Abstract
We have identified a gene encoding RGS domain-containing protein kinase (RCK1), a novel regulator of G protein signaling domain-containing protein kinase. RCK1 mutant strains exhibit strong aggregation and chemotaxis defects. rck1 null cells chemotax ∼50% faster than wild-type cells, suggesting RCK1 plays a negative regulatory role in chemotaxis. Consistent with this finding, overexpression of wild-type RCK1 reduces chemotaxis speed by ∼40%. On cAMP stimulation, RCK1 transiently translocates to the membrane/cortex region with membrane localization peaking at ∼10 s, similar to the kinetics of membrane localization of the pleckstrin homology domain-containing proteins CRAC, Akt/PKB, and PhdA. RCK1 kinase activity also increases dramatically. The RCK1 kinase activity does not rapidly adapt, but decreases after the cAMP stimulus is removed. This is particularly novel considering that most other chemoattractant-activated kinases (e.g., Akt/PKB, ERK1, ERK2, and PAKa) rapidly adapt after activation. Using site-directed mutagenesis, we further show that both the RGS and kinase domains are required for RCK1 function and that RCK1 kinase activity is required for the delocalization of RCK1 from the plasma membrane. Genetic evidence suggests RCK1 function lies downstream from Gα2, the heterotrimeric G protein that couples to the cAMP chemoattractant receptors. We suggest that RCK1 might be part of an adaptation pathway that regulates aspects of chemotaxis in Dictyostelium.
INTRODUCTION
G protein-coupled receptors (GPCRs) mediate the signal transduction of diverse extracellular stimuli, including hormones, chemokines, neurotransmitters, light, and odorants (Carman and Benovic, 1998; Marinissen and Gutkind, 2001). Binding of an agonist to its GPCR results in the dissociation of a heterotrimeric G protein into the Gα and Gβγ subunits, which in turn regulate downstream effector molecules (Hamm, 1998). The GPCR-mediated signaling then rapidly adapts. This adaptation is achieved at the level of GPCR by receptor desensitization, at the level of the G protein by its inactivation, or through the independent adaptation of the downstream effector pathways.
Regulators of G protein signaling (RGS) proteins bind to the GTP form of the Gα subunit of heterotrimeric G proteins and can act as Gα subunit GTPase-activating proteins (GAPs), in which case they increase the intrinsic Gα subunit GTPase activity, thus attenuating signaling (Hepler, 1999; Sierra et al., 2000). Although most of the RGS proteins tested are GAPs for Gα proteins, in some cases RGS proteins carry other effector domains (Zhong and Neubig, 2001) that provide additional functions. In such proteins, the RGS domain can serve as a Gα recognition domain with or without GAP activity. For example, the RGS domains in p115-Rho-GEF and Axin play active roles in transmitting receptor signals to downstream effectors (Kozasa et al., 1998; Spink et al., 2000). d-AKAP2, a protein kinase A (PKA)-anchoring protein containing a putative RGS domain, may provide a link between PKA and the plasma membrane signaling machinery (Huang et al., 1997). Seven G protein receptor kinase (GRK) family members contain an N-terminal atypical RGS domain (Siderovski et al., 1996; Penn et al., 2000). Although very little is known about the function of the RGS domain in most of these proteins, it has been shown that the RGS domain of GRK2 binds to Gαq/11 and inhibits Gαq-mediated activation of phospholipase C-β (Carman et al., 1999).
In Dictyostelium, the transition from vegetative growth to multicellular development is triggered by starvation and subsequent production of the chemoattractant and morphogen extracellular cAMP (Aubry and Firtel, 1999). cAMP binds to and activates the G protein-coupled serpentine receptors (cARs) that initiate a series of integrated G protein-dependent and -independent signal transduction events that control developmental responses, including chemotaxis of cells toward the chemoattractant cAMP (Chen et al., 1996; Parent and Devreotes, 1996; Aubry and Firtel, 1999; Chung and Firtel, 1999). cAR1 is the major cAR that mediates chemotaxis to cAMP and the activation of adenylyl cyclase during aggregation and is coupled to the heterotrimeric G protein containing the Gα protein subunit Gα2.
Our understanding of chemotaxis in Dictyostelium derives from careful genetic and biochemical analysis of signaling components regulating this pathway combined with the ability to perform single-cell chemotaxis assays. Recent studies have focused on phosphatidylinositol 3-kinase (PI3K) and its downstream effector pathways that are required for regulating cell polarity and motility (Parent and Devreotes, 1999; Firtel and Chung, 2000; Chung et al., 2001a; Comer and Parent, 2002; Funamoto et al., 2002; Iijima et al., 2002; Iijima and Devreotes, 2002). Through the use of gene knockouts and green fluorescent protein (GFP) fusions, the function and changes in the subcellular localization have provided key insights into the pathways controlling chemotaxis. In addition to PI3K and its effectors, other pathways important for chemotaxis include the mitogen-activated protein kinase kinase 1/extracellular signal-regulated kinase 1 (ERK1) mitogen-activated protein (MAP) kinase cascade and interacting proteins; Ras; PAKa; and myosin heavy chain kinases that regulate myosin II assembly; the PKA regulatory pathway; guanylyl cyclase; Rac family GTPases; and the actin polymerization machinery and actin binding proteins (Parent and Devreotes, 1999; van Es and Devreotes, 1999; Firtel and Chung, 2000; Chung et al., 2001a). In addition to the pathways controlling chemotaxis, Dictyostelium cells must integrate other chemoattractant-mediated pathways such as those controlling the activation of adenylyl cyclase.
The adaptation of signaling pathways plays an equally essential role in the regulation of cellular responses. All of the previously examined cAR1/Gα-mediated pathways rapidly adapt, often with very different kinetics. For example, chemoattractant-stimulated adenylyl cyclase activity peaks at 60–90 s, whereas the first peak of actin polymerization is at ∼5 s; ERK1 activation is at ∼15 s; ERK2 activation is at ∼50 s; PI3K-dependent pleckstrin homology (PH) domain localization and cGMP accumulation are maximal at ∼8 and 10 s, respectively; and Akt/PKB and PAKa activities are maximal at 15 and 30 s after stimulation, respectively (van Es and Devreotes, 1999; Firtel and Chung, 2000). Except for the role of the phosphatidylinositol 3′ lipid phosphatase PTEN, which is required for the adaptation of the chemoattractant-mediated PI3K pathways, little is known about the adaptation pathways for any of the responses (Funamoto et al., 2002; Iijima and Devreotes, 2002). As in other systems, the C-terminal tail of the G protein-coupled receptor cAR1 is phosphorylated; however, this seems to have little impact on the overall adaptation of cAR1-mediated pathways (Caterina et al., 1995). Cells expressing a cAR1 receptor lacking the tail phosphorylation sites do not exhibit a ligand-mediated reduction in ligand affinity and are partially defective in the activation of STATa, but show a normal adaptation of adenylyl and guanylyl cyclase (Vaughan and Devreotes, 1988; Caterina et al., 1995; Kim et al., 1997).
Herein, we describe RGS domain-containing protein kinase (RCK1), a novel kinase containing an RGS domain. We demonstrate that rck1 null cells chemotax more rapidly than wild-type cells, whereas overexpression of RCK1 leads to reduced chemotaxis, suggesting that RCK1 plays a negative regulatory role in controlling chemotaxis. Furthermore, we show that RCK1 kinase activity is stimulated in response to cAMP. Unlike other kinases and signaling pathways activated by chemoattractant stimulation, RCK1 activity remains elevated unless the stimulus is removed. These findings suggest that RCK1 might be a candidate for controlling the adaptation of some chemoattractant-mediated pathways.
MATERIALS AND METHODS
Materials
The cAR1 antibody was a gift from the Devreotes laboratory (John Hopkins University School of Medicine, Baltimore, MD). Sodium orthovanadate, β-glycerophosphate, aprotinin, and leupeptin were purchased from Sigma-Aldrich (St. Louis, MO). Histone H2B and myelin basic protein (MBP) were purchased from Roche Diagnostics (Indianapolis, IN). [32P]γ-ATP was from ICN Pharmaceuticals (Costa Mesa, CA). Protein A-Sepharose CL-4B was from Amersham Biosciences (Piscataway, NJ. Protein G plus agarose was from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell Culture and Development of Dictyostelium Cells
Wild-type Dictyostelium strain Kax-3 was used to generate various mutant strains. Cells were grown in nutrient medium (Watts and Ashworth, 1970) at 23°C either in shaking culture or on a Petri dish. For the developmental study, cells were washed twice in 12 mM Na+/K+ phosphate buffer and plated at various densities on agar plates.
