type SSL1 allele. Our findings indicate that the ssl1-t mutant functions as a gain-of-function allele during suppression, even though Ssl1p function is probably reduced by truncation of Ssl1p. Most likely, in cells expressing both Ssl1p and Ssl1p-t, both forms are present in a fraction of the Ssl1p-containing complexes. We were unable to detect a physical association between Dbp5p and Ssl1p but we believe this is due to technical limitations. We were unable to obtain a functioning anti-Ssl1p antibody. We produced functional tagged forms of Ssl1p, but they seemed to be present at levels much below those of the other subunits of TFIIH studied (Estruch and Cole, unpublished data). Together, our data suggests that suppression of dbp5 defects by ssl1-t reflects the functioning and activity of a partially defective TFIIH complex containing Ssl1p-t. One possibility is that Ssl1p-t causes reduced rates of some early events in transcription involving TFIIH, such that the reduced activity of mutant forms of Dbp5p becomes sufficient for gene expression.
To address the question of how specific are the genetic interactions between rat8, ssl1, and bur6, we also explored genetic interactions between rat8 mutants and mutations not identified in our screens but that are also involved in the control of transcription: srb4 and mot1. Both Srb4p and Mot1p seem to act during transcription initiation but are thought to perform very different functions. Srb4p is an essential component of the mediator subcomplex of the RNA polymerase II holoenzyme (Kim et al., 1994; Myer and Young, 1998; Malik and Roeder, 2000). The mediator is composed of ∼20 polypeptides, is required for activated transcription, and also plays a positive role both in basal transcription and in the phosphorylation of RNA polymerase II by the TFIIH-associated CTD kinase (Kim et al., 1994). We found that, like ssl1, mutations in SRB4 are able to suppress the growth defect at 34°C of rat8-2 (Figure 3). The functions of Mot1p seem to be similar to those of Bur6p, and in fact, it was identified as Bur3p in the genetic screen that identified BUR6 (Prelich and Winston, 1993). Like Bur6p, Mot1p also interacts with TBP and is thought to repress transcription from some genes but to activate expression from others (Prelich and Winston, 1993; Muldrow et al., 1999). Our results show that there is a genetic interaction between MOT1 and RAT8, because a rat8-2 mot1-301 double mutant strain was completely unable to grow at 30°C (Figure 3). Thus, the genetic interactions between RAT8 and both SRB4 and MOT1, and the previously mentioned interactions with SSL1 and BUR6, follow a common pattern: combining rat8-2 mutations with mutations that affect positively-acting transcription factors partially suppresses the growth and mRNA export defects of rat8 ts strains, whereas combining rat8-2 with mutations which affect factors that play a repressing role in transcription led to synthetic lethality. Consistent with this reasoning, we also found that the synthetic lethality seen in rat8-2 bur6-1 and rat8-2 mot1-301 cells could be suppressed by conditions that reduce the rate of transcription such as low growth temperature, expression of the ssl1-t truncation, or addition of 6-AU (Figure 8).
We also examined genetic interactions between rat8 mutants and mutants affecting three factors thought to be involved in transcription elongation, Elp3p, Spt6p, and Hpr1p. No suppression of rat8 defects were found when we combined the rat8-2 allele with loss-of-function mutations affecting these elongation factors, showing that not every mutation that negatively affects transcription is able to suppress mutations in rat8. This supports the idea that suppression reflects specific events and interactions involving Rat8p that occur early during transcription, before elongation.
An alternate interpretation consistent with some of our data is that the maximum rate for export of mRNA through NPCs is reduced when Dbp5p function is compromised, but that the growth defects seen in dbp5 mutant strains can be suppressed if the amount of RNA requiring export is reduced. In contrast, according to this explanation, the growth defects in dbp5 mutant strains would be exacerbated by conditions that acted to increase the amount of mRNA needing export. This reasoning suggests that the growth defect is caused by an inability to export mRNP as fast as it is produced. Although there may be a regulatory relationship such that accumulation of nuclear poly(A)+ mRNA leads to decreased growth, the genetic and molecular analyses presented herein suggest that the interactions between Rat8p/Dbp5p and components of the transcription machinery are specific and direct and reflect interactions that are functionally important for mRNA export.