Library Construction and Screening
The C-terminal half of RCK1 was obtained by screening a 12- to 16-h developmental λZAP library using a cDNA fragment corresponding to the kinase domain of a putative Dictyostelium kinase as a probe (Dictyostelium database sequence IICCP2E13713). To obtain the N terminus of RCK1, a mini-genomic library was constructed. Briefly, the genomic DNA from Kax-3 cells was digested with HindIII, which cuts upstream of RCK1, and SpeI, which cuts immediately after the RGS domain. The DNA was size fractionated on an agarose gel. DNA fragments of the right size were recovered, ligated into a vector, and transformed into Escherichia coli. All library screenings were done using standard protocols (Sambrook et al., 1989) with slight modifications that reduced the stringency by lowering the concentration of formamide to 40% and by performing the hybridizations at 37°C.
Generation of rck1 Null Cells and Cells Expressing Wild-Type and Mutant RCK1s
The rck1 null mutant was obtained by a gene replacement technique based on that of Manstein et al. (1989) but with a blasticidin resistance cassette (Sutoh, 1993). The targeting construct was made by inserting a Bsr cassette in the second BglII site of RCK1.
To make wild-type RCK1 expression constructs, RCK1 was tagged at the N terminus with either the myc epitope or the myristoylation signal of chicken c-Src. Myristoylation-site-tagged RCK1 was made by cloning the RCK1 coding region in-frame downstream of the myristoylation signal in premade vectors containing this tag (Meili et al., 1999). The myc-tagged RCK1 was made similarly except that a myc tag instead of myristoylation-site tag was incorporated in the linker. The primers used to amplify the RCK1 coding region are 5′-GTTTTTGATATCTGAAATGAAAACATCAAAGGATAGT and 5′-GTTTTCTCGAGTATTAT TATTTATTTTTTGGCTGTGC. For making RCK1 mutations, site-directed mutagenesis was done by a two-step polymerase chain reaction approach by using overlapping primers containing the desired mutations (Li and Wilkinson, 1997). The DNA fragments containing mutations were used to replace the corresponding fragments in the wild-type RCK1 constructs. All constructs were sequenced and subcloned into Exp4 (+) expression vectors and transformed KAx-3 or rck1 null cells.
All transformations were done as described previously (Nellen et al., 1987). Transformants were selected with 10–20 μg/ml G418, and clones were isolated by plating cells on DM agar (12 mM Na+/K+ phosphate buffer pH 6.1, 1% Bacto peptone, 0.2% d-glucose, 1.5% agar) containing 40 μg/ml G418. In the case of making rck1 null cells, cells were selected with 7.5 μg/ml blasticidin.
Chemotaxis Assay
Aggregation-competent cells were made by pulsing cells with cAMP as described previously (Meili et al., 1999) except that cells were pulsed at 6-min intervals. Pulsed cells of 4 × 105cells/ml were plated onto a small dish with a hole covered by a 0.17-mm glass coverslip at 40 μl/dish and allowed to adhere for 15 min. A micropipette filled with 150 μM cAMP was positioned close to cells by using a micromanipulator (Eppendorf Patchman, Fisher Scientific, Pittsburgh, PA), and the response of cells was recorded at 6-s intervals with a time-lapse video recorder and NIH Image software. Cell movement was analyzed using the DIAS program (Wessels et al., 1998).
Immunofluorescence Microscopy
Pulsed cells were allowed to settle onto glass coverslips for 30 min and stimulated for different time periods by submerging the coverslips into 20 μM cAMP solution. Cells were quickly fixed and permeabilized in 100% cold methanol for 20 min at −20°C. Cells were then washed and incubated with 0.5 μg/ml anti-myc monoclonal antibody (Invitrogen, Carlsbad, CA) in phosphate-buffered saline containing 0.5% bovine serum albumin and 0.05% Tween 20 for 1 h. Cells were incubated with fluorescein isothiocyanate-labeled anti-mouse IgG antibody for 1 h, washed, and mounted for observation with a 60× oil immersion lens on a Microphot-FX microscope (Nikon, Tokyo, Japan).
Kinase Assay
Log phase cells were pulsed as usual. After pulsing, cells were collected by centrifugation and resuspended at a density of 2–3 × 107cells/ml. Cells were resensitized by bubbling air through the cell suspension for 10 min (Devreotes et al., 1987; Ma et al., 1997). The kinase assay was performed similarly to the assay for chemoattractant-stimulated Akt/PKB activity described previously (Meili et al., 1999). Cells were then stimulated with 100 μl of cAMP and 500 μl of cells were taken at each time point and lysed immediately by mixing with an equal volume of 2× lysis buffer containing 50 mM Tris, pH 7.6, 200 mM NaCl, 20 mM NaF, 2 mM vanadate, 50 mM β-glycerophosphate, 6 mM sodium pyrophosphate, 2 mM EDTA, 2 mM EGTA, 4 μg/ml leupeptin, 4 μg/ml aprotinin, 2% NP-40, 20% glycerol, and 2 mM dithiothreitol. The lysate was precleared by centrifugation. One microliter of antibody (1 μg/μl) was added and incubated with agitation in a cold room for 1 h. The formed immune complexes were collected with 50 μl of a 1:1 slurry of either protein A or protein G beads in lysis buffer by incubation under agitation for 1 h at 4°C. The beads were washed twice with lysis buffer and twice with kinase buffer (25 mM MOPS pH 7.4 at room temperature, 25 mM glycerophosphate, 20 mM magnesium chloride, and 1 mM dithiothreitol). The kinase activity was measured using either MBP or Histone H2B protein as substrate in a reaction in 60 μl of kinase buffer containing 5 μCi of [32P]γ-ATP, 5 μM cold ATP, and 5 μg of MBP. The reactions were stopped by adding 20 μl of 4× SDS buffer and boiling for 5 min. Samples were then resolved by SDS-PAGE (12.5%), blotted onto polyvinylidene difluoride membrane (Millipore, Bedford, MA), and exposed to film.
RESULTS
Identification of RCK1
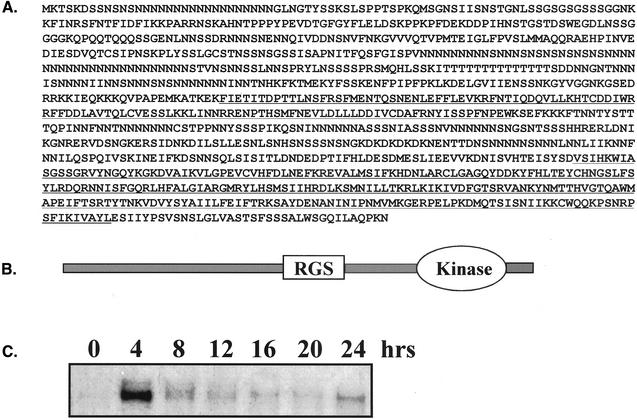
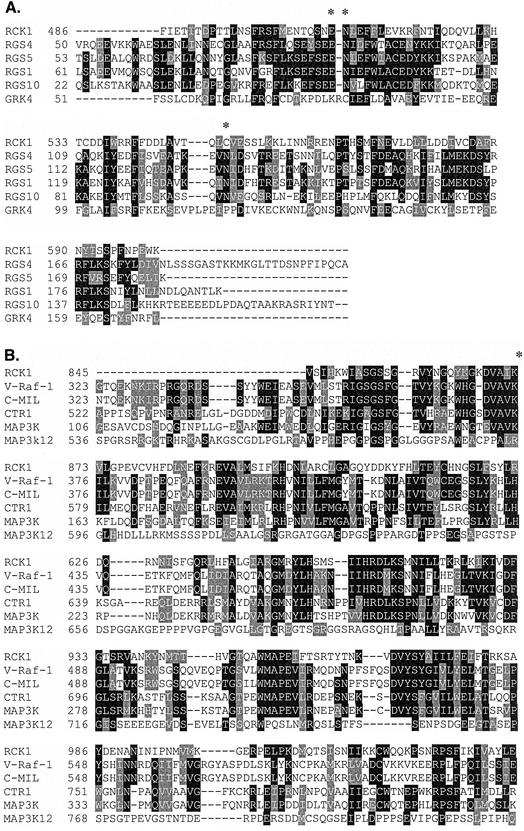
RCK1 was identified by a low-stringency hybridization screen as described in MATERIALS AND METHODS. The RCK1 open reading frame encodes a protein of 1123 amino acids (Figure 1A; GenBank accession no. AY163574). Structure analysis using SMART predicted an RGS domain in the middle of the open reading frame and a dual-specificity kinase at the C terminus of the protein (Figure 1B). The long N-terminal domain does not show homology to any other proteins in the databases. The RGS domain of the Dictyostelium RCK1 exhibits a moderate homology of ∼40–50% to known RGS domains, with highest homology to that of Xenopus RGS4 (25% identity and 49% homology; Figure 2A). The RCK1 kinase domain exhibits considerable homology to a family of mitogen-activated protein kinase kinase kinases that includes MAP3KΔ-1, murine Raf-1, and Arabidopsis CTR1 (Figure 2B). Some GRKs, which are important players in receptor desensitization and adaptation (Ferguson and Caron, 1998; Penn et al., 2000), contain an RGS domain in addition to the receptor kinase domain; however, these kinase domains are not homologous to that of RCK1.
Figure 1.
Sequence analysis of Dictyostelium RCK1. (A) Deduced amino acid sequence of RCK1 from the 3990 base pairs DNA clone. The underlined region in the middle is the RGS domain. The kinase domain at the C terminus is also underlined. (B) Schematic diagram of RCK1 domain structure. (C) Northern blot showing the developmental time course of RCK1 expression. Total RNA of 8 μg/sample was resolved on a 1.0% denaturing agarose gel, blotted, and probed as described previously (Datta and Firtel, 1987). The 0-h time point is for vegetative cells.
Figure 2.
Sequence comparison of RCK1 with other proteins. (A) Alignment of the RGS domain of RCK1 with that of Xenopus RGS4, human RGS5, mouse RGS1, human RGS10, and mouse GRK4. The GenBank accession numbers for these genes are Xenopus RGS4, AF263451; human RGS5, XM_002185; mouse RGS1, NM_015811; human RGS10, XM_049798; and mouse GRK4, AF040745. The positions marked by “*” are those mutated in RCK1 in this study. (B) Comparison of the kinase domain of RCK1 with that of other kinases including mouse v-raf-1, chick C-MIL, Arabidopsis CTR1, Arabidopsis MAP3K Delta-1 (MAP3K in the figure), and human MAP3K12. The GenBank accession numbers for these genes are mouse v-raf-1, NM_029780; chick C-MIL, X07017; Arabidopsis CTR1, L08790; Arabidopsis MAP3K Delta-1, Y14199; and human MAP3K12, XM_006800.
To examine the timing of RCK1 expression during the Dictyostelium developmental cycle, we performed RNA blot analysis. As depicted in Figure 1C, RCK1 shows a very low level of mRNA expression in vegetative cells, which increases dramatically at 4 h after starvation and then rapidly falls to very low levels throughout the remainder of development.
Analysis of the Phenotype of rck1 Null Cells
rck1 null strains were generated by gene disruption via homologous recombination as described in MATERIALS AND METHODS and confirmed by Southern blot analysis (our unpublished data). Northern blot analysis of rck1null cells revealed no detectable RCK1 mRNA (our unpublished data). Seven independent rck1 null strains were identified, and initial studies indicated that all strains had an indistinguishable developmental phenotype. A single clone was selected for subsequent analysis.
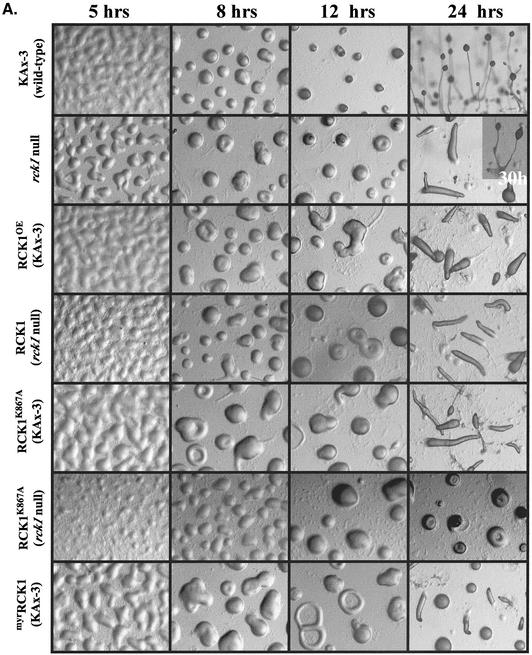
Compared with wild-type cells, rck1 null cells formed multicellular aggregates slightly faster when the cells were plated on a nonnutrient agar surface. In addition, the mounds were larger than those of wild-type cells at a number of plating densities (Figure 4A; other densities not shown). By 5 h after starvation, rck1 null cells had already formed loose mounds; however, the development of rck1 null cells was significantly delayed after mound formation and slugs were identified only at ∼24 h. Normally shaped fruiting bodies formed at ∼30 h. Expression of RCK1 from the constitutive Actin 15 promoter in wild-type cells resulted in significantly delayed development. Aggregation and mound formation occurred more slowly than in wild-type cells, and slugs were not formed until ∼24 h in development. Normally shaped fruiting bodies developed at 30–36 h. Expression of this construct in rck1null cells resulted in a similar although slightly less severe phenotype. Because Dictyostelium transformants obtained using G418 selection have a high copy number of the vector, levels of expression of the exogenous protein from the Actin 15 promoter are usually higher than those of the endogenous protein. This probably accounts for the similarity of the phenotypes of rck1 null and wild-type strains expressing exogenous RCK1. We cannot rule out the possibility that differences in the temporal pattern of expression of exogenous RCK1 from the constitutive Actin 15 promoter compared with the highly regulated developmental pattern of expression of endogenous RCK1, rather than the expression level of the protein, account for the observed developmental defect of RCK1OE cells. We also cannot discount that the myc tag may result in abnormal protein function, although we expect that it is unlikely that the myc tag is responsible for the defects, as the kinase activity of myc-RCK1 is rigorously activated in response to cAMP stimulation (see below). These analyses indicated that both the lack of expression and the overexpression of RCK1 lead to developmental defects.
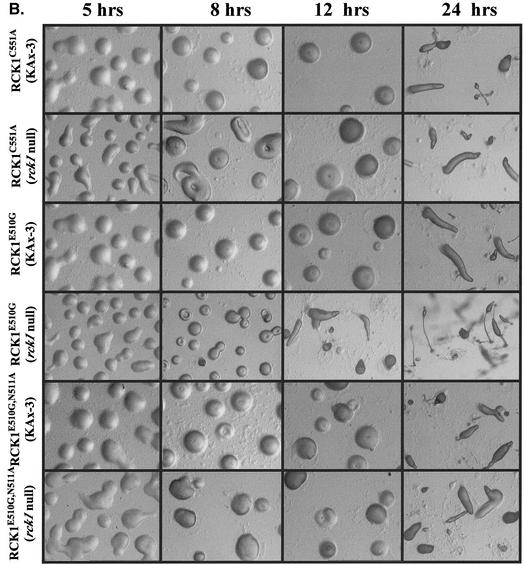
Figure 4.
Developmental phenotypes of rck1 null cells and cells expressing wild-type and mutant RCK1. Cells grown in axenic medium were washed twice with Na+/K+ phosphate buffer and plated on nonnutrient agar plates. Photographs were taken at various developmental stages. (A) Development phenotypes of rck1 null, RCK1OE, myrRCK1, and RCK1K867A cells. Wild-type RCK1 and mutant RCK1K867A were expressed in both wild-type and null backgrounds. (B) Development phenotypes of RCK1 mutants with mutations in the RGS domain in both wild-type and null backgrounds. All photographs were taken at the same magnification..
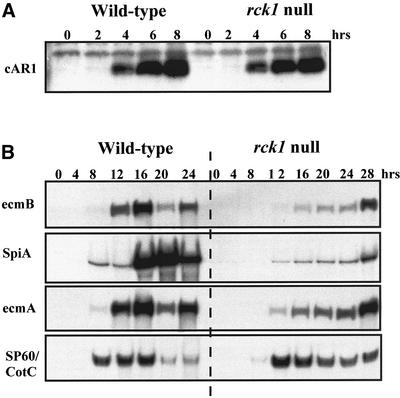
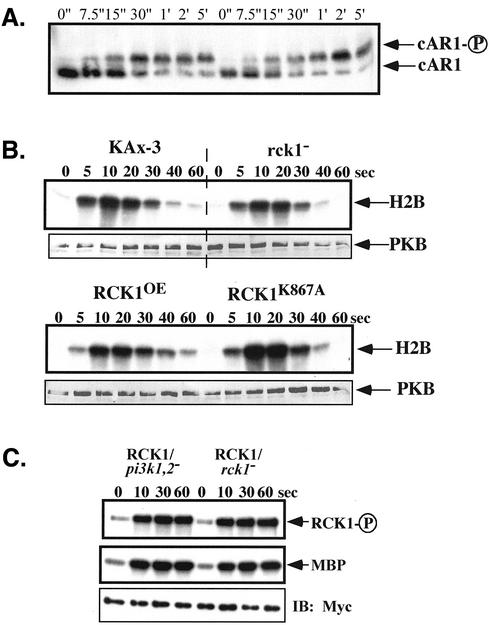
To better understand the temporal delay exhibited by rck1 null cells, we examined the expression of a series of developmental markers that are expressed during the aggregation and multicellular stages in development. Figure 3A shows the expression of the cAR1 protein. The protein is visibly induced by 4 h of development and is maximal at 6–8 h. In rck1 null cells, cAR1 expression is similar to that in the wild-type cells.