If Dbp5p were to interact with the transcriptional machinery during initiation, one would expect that it might be possible to show a direct interaction using chromatin immunoprecipitation. We have made multiple unsuccessful attempts to detect an association between Rat8p/Dbp5p and chromatin using published protocols, yet both positive and negative controls behaved as expected and reported by others (Heath and Cole, unpublished data). These negative results, in combination with the physical and genetic interactions found between Rat8p/Dbp5p and general transcription factors, are difficult to interpret. It may mean that Rat8p/Dbp5p interacts only very briefly with the transcription machinery. Alternatively, conditions used for chromatin immunoprecipitation, including use of formaldehyde as the cross-linking agent, may not be able to detect Dbp5p even if it is chromatin-associated. In fact, a recent study provides evidence for the association of Dbp5p with pre-mRNA during transcription. It was reported that the Dbp5p homolog in C. tentans (Ct-Dbp5) binds to the Balbiani ring mRNP (BR-mRNP) cotranscriptionally along the entire length of the BR-mRNP, accompanies these mRNPs to and through the nuclear pore and some remains associated with the BR-mRNPs in the cytoplasm (Zhao et al., 2002). These studies of the BR-mRNP, however, provide no information about the mechanism by which Ct-Dbp5p becomes associated with the BR-mRNP, nor do they show whether the interaction between Ct-Dbp5p and the BR-mRNP reflects a direct interaction with the BR-mRNA or interactions with protein components of the BR-mRNP.
Evidence for a link between mRNA biogenesis and mRNA export has been accumulating for many years. Because all pre-mRNA processing steps must be completed properly for mRNA export to occur, mRNA export likely depends on effective recruitment of pre-mRNA processing factors to the CTD of RNA pol II. However, a more direct link between transcription and export was proposed, based on the finding that the THO complex, involved in transcriptional elongation, recruits Yra1p and Sub2p to mRNA (Jimeno et al., 2002; Strasser et al., 2002). Both Yra1p and Sub2p are required for mRNA export. Our studies indicate that a sub2 mutant can be also be suppressed by ssl1-t (Figure 2). Interestingly, mutation of RAD3, which encodes another subunit of TFIIH, also partially suppresses the growth and mRNA export defects of a sub2 mutant strain (Jensen et al., 2001a).
The studies reported herein suggest an association of Dbp5p with mRNA at a very early step during transcription. Because Dbp5p is a DEAD-box protein, it is tempting to speculate that this early recruitment might be involved in rearranging RNA–protein interactions that form early during transcription. Soon after the nascent mRNA chain is emerge from the exit channel of the RNA polymerase II holoenzyme (∼25 nucleotides), the 5′ end of the mRNA is capped through the action of three enzymes: a pyrophosphatase, a guanylyl transferase, and a methyl transferase. The first two of these interact initially with the pol II CTD and subsequently modify the 5′ cap of the nascent pre-mRNA. Cet1p, the yeast pyrophosphatase enzyme involved in 5′ capping, was recently shown to function as a repressor of transcriptional reinitiation (Myers et al., 2002), suggesting that there is communication between the transcription initiation and RNA-processing machineries. Possibly, the capping machinery represses reinitiation until it is clear that capping has been performed successfully. Once the cap has been synthesized, the enzyme complex is replaced by the nuclear cap-binding complex, which accompanies the mRNA during export. One possibility is that Dbp5p participates in rearranging or releasing the capping enzymes, thereby allowing the cap-binding complex to bind to the mRNA and also allowing transcriptional reinitiation to occur.
Strasser and Hurt (2001) have proposed a model in which Sub2p is involved in recruiting Yra1p to mRNPs and, in a later step, Mex67p might replace Sub2p, targeting mRNA for export. We have also found genetic interactions between RAT8 and both YRA1 and SUB2 (Estruch, Hodge and Cole, unpublished data), suggesting a scenario where Dbp5p might facilitate release of Sub2p from the hnRNP complex, thereby allowing Mex67p to associate with Yra1p on the mRNP.
ACKNOWLEDGMENTS
We thank T. Donahue, A. Aguilera, G. Prelich, R. Young, D. Reinberg, J. Svejstrup, A. Ballis, and E. Hurt for providing plasmids and yeast strains and L. Myers for the antibody against Tfb1. This research was supported by a grant (to C.N.C.) from the National Institutes of General Medical Sciences, National Institutes of Health (GM-33998). F.E. is a recipient of a fellowship from the Ministerio de Educacion, Cultura y Deporte (Spain). We thank Larry Myers for valuable discussions and the members of the Cole laboratory for helpful discussions and for critically reading the manuscript.