Figure 3.
Expression of developmental markers in wild-type KAx-3 and rck1 null cells. (A) Western blot analysis of cAR1 expression in response to pulsing. Cells were pulsed with 30 nM cAMP and samples were taken at indicated time points, lysed, and subjected to Western blot analysis by using polyclonal cAR1 antibodies. (B) Northern blot analysis of total RNA prepared from wild-type KAx-3 and rck1 null cells at different stages of development as indicated. Blots were hybridized with probes for ecmA (prestalk specific), ecmB (pstB and ALC specific), spiA (spore specific), and SP60/CotC (prespore specific).
We also examined cell-type–specific markers, including the prespore-specific gene SP60/CotC, the spore-specific gene SpiA, and two prestalk-specific genes, ecmA and ecmB (Figure 3B). SP60/CotC exhibits a delay of induction of expression by ∼4 h in rck1 null cells compared with the time of expression in wild-type cells. In rck1null cells, SP60/CotC RNA levels do not decrease rapidly at 16 h, as they do in wild-type cells, but remain at high levels through 28 h in development. The prestalk-specific genes ecmA and ecmB also have delayed developmental expression. This delay is also exhibited by SpiA, a spore-specific marker. These findings are consistent with the observed delays in the morphogenesis of this strain.
Effect of Point Mutations within RGS and Kinase Domains on Morphogenesis
To better understand the potential role of the subdomains of RCK1 in regulating RCK1 function, we made a series of amino acid substitutions in the RGS domain in residues known to interact with Gα subunits in RGS family members and the kinase domain. The mutant proteins were expressed as myc-tagged proteins in both wild-type and rck1 null cells and the effects on RCK1 translocation to the plasma membrane, RCK1 kinase activity, chemotaxis, and multicellular development were measured. Western blot analysis of clones showed that all expressed similar levels of RCK1 protein (our unpublished data; Figure 7). Expression of kinase-dead RCK1 (RCK1K867A) in wild-type cells resulted in a phenotype that was very similar to that of rck1 null cells. Aggregation occurred slightly more rapidly than in wild-type cells, but the cells arrested temporarily at the mound stage, with fruiting bodies appearing at ∼30 h (Figure 4A, 30-h time point is not shown). Expression of the kinase-dead RCK1 in rck1 null cells produced an even more severe phenotype than that of rck1 null cell (Figure 4A). Cells formed aggregates more slowly than wild-type cells and remained arrested at the mound stage with no formation of slugs and fruiting bodies within the time course of the experiments. Our results suggest that RCK1K867A functions as a dominant interfering (negative) protein. The stronger phenotype observed from the expression of RCK1K867A in rck1 null cells suggests that the kinase-dead RCK1 may interfere with a binding partner. The results also indicate that the RCK1 kinase domain is essential for RCK1 function.
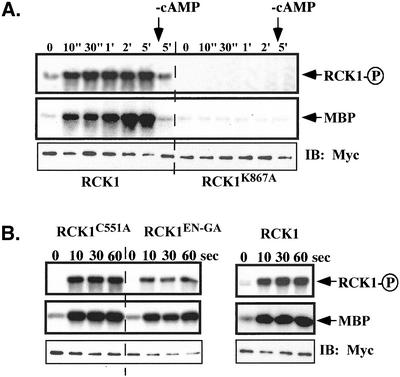
Figure 7.
Activation of RCK1 by cAMP stimulation. Aggregation-competent cells made by pulsing (see MATERIALS AND METHODS) were stimulated with cAMP and aliquots were taken at the time points indicated. Cells were lysed, RCK1 or its mutant was immunoprecipated with anti-myc monoclonal antibody, and kinase activity was measured as described in MATERIALS AND METHODS. (A) RCK1 kinase activity using MBP as a substrate before (0") and after stimulation (10"-5′). The kinase-dead mutant RCK1 K867A was used as a control and no activation was observed in these cells. To test the activity of RCK1 after the removal of cAMP, a sample was taken at the 1-min time point and 0.05 unit of PDE (phosphodiesterase; Sigma-Aldrich) was added. Cells were quickly washed twice with 12 mM Na+/K+ phosphate buffer and lysed at 5 min after addition of cAMP. The RCK1 kinase activity was assayed in the same experiment (marked −cAMP). Top, autophosphorylated RCK1. Middle, RCK1-phosphorylated MBP. To examine the immunoprecipitated RCK1 protein level, a Western blot was carried out on the same blot used for the kinase assay (bottom). (B) Kinase assay on RCK1 with mutations in the RGS domain including C551A and EN510-511GA. Top, autophosphorylated RCK1. Middle, RCK1-phosphorylated MBP. Bottom, immunoprecipitated RCK1 protein level. Wild-type RCK1 was used as a control.
To obtain insight into the role of the RGS domain, we made mutations of residues previously demonstrated to play a central role in the interactions of other RGS domains with Gα subunits and/or RGS GAP activity. The crystal structure of the RGS4-Gαi1 complex shows that Glu87, Asn88, and Asn128 in RGS4 are involved in the interaction with Gαi1 (Tesmer et al., 1997). Cysteine 551 (Cys551) is present in a highly conserved domain and corresponds in position to a conserved Asn/Ser residue in mammalian RGS proteins (Asn128 in RGS4 and Asn130 in RGS16). Mutation of this residue in RGS4 and RGS16 significantly reduces RGS GAP activity and Gα binding (Posner et al., 1999; Wieland et al., 2000). Residue Glu89 in RGS16 (corresponding to Glu87 in RGS4) interacts with Lys210 in Gαi1. Mutation of Glu89 to Lys does not affect GAP activity but reduces affinity of RGS16 to Gαi1 by ∼100-fold, whereas mutating Asn90 to Glu in RGS16 (corresponding to Asn88 in RGS4) strongly attenuates RGS16's GAP activity (Wieland et al., 2000). A separate study showed that a double mutation in RGS16 (E89G and N90A) eliminates RGS16 binding to several Gα protein subunits (Chen et al., 1997).
To examine the role of the RGS domain, we made the corresponding substitutions in RCK1 and examined the effects of expressing the mutant proteins in wild-type and rck1 null cells on development. Western blot analysis shows that the RCK1 constructs are expressed at similar levels (our unpublished data; see below). Wild-type cells expressing RCK1E510G or RCK1E510G,N511A (corresponding to RGS16E89G andRGS16N90A) exhibited normal kinetics of aggregation, but the cells arrested for a period of time at the mound stage and were at the slug or early culminant stage by 24 h, similar to the phenotype of wild-type cells overexpressing wild-type RCK1 (Figure 4B). A similar effect was observed for RCK1C551A. Expression of RCK1E510G in rck1 null cells seemed to complement, at least at the morphological level, normal Dictyostelium morphogenesis. The cells aggregated with approximately the same kinetics as wild-type cells and showed only a minimal delay after mound formation. Slugs were present at 16 h and culminants were present at 24 h, although these were slightly delayed compared with wild-type cells developed in parallel on the same plates. The observation that RCK1E510G expression in rck1 null cells resulted in an almost wild-type phenotype, which was not seen for wild-type RCK1 expressed at the same level, suggests that the RCK1E510G mutation might have reduced RCK1 activity. Neither RCK1E510G,N511A nor RCK1C551Acomplemented the rck1 null phenotype; however, because both mutant proteins exhibited nearly wild-type activation of kinase activity on cAMP stimulation (see below), we expect that the phenotypes of these strains are due to overexpression of a functional RCK1 protein, similar to observations of the overexpression of wild-type RCK1.
rck1 Null and RCK1 Overexpressing (RCK1OE) Cells Exhibit Chemotaxis Defects
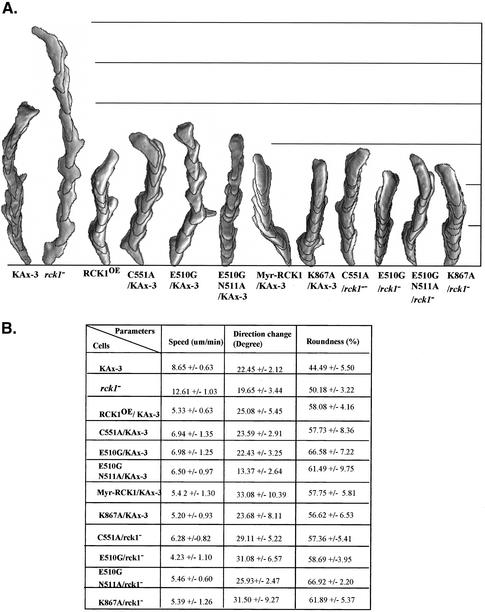
One possible explanation for the developmental defects exhibited by the RCK1 mutant strains is defects in the ability to chemotax and/or undergo morphogenesis. We therefore examined the ability of wild-type and RCK1 mutant strains to chemotax toward a micropipette containing cAMP. Using DIAS computer software, we analyzed the cells' rate of movement, directional change, and roundness (Soll and Voss, 1998; Wessels et al., 1998). One way of visualizing these data is to examine overlapping images of cells that have moved over an interval of time (Figure 5). A change in the speed of movement toward the chemoattractant source can be readily observed by the increase in the distance between images taken at 1-min intervals and those seen in wild-type cells. The shape of the cell provides an indication of lateral pseudopodia and/or loss of cell polarity. As shown in Figure 5, wild-type cells move in an essentially straight line toward the chemoattractant source with a speed of ∼8.6 μm/min (as measured by the rate of movement of the cell's centroid over time). rck1 null cells exhibit an increased rate of movement to ∼12.6 μm/min, whereas RCK1-overexpressing cells have a significant decrease in speed (5.3 μm/min). These cell movement defects are interesting, considering rck1 null and RCK1OE cells displayed similar developmental defects. Overexpression of RCK1 carrying the point mutations in the RGS domain in wild-type cells led to a loss in chemotaxis speed, although the decrease in speed was not as great as that caused by the overexpression of wild-type RCK1, suggesting that the RGS1 point mutants have decreased activity compared with the wild-type protein. As expected from the developmental phenotype, overexpression of the kinase-dead RCK1 (RCK1K867A) resulted in reduced speed. It is possible that like RCK1K867A, RCK1 proteins carrying point mutations within the RGS domain may exhibit similar dominant negative phenotypes. Overexpression of RCK1 may also inhibit chemotaxis by affecting the activation of other downstream pathways through the RGS domain acting as a GAP for a Gα protein subunit required during aggregation. Interestingly, expression of the mutant RCK1 proteins had little effect on directional change, in contrast to many known signaling proteins that affect chemotaxis and often cause a decrease in the efficiency (chemotaxis index) with which cells move toward the needle (Kitayama et al., 1998; Peracino et al., 1998; Firtel and Chung, 2000; Wessels et al., 2000). There is some increase in roundness of the mutant strains compared with the wild type, suggesting some decrease in cell polarity.
Figure 5.
Chemotaxis analysis of rck1 null cells and cells expressing wild-type or mutant RCK1. Cells were washed and pulsed every 6 min with 30 nM cAMP (see MATERIALS AND METHODS; Meili et al., 1999). Cells were plated on a Petri dish specifically made for observing cells with an inverted microscope and placed close to a micropipette filled with 150 μM cAMP. Cell movement was recorded with NIH Image software and analyzed using the DIAS program. A comparison of cell shape, direction change, and speed of movement of different cells is depicted. (A) A representative cell from each type of cells is shown. The image was obtained by superimposing nine frames with 1-min intervals between frames. (B) Computed data indicate the speed, direction change, and polarity of cells.
RCK1 Translocates to Plasma Membrane in Response to Chemoattractant Stimulation
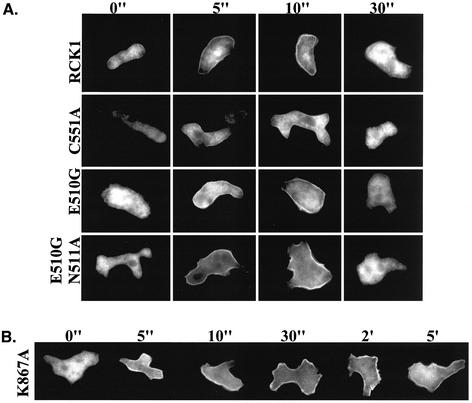
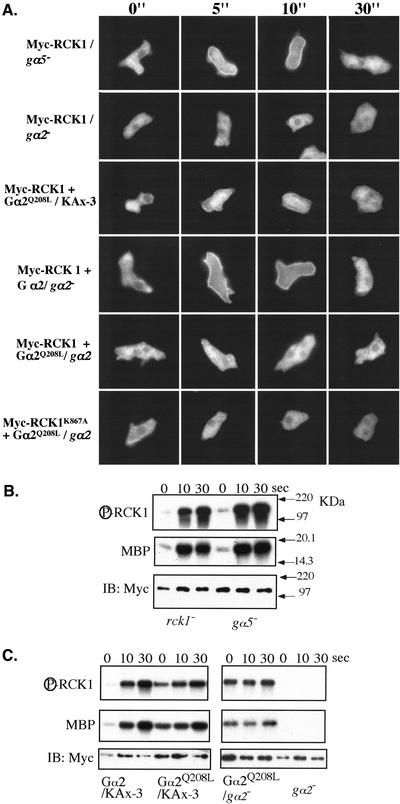
Due to the presence of an RGS domain, it is possible that RCK1 interacts with a Gα heterotrimeric G protein subunit. Because many Gα protein subunits are modified by fatty acid residues in a variety of systems (Wedegaertner, 1998) and are localized at the plasma membrane, we wanted to know the subcellular localization of RCK1 and whether this changes in response to chemoattractant stimulation. To study RCK1's subcellular localization, we made two types of RCK1-tagged fusions. In the first, RCK1 was fused to GFP at either the N or C terminus of the protein. When we examined the localization of RCK1-GFP in unstimulated and stimulated cells, the majority of the protein was nuclear (our unpublished data). As this was unexpected, we wanted to further examine RCK1 localization by using a myc-tagged RCK1 by examining immunostained cells before or after cAMP stimulation by indirect immunofluorescence. Figure 6 shows that myc-RCK1 is cytosolic in unstimulated cells and rapidly translocates to the plasma membrane, with membrane localization peaking at ∼10 s. myc-RCK1 then delocalizes from the plasma membrane by 30 s. These observations are similar to previous findings for PI3K and PH domain-containing proteins such as CRAC, Akt/PKB, and PhdA, which localized to the plasma membrane in response to the production of PI3K-dependent lipid binding sites (Parent et al., 1998; Meili et al., 1999; Funamoto et al., 2001, 2002). However, unlike these proteins, which localize to the leading edge in chemotaxing cells, we did not observe polarized localization of RCK1 in aggregating cells (our unpublished data).
Figure 6.
Translocalization of RCK1 after cAMP stimulation. (A) Localization of wild-type RCK1 and mutants C551A, E510G, and EN510-511GA before (0") and after cAMP stimulation. Cells expressing myc-tagged RCK1 or its mutants were pulsed for 4.5 h, stimulated with 20 μM cAMP, fixed at the time indicated, and stained for myc-RCK1 as described in MATERIALS AND METHODS. (B) Localization of the RCK1 kinase-dead mutant RCK1K867A before (0") and after cAMP stimulation.
To investigate which domains of RCK1 might be involved in chemoattractant-mediated RCK1 subcellular localization, we expressed myc fusions of the region N-terminal to the RGS domain, the RGS domain, and the kinase domain. All of these proteins were stably expressed and were cytosolic before or after stimulation, suggesting the entire protein is required for localization and/or the fusion proteins do not fold properly. To further investigate the role of the RGS domain, we examined the subcellular localization of myc-RCK1C551A, myc-RCK1E510G, and myc-RCK1E510G,N511A. As shown in Figure 6, myc-RCK1E510G,N511A and myc-RCK1E510G displayed kinetics of localization and delocalization from the plasma membrane similar to those of wild-type RCK1. In contrast, RCK1C551A showed a very weak membrane localization after cAMP stimulation. The observation that RCK1C551A translocates very weakly suggests the RGS domain may play a key role in RCK1's change in subcellular localization.
We also examined the subcellular localization of the kinase-dead RCK1K867A. Although this protein localized to the plasma membrane with the same kinetics as wild-type RCK1, the protein remained associated with the plasma membrane for up to 5 min after chemoattractant stimulation (Figure 6B). The inability of the kinase-dead mutation to delocalize suggests that the kinase function of RCK1 is directly involved in delocalizing RCK1 from the plasma membrane, possibly by affecting its interaction with a Gα protein subunit.
Because RCK1 is targeted to the plasma membrane, we tested the developmental consequences of expressing a myristylated-tagged RCK1 (myrRCK1) from the Actin 15 promoter in wild-type cells. These cells exhibited a developmental phenotype similar to that of RCK1OE but with even more delayed development and ∼50% of the aggregates arrested at the mound stage at 24 h of development (Figure 4A). Wild-type cells expressing myrRCK1 also exhibited chemotaxis defects with a decrease in speed similar to RCK1OE cells (Figure 5).