Footnotes
DOI: 10.1091/mbc.E02–09–0602.
REFERENCES
- Amberg DC, Goldstein AL, Cole CN. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 1992;6:1173–1189. doi: 10.1101/gad.6.7.1173. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted and intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Cole CN, Heath CV, Hodge CA, Hammell CM, Amberg DC. Analysis of RNA export. Methods Enzymol. 2002;351:568–587. doi: 10.1016/s0076-6879(02)51869-3. [DOI] [PubMed] [Google Scholar]
- Damelin M, Silver PA. Mapping interactions between nuclear transport factors in living cells reveals pathways through the nuclear pore complex. Mol Cell. 2000;5:133–140. doi: 10.1016/s1097-2765(00)80409-8. [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Exinger F, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Henry NL, Kornberg RD. Relationship of CDK-activating kinase and RNA polymerase II CTD kinase TFIIH/TFIIK. Cell. 1994;79:1103–1109. doi: 10.1016/0092-8674(94)90040-x. [DOI] [PubMed] [Google Scholar]
- Feaver WJ, Svejstrup JQ, Bardwell L, Bardwell AJ, Buratowski S, Gulyas KD, Donahue TF, Friedberg EC, Kornberg RD. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75:1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- Fan HY, Merker RJ, Klein HL. High-copy-number expression of Sub2p, a member of the RNA helicase superfamily, suppresses hpr1-mediated genomic instability. Mol Cell Biol. 2001;21:5459–5470. doi: 10.1128/MCB.21.16.5459-5470.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadbois EL, Chao DM, Reese JC, Green MR, Young RA. Functional antagonism between RNA polymerase II holoenzyme and global negative regulator NC2 in vivo. Proc Natl Acad Sci USA. 1997;94:3145–3150. doi: 10.1073/pnas.94.7.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol. 2001;11:1716–1721. doi: 10.1016/s0960-9822(01)00532-2. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Holstege FC, Young RA, Struhl K. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goppelt A, Meisterernst M. Characterization of the basal inhibitor of class II transcription NC2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:4450–4455. doi: 10.1093/nar/24.22.4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goppelt A, Stelzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- Hammell CM, Gross S, Zenklusen D, Heath CV, Stutz F, Moore C, Cole CN. Coupling of termination, 3′ processing, and mRNA export. Mol Cell Biol. 2002;22:6441–6457. doi: 10.1128/MCB.22.18.6441-6457.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Wada T, Handa H, Winston F. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P, McCarthy T, Rosbash M, Parker R, Jensen TH. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature. 2001;413:538–542. doi: 10.1038/35097110. [DOI] [PubMed] [Google Scholar]
- Hillerin P, Parker R. Defects in the mRNA export factors Rat7p, Gle1p, Mex67p, and Rat8p cause hyperadenylation during 3′-end formation of nascent transcripts. RNA. 2001;7:753–764. doi: 10.1017/s1355838201010147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1-1 cells. EMBO J. 1999;18:5778–5788. doi: 10.1093/emboj/18.20.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Jensen TH, Boulay J, Rosbash M, Libri D. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr Biol. 2001a;11:1711–1715. doi: 10.1016/s0960-9822(01)00529-2. [DOI] [PubMed] [Google Scholar]
- Jensen TH, Patricio K, McCarthy T, Rosbash M. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol Cell. 2001b;7:887–898. doi: 10.1016/s1097-2765(01)00232-5. [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondon AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II, and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuldell NH, Buratowski S. Genetic analysis of the large subunit of yeast transcription factor IIE reveals two regions with distinct functions. Mol Cell Biol. 1997;17:5288–5298. doi: 10.1128/mcb.17.9.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Kim YJ. Requirement for a functional interaction between mediator components Med6 and Srb4 in RNA polymerase II transcription. Mol Cell Biol. 1998;18:5364–5370. doi: 10.1128/mcb.18.9.5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Krebber H, Silver PA. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 2001;15:1771–1782. doi: 10.1101/gad.892401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire M, Xie J, Meisterernst M, Collart MA. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol Microbiol. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Linder P. Quick guide: DEAD-box proteins. Curr Biol. 2000;10:R887. doi: 10.1016/s0960-9822(00)00857-5. [DOI] [PubMed] [Google Scholar]