RCK1 Kinase Activity Is Activated in Response to Chemoattractant Stimulation
We examined whether RCK1 kinase activity changes in response to chemoattractant stimulation. Cells expressing wild-type myc-tagged RCK1 were lysed before and at specific times after chemoattractant stimulation. myc-RCK1 was immunoprecipitated and the immune precipitate assayed using MBP as a substrate as described previously (Meili et al., 1999). As depicted in Figure 7A, chemoattractant stimulation leads to rapid activation of RCK1 kinase activity. In contrast to many pathways that are stimulated in response to chemoattractants in Dictyostelium, RCK1 activity remains elevated throughout the time course of the experiment (5 min; see DISCUSSION). RCK1 activity only decreased on removal of chemoattractant stimulation. A control experiment using myc-tagged kinase-dead RCK1 (RCK1K867A) showed no activity, suggesting that the kinase activity in the immunoprecipitate from wild-type–stimulated cell extracts is not due to a coimmunoprecipitation of another kinase.
In addition to the band of phosphorylated MBP, we observed a phosphorylated band corresponding to the size of RCK1 (∼130 kDa) and at the same mobility as myc-RCK1 on the Western blot. As with MBP, the phosphorylation of this protein increased upon chemoattractant stimulation. Additional support for the proposition that the labeled ∼130-kDa band is due to RCK1 autophosphorylation comes from the observation that this band is absent in the kinase-dead RCK1 control experiments. We cannot completely exclude the possibility that a kinase associated only with wild-type RCK1 is responsible for this phosphorylation. As with MBP, RCK1 phosphorylation remains elevated throughout the time course of the experiment and rapidly decreases upon removal of cAMP.
To examine the effect of mutations in the RGS domain on activation of RCK1 kinase activity, myc-tagged RCK1C55A and RCK1E510G,N511A were also assayed. All proteins exhibited a similar increase in the phosphorylation of kinase activity against MBP in response to chemoattractant stimulation, with the level of activity of the kinases appearing to be proportional to the level of expression of the protein, as determined by Western blot analysis. The results with RCK1C551A suggest that membrane localization of RCK1 is not essential for RCK1 kinase activation or that the weak membrane localization that is observed is sufficient for activation.
Effect of RCK1 Mutations on Aggregation-stage Signaling Pathways
We investigated the effect of RCK1 mutations on several signal transduction pathways known to be important for signal perception, adaptation, and chemotaxis. PI3K and its downstream effector Akt/PKB are important for cell polarization and directional movement (Meili et al., 1999; Chung et al., 2001b; Funamoto et al., 2001, 2002; Iijima and Devreotes, 2002). To investigate whether PI3K lies upstream from the activation of the RCK1 kinase, RCK1 was expressed in pi3k1/pi3k2 double knockout cells and we examined the RCK1 activity. As shown in Figure 8A, RCK1 kinase activity was indistinguishable from RCK1 activity expressed in rck1 null cells. Similarly, the activation of Akt/PKB in rck1 null and RCK1 overexpressing cells was very similar to that of wild-type cells (Figure 8B). Cells expressing the dominant negative, kinase-dead RCK1 exhibit a slightly elevated level of PKB activation compared with the other cell types. Overall, the results suggest that RCK1 does not directly interact with the PI3K pathways.
Figure 8.
(A) SDS-PAGE mobility shift assay on cAR1 phosphorylation in rck1 null cells. Cells were pulsed every 6 min for 4.5 h with 30 nM cAMP and stimulated with 100 μM of cAMP. Aliquots of cells were taken at indicated time points after adding cAMP and lysed in the same lysis buffer used for the kinase assay except that the 200 mM NaCl was omitted. SDS sample buffer was added and samples were loaded on a 12% Low-Bis SDS-PAGE gel (0.6% Bis) without boiling. Western blotting was performed using cAR1 antibodies at 1:1000 dilution. (B) PKB kinase assay in rck1 null cells and cells overexpressing wild-type and kinase-dead RCK1. Cells were pulsed with 30 nM cAMP for 4.5 h at 6-min intervals and stimulated with 100 nM cAMP. Aliquots of cells were taken at indicated time points and the PKB assay was performed as described in the Materials and Methods. For Western blotting, the same blot was probed with a rabbit polyclonal anti-PKB antibody and detected using an alkaline phosphatase-conjugated secondary antibody. (C) RCK1 activation in pi3k1/2 null cells. The RCK1 kinase activity assay was performed in pi3k1/2 null cells expressing myc-RCK1 the same way described above.
The cAR1 is a receptor for the chemoattractant cAMP and plays a major part in controlling aggregation responses in Dictyostelium, including chemotaxis toward cAMP. cAR1 is phosphorylated in response to cAMP stimulation, which is detected by a mobility shift change on SDS-PAGE (Vaughan and Devreotes, 1988). This phosphorylation has been linked to a decrease in affinity of the receptor for cAMP (Caterina et al., 1995). Although the RCK1 kinase domain does not exhibit high homology to the receptor kinase of the RGS-domain–containing receptor kinases, we wanted to examine whether RCK1 might be responsible for cAR1 phosphorylation and/or have an effect on the kinetics or level of cAR1 phosphorylation. The results shown in Figure 8A indicate that cAR1 is phosphorylated in rck1 null cells, although the kinetics of phosphorylation is slightly delayed compared with those of cAR1 in wild-type cells.
Possible Genetic Interactions of RCK1 with Gα Subunits
We imagine that the RCK1 RGS domain interacts with one or more Gα subunits. To examine this possibility, we determined whether RCK1 membrane localization and activation in response to chemoattractant stimulation were altered after cAMP stimulation in various Gα mutant backgrounds. Gα2 is the Gα subunit that interacts with cARs to mediate G protein-dependent, cAMP-stimulated pathways (Parent and Devreotes, 1996; Aubry and Firtel, 1999). As shown in Figure 9A, myc-RCK1 does not translocate to the membrane in gα2 null cells expressing exogenous cAR1. (Because endogenous cAR1 expression is dependent upon Gα2 function, cells are cotransformed with a cAR1 expression vector to provide necessary receptor levels.) We also examined RCK1 subcellular localization in gα2 null and wild-type cells expressing constitutively active Gα2Q208L. Wild-type cells expressing Gα2Q208L exhibited weaker RCK1 translocation, whereas gα2 null cells expressing Gα2Q208L showed no translocation. Gα5 is also potentially involved in the chemoattractant-mediated signaling pathway, because it affects the developmental timing of morphogenesis. In contrast to observations in gα2null cells, RCK1 translocated normally in gα5 null cells.
Figure 9.
(A) RCK1 translocation in various Gα backgrounds. Myc-tagged RCK1 or its mutant was either transfected alone or cotransfected with wild-type or constitutively active Gα2 (Gα2 Q208L) into various gα null cells. After establishment of positive clones, cells were pulsed for 4.5 h, stimulated with 20 μM cAMP, fixed at the time indicated, and stained for myc-RCK1 as described in MATERIALS AND METHODS. (B and C) Kinase assay of RCK1 in various Gα backgrounds as indicated. Top, autophosphorylated RCK1. Middle, RCK1-phosphorylated MBP. The immunoprecipitated RCK1 protein levels were examined by Western blot analysis on the same blot used for the kinase assay (bottom).
RCK1 kinase activation was also examined in several Gα mutant strains. As shown in Figure 9, B and C, RCK1 kinase activation was normal in gα5 cells but was not activated in gα2 null cells expressing exogenous cAR1. In contrast, RCK1 kinase activity exhibited a very high basal activity (activity in the absence of cAMP stimulation) in KAx-3 cells expressing Gα2Q208L. Activity increased an additional twofold in response to cAMP stimulation in contrast to the 6- to 10-fold activation in KAx-3 expressing wild-type Gα2 from the exogenous Actin 15 promoter. gα2 null cells expressing Gα2Q208L exhibited the high basal activity, but the activity did not increase upon cAMP stimulation, suggesting the increase observed in wild-type cells expressing Gα2Q208L may be due to the activation of the endogenous Gα protein. Our results with gα2 null cells and wild-type and gα2 null cells expressing Gα2Q208L suggest that Gα2 may interact directly with RCK1 and regulate its function.