- Linder P, Daugeron MC. Are DEAD-box proteins becoming respectable helicases? Nat Struct Biol. 2000;7:97–99. doi: 10.1038/72464. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. Pre-mRNA splicing, and mRNA export linked by direct interactions between UAP56, and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- Malik S, Roeder RG. Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) EMBO J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Meisels E, Gileadi O, Corden JL. Partial truncation of the yeast RNA polymerase II carboxyl-terminal domain preferentially reduces expression of glycolytic genes. J Biol Chem. 1995;270:31255–31261. doi: 10.1074/jbc.270.52.31255. [DOI] [PubMed] [Google Scholar]
- Muldrow TA, Campbell AM, Weil PA, Auble DT. MOT1 can activate basal transcription in vitro by regulating the distribution of TATA binding protein between promoter and nonpromoter sites. Mol Cell Biol. 1999;19:2835–2845. doi: 10.1128/mcb.19.4.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myer VE, Young RA. RNA polymerase II holoenzymes and subcomplexes. J Biol Chem. 1998;273:27757–27760. doi: 10.1074/jbc.273.43.27757. [DOI] [PubMed] [Google Scholar]
- Myers LC, Lacomis L, Erdjument-Bromage H, Tempts P. The yeast capping enzyme represses RNA polymerase II transcription. Mol Cell. 2002;10:883–894. doi: 10.1016/s1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- Nasmyth KA, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19:753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Nonet M, Sweetser D, Young RA. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2alpha homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prelich G, Winston F. Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics. 1993;135:665–676. doi: 10.1093/genetics/135.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- Reed R, Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- Reed R, Magni K. A new view of mRNA export: separating the wheat from the chaff. Nat Cell Biol. 2001;3:E201–E204. doi: 10.1038/ncb0901-e201. [DOI] [PubMed] [Google Scholar]
- Segref A, Sharma K, Doye V, Hellwig A, Huber J, Luhrmann R, Hurt E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr255p. EMBO J. 1999;18:5761–5777. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19:410–420. doi: 10.1093/emboj/19.3.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413:648–652. doi: 10.1038/35098113. [DOI] [PubMed] [Google Scholar]
- Strasser K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- Stutz F, Bachi A, Doerks T, Braun IC, Seraphim B, Wilm M, Bork P, Izaurralde E. REF, an evolutionarily conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner NK, Linder P. DExD/H box RNA helicases. from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J. 1998;17:2651–2662. doi: 10.1093/emboj/17.9.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valay JG, Simon M, Dubois MF, Bensaude O, Facca C, Faye G. The KIN28 gene is required both for RNA polymerase II mediated transcription and phosphorylation of the Rpb1p CTD. J Mol Biol. 1995;249:535–544. doi: 10.1006/jmbi.1995.0316. [DOI] [PubMed] [Google Scholar]
- Wang Z, Buratowski S, Svejstrup JQ, Feaver WJ, Wu X, Kornberg RD, Donahue TF, Friedberg EC. The yeast TFB1 and SSL1 genes, which encode subunits of transcription factor IIH, are required for nucleotide excision repair and RNA polymerase II transcription. Mol Cell Biol. 1995;15:2288–2293. doi: 10.1128/mcb.15.4.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis K. Nucleocytoplasmic transport: cargo trafficking across the border. Curr Opin Cell Biol. 2002;14:328–335. doi: 10.1016/s0955-0674(02)00337-x. [DOI] [PubMed] [Google Scholar]
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- Yoon H, Miller SP, Pabich EK, Donahue TF. SSL1, a suppressor of a HIS4 5′-UTR stem-loop mutation, is essential for translation initiation and affects UV resistance in yeast. Genes Dev. 1992;6:2463–2477. doi: 10.1101/gad.6.12b.2463. [DOI] [PubMed] [Google Scholar]
- Zhao J, Jin SB, Bjorkroth B, Wieslander L, Daneholt B. The mRNA export factor Dbp5 is associated with Balbiani ring mRNP from gene to cytoplasm. EMBO J. 2002;21:1177–1187. doi: 10.1093/emboj/21.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]