DISCUSSION
RCK1 Is Required for Chemotaxis and Morphogenesis
We have identified a novel kinase, RCK1, that contains an RGS and a kinase domain. RCK1 is preferentially expressed during the aggregation stage in development and our analyses of the rck1 null strain demonstrated that RCK1 plays a critical role controlling chemotaxis and morphogenesis. rck1 null cells exhibit an ∼50% increase in the speed of directional movement during chemotaxis, whereas RCK1-overexpressing (RCK1OE) cells show an ∼40% decrease in the speed of cell movement. This finding is particularly novel because other chemotaxis/motility null mutants described in the literature for Dictyostelium exhibit a decrease in chemotaxis. The observations of an increase in the speed of chemotaxis of the rck1 null strain combined with a decrease in the speed of chemotaxis in the RCK1OE strain suggest that RCK1 plays a negative role in controlling chemotaxis and/or motility. We also found that expression of RCK1 in rck1 null cells leads to a phenotype similar to that observed for RCK1OE cells. However, this observation was not unexpected, considering RCK1OE cells are presumed to have an elevated level of expression of RCK1 compared with wild-type cells because of higher levels of RCK1 RNA transcripts. Although we think it is unlikely, we cannot exclude the possibility that the temporally altered expression of RCK1 from the Actin 15 promoter may account for the RCK1OE phenotypes. We found that the rck1 null and RCK1OE strains exhibit a similar multicellular morphogenesis defect after the formation of the multicellular aggregate. The rck1 null strains exhibited slightly faster aggregation and produced larger amounts, but had a significant delay of development with a prolonged mound stage. Time-lapse video analysis of the rck1 null cell development revealed normal cAMP wave patterns compared with those of wild-type cells (our unpublished observation), suggesting that the production and propagation of cAMP is probably not affected in this strain. Because morphogenesis is partially mediated by the differential sorting of prestalk cells within the multicellular aggregate to apical cAMP signals (Weijer, 1999; Clow et al., 2000), we expect that the observed chemotaxis defects of the RCK1OE and rck1 null strains are the basis of the inability of these cells to properly undergo morphogenesis. It is unexpected that cells exhibiting a faster than wild-type speed of chemotaxis experience a delay in morphogenesis. It is possible that the morphogenesis defect is due to an effect of RCK1 on cytoskeletal processes distinct from those controlling leading edge responses.
Cells expressing myrRCK1 exhibited a similar developmental phenotype to RCK1OE cells but with an even longer delay. The chemotaxis analysis showed that these two types of cells showed very similar chemotaxis defects with the only noticeable difference being that myrRCK1 cells exhibit a greater directional change (33.08 ± 10.39 vs. 25.08 ± 5.45). It is possible that the difference in direction change may be due to the constitutive localization of myrRCK1 on the plasma membrane and that this difference may account for the difference in developmental phenotype between these two cell lines.
The strongest morphogenesis defects are exhibited by expressing the kinase-dead RCK1 (RCK1K867A) in an rck1 null background. We expect these defects are due to a dominant negative effect of expressing a protein that may interact with some effectors through its RGS or kinase domain but is unable to properly phosphorylate its substrates. Because RCK1 kinase activity is necessary for the release of RCK1 from the plasma membrane/cortex, we propose that this inability to delocalize from the plasma membrane may lead to the strong chemotaxis and morphogenesis phenotypes in this strain. We expect that the RGS domain interacts with one or more of the Gα protein subunits found in Dictyostelium and that the kinase-dead protein may continue to interact with a particular α subunit and either block other effectors or rapidly down-regulate Gα protein function by acting as a GTPase-activating enzyme.
RCK1 Translocates to Plasma Membrane in Response to Chemoattractant Stimulation
We demonstrate that RCK1 rapidly and transiently translocates to the plasma membrane in response to chemoattractant stimulation. We show that this translocation does not require a functional kinase domain, but seems to require a functional RGS domain. We tested several RGS domain mutants that had been previously demonstrated in other systems to affect RGS protein function. One of these mutants, RCK1C551A, shows a significant reduction in chemoattractant-mediated membrane translocation. Because a mutation of a conserved Asn/Ser residue in another RGS domain, which corresponds to C551 in the RGS domain of RCK1, affects the interaction of the RGS domain with Gα protein subunits, we suggest that RGS domain interaction with a Gα protein mediates RCK1 membrane translocation. We tested the ability of the various subdomains of RCK1, including the RGS domain, to translocate to the plasma membrane in response to chemoattractant stimulation. All of these constructs remain cytosolic. We cannot distinguish between the RGS domain alone being insufficient for the translocation and potential problems with the folding of the deleted protein.
Gα2 is known to mediate G protein-dependent functions downstream from the cAMP receptors. RCK1 does not translocate in gα2 null cells, suggesting that RCK1 translocation may be mediated through RCK1's binding to Gα2GTP, although we do not have any direct evidence that RCK1 binds to Gα2GTP. cAMP-mediated chemotaxis requires the expression of a variety of gene products whose expression is dependent on cAMP signaling and Gα2 function. Although we use a gα2 null strain coexpressing exogenous cAR1 (Insall et al., 1994), it is possible that this strain may not be fully competent to respond to cAR1-mediated pathways even if they are Gα2 dependent. Thus, the absence of RCK1 translocation to the plasma membrane in this strain may be due to secondary effects. These data suggest involvement of Gα2 in mediating RCK1 function, but we do not have any data indicating whether the RCK1 RGS domain has any GAP activity against Gα2GTP or another Gα subunit.
Previous studies revealed that a variety of signaling molecules, including PH domain-containing proteins that bind to the PI3K products phosphatidylinositol 3,4,5-triphosphate and phosphatidylinositol 4,5-triphosphate as well as PI3K1 and PI3K2, also localize to the plasma membrane in response to global stimulation (Parent et al., 1998; Meili et al., 1999; Funamoto et al., 2001, 2002; Iijima and Devreotes, 2002). In contrast to these proteins, RCK1 does not localize to the leading edge in chemotaxing cells (cells responding to a directional chemoattractant gradient) but is found more uniformly distributed along the plasma membrane. Presently, the mechanism by which PI3K1 and PI3K2 localize to the leading edge in chemotaxing cells is unknown. We know, however, that PI3K is uniformly localized along the plasma membrane. Through the use of a myristoylation membrane-targeting sequence and through the analysis of pten null cells in Dictyostelium, it was discovered PI3K function is activated along the lateral sides as well as the leading edge, indicating that chemoattractant-mediated pathways can be activated along the lateral sides as well as the leading edge of cells (Funamoto et al., 2002; Iijima and Devreotes, 2002). The presence of RCK1 along the perimeter of chemotaxing cells suggests that the signaling pathway that mediates RCK1 membrane localization, possibly the activation of Gα2, is activated along the entire perimeter of cells in a chemoattractant gradient.
RCK1 Is Activated in Response to Chemoattractant Stimulation
RCK1 kinase activity is also rapidly activated in response to chemoattractant stimulation, with activity levels plateauing by 10 s. This kinetics is very similar to that of the activation of Akt/PKB and guanylyl cyclase. However, in contrast to these pathways and all other previously examined chemoattractant-mediated pathways in Dictyostelium, RCK1 activity does not rapidly adapt, but remains high throughout the time course of the experiment and only decreases after the removal of cAMP (Parent and Devreotes, 1996; Aubry and Firtel, 1999). The cAR1 receptor is phosphorylated in response to cAMP stimulation and, like RCK1 kinase activity, remains phosphorylated as long as the ligand is present. In this regard, the continued elevated RCK1 kinase activity is similar to the levels of the phosphorylated form of the cAR1 receptor.
Interestingly, RCK1 kinase activity may not require continuous localization of RCK1 on the plasma membrane. In fact, RCK1 kinase activity is required for the delocalization of RCK1 from the membrane. Studies with RCK1C551A, which localizes poorly to the plasma membrane in response to global stimulation, result in the same level of “autophosphorylation” and kinase activity against MBP obtained with wild-type protein expressed at the same level. This finding suggests that either membrane localization of RCK1 is not required for its activation or the low level of association observed in our studies is sufficient to mediate the activation response. If RCK1's translocation were required for kinase activation, we would have expected this mutant to exhibit some decrease in the level of kinase activity. It is possible that the membrane localization targets RCK1 to its putative substrate. Furthermore, we expect that the rapid delocalization of the RCK1 protein from the plasma membrane, possibly due to its interaction with a Gα protein effector, may be critical in controlling RGS1 function.
Regulation of RCK1
The heterotrimeric G protein containing the Gα2 subunit couples to the cAMP chemoattractant subunits and controls all known cAMP-mediated, G protein-dependent signaling pathways. We found that RCK1 neither is activated nor translocated to the plasma membrane in gα2 null cells. Because RCK1 translocation seems to be dependent on the RGS domain (with the caveats discussed above), these data suggest that Gα2GTP may interact with the RGS domain, although we have no evidence from binding studies to support this suggestion. We found that RCK1 cells expressing constitutively active Gα2, which should be in the GTP-bound state, have a very elevated basal kinase activity as determined by the kinase assay using MBP as a substrate and RCK1 autophosphorylation activity (in the absence of chemoattractant stimulation). However, we do not observe constitutive localization of RCK1 to the plasma membrane. Rather, the translocation of RCK1 was reduced in wild-type cells expressing constitutively active Gα2 and absent in rck1 null cells expressing constitutively active Gα2. This might be expected, considering our data suggest that RCK1 kinase activity mediates or is required for the delocalization of RCK1 from the membrane. Previous studies showed that many cAMP-stimulated pathways are constitutively adapted in cells expressing Gα2Q208L and cannot be further stimulated by cAMP, and these cells are unable to aggregate (Okaichi et al., 1992). Similarly, activation of pathways ranging from guanylyl and adenylyl cyclase to Akt/PKB all rapidly adapt when cells are stimulated by cAMP, even in the continued presence of ligand (Parent and Devreotes, 1996; Firtel and Chung, 2000). However, in wild-type cells expressing constitutively active Gα2 (Gα2Q208L), which also express endogenous Gα2, cAMP stimulation results in an additional increase in RCK1 kinase activity. This is not observed in gα2 null cells expressing constitutively active Gα2 (Gα2Q208L), possibly because these cells are generally not very responsive. We suggest that Gα2Q208L leads to the constitutive stimulation of RCK1 kinase activity and this pathway does not adapt, consistent with our observation that RCK1 kinase activity remains elevated as long as the cells are being stimulated by a chemoattractant. In contrast, RCK1 protein translocation may be constitutively adapted, as are some other signaling pathways. We also examined another Gα subunit, Gα5. We observed no effect on RCK1 membrane localization or kinase activity.
Our results indicate that RCK1 plays a key role in controlling cAMP-mediated chemotaxis. RCK1 may function as a “governor,” controlling the rate of chemotaxis or acting as part of the adaptation pathway. The idea of a possible function of RCK1 in adaptation is consistent with some of our observations. First, rck1 null cells chemotax faster than wild-type cells. This could be the due to extended signaling if RCK1 is involved in adaptation. Second, RCK1 activation does not adapt in the presence of a continuous signal. The kinase activity of RCK1 peaks after 10 s and only decreases when cAMP is removed. The continuous stimulated RCK1 kinase activity might be the signal to block stimulation of some cAMP-induced pathways. RCK1 might function with or in parallel with Gα9, which was recently demonstrated to be important for regulating adaptation of some signaling pathways during aggregation (Brzostowski et al., 2002). Although individual rck1 null cells seem to chemotax more efficiently in a micropipette chemotaxis assay, the cells exhibit aggregation defects, suggesting that RCK1 plays an important part in controlling a physiological response that is more closely related to the natural biology of the organism. We have examined a number of chemoattractant-mediated signaling pathways, but none of these were detectably affected in our assays. The identification of the RCK1 substrates should elucidate the pathway by which RCK1 functions.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0550. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0550.
REFERENCES
- Aubry L, Firtel R. Integration of signaling networks that regulate Dictyostelium differentiation. Annu Rev Cell Dev Biol. 1999;15:469–517. doi: 10.1146/annurev.cellbio.15.1.469. [DOI] [PubMed] [Google Scholar]
- Brzostowski JA, Cynthia Johnson C, Kimmel AR. Gα-mediated inhibition of developmental signal response. Curr Biol. 2002;12:1199–1208. doi: 10.1016/s0960-9822(02)00953-3. [DOI] [PubMed] [Google Scholar]
- Carman CV, Benovic JL. G-protein-coupled receptors: turn-ons and turn-offs. Curr Opin Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. Selective regulation of Gα(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem. 1999;274:34483–3492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Devreotes PN, Borleis J, Hereld D. Agonist-induced loss of ligand binding is correlated with phosphorylation of cAR1, a G protein-coupled chemoattractant receptor from Dictyostelium. J Biol Chem. 1995;270:8667–8672. doi: 10.1074/jbc.270.15.8667. [DOI] [PubMed] [Google Scholar]
- Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- Chen MY, Long Y, Devreotes PN. A novel cytosolic regulator, Pianissimo, is required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase in Dictyostelium. Genes Dev. 1997;11:3218–3231. doi: 10.1101/gad.11.23.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Firtel RA. PAKa, a putative PAK family member, is required for cytokinesis and the regulation of the cytoskeleton in Dictyostelium discoideum cells during chemotaxis. J Cell Biol. 1999;147:559–576. doi: 10.1083/jcb.147.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CY, Funamoto S, Firtel RA. Signaling pathways controlling cell polarity and chemotaxis. Trends Biochem Sci. 2001a;26:557–566. doi: 10.1016/s0968-0004(01)01934-x. [DOI] [PubMed] [Google Scholar]
- Chung CY, Potikyan G, Firtel RA. Control of cell polarity and chemotaxis by Akt/PKB and PI3 kinase through the regulation of PAKa. Mol Cell. 2001b;7:937–947. doi: 10.1016/s1097-2765(01)00247-7. [DOI] [PubMed] [Google Scholar]
- Clow PA, Chen TLL, Chisholm RL, McNally JG. Three-dimensional in vivo analysis of Dictyostelium mounds reveals directional sorting of prestalk cells and defines a role for the myosin II regulatory light chain in prestalk cell sorting and tip protrusion. Development. 2000;127:2715–2728. doi: 10.1242/dev.127.12.2715. [DOI] [PubMed] [Google Scholar]
- Comer FI, Parent CA. PI 2-kinases and PTEN: how opposites chemoattract. Cell. 2002;109:541–544. doi: 10.1016/s0092-8674(02)00765-1. [DOI] [PubMed] [Google Scholar]
- Datta S, Firtel RA. Identification of the sequences controlling cyclic AMP regulation and cell-type-specific expression of a prestalk-specific gene in Dictyostelium discoideum. Mol Cell Biol. 1987;7:149–159. doi: 10.1128/mcb.7.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P, Fontana D, Klein P, Sherring J, Theibert A. Transmembrane signaling in Dictyostelium. Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Ferguson SS, Caron MG. G protein-coupled receptor adaptation mechanisms. Semin Cell Dev Biol. 1998;9:119–127. doi: 10.1006/scdb.1997.0216. [DOI] [PubMed] [Google Scholar]
- Firtel RA, Chung CY. The molecular genetics of chemotaxis: sensing and responding to chemoattractant gradients. Bioessays. 2000;22:603–615. doi: 10.1002/1521-1878(200007)22:7<603::AID-BIES3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phosphoinositides by PI 3-kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- Funamoto S, Milan K, Meili R, Firtel R. Role of phosphatidylinositol 3′ kinase and a downstream plekstrin homology domain-containing protein in controlling chemotaxis in Dictyostelium. J Cell Biol. 2001;153:795–810. doi: 10.1083/jcb.153.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm HE. The many faces of G protein signaling. J Biol Chem. 1998;273:669–672. doi: 10.1074/jbc.273.2.669. [DOI] [PubMed] [Google Scholar]
- Hepler JR. Emerging roles for RGS proteins in cell signaling. Trends Pharmacol Sci. 1999;20:3763–3782. doi: 10.1016/s0165-6147(99)01369-3. [DOI] [PubMed] [Google Scholar]
- Huang LJ, Durick K, Weiner JA, Chun J, Taylor SS. D-AKAP2, a novel protein kinase A anchoring protein with a putative RGS domain. Proc Natl Acad Sci USA. 1997;94:11184–11189. doi: 10.1073/pnas.94.21.11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Devreotes P. Tumor suppressor PTEN mediates sensing of chemoattractant gradients. Cell. 2002;109:599–610. doi: 10.1016/s0092-8674(02)00745-6. [DOI] [PubMed] [Google Scholar]
- Iijima M, Huang YE, Devreotes P. Temporal and spatial regulation of chemotaxis. Dev Cell. 2002;3:469–478. doi: 10.1016/s1534-5807(02)00292-7. [DOI] [PubMed] [Google Scholar]
- Insall RH, Soede RDM, Schaap P, Devreotes PN. Two cAMP receptors activate common signaling pathways in Dictyostelium. Mol Biol Cell. 1994;5:703–711. doi: 10.1091/mbc.5.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Soede RD, Schaap P, Valkema R, Borleis JA, Van Haastert PJ, Devreotes PN, Hereld D. Phosphorylation of chemoattractant receptors is not essential for chemotaxis or termination of G-protein-mediated responses. J Biol Chem. 1997;272:27313–27318. doi: 10.1074/jbc.272.43.27313. [DOI] [PubMed] [Google Scholar]
- Kitayama C, Dai J, Ting-Beall HP, Titus MA, Uyeda QP. Generation and phenotypic analysis of a myosin triple mutant (myosin II-/myoA-/B-) Mol Biol Cell. 1998;9:387. [Google Scholar]
- Kozasa T, Jiang X, Hart MJ, Sternweis PM, Singer WD, Gilman AJ, Bollag G, Sternweis PC. p115 RhoGEF, a GTPase activating protein for Galpha12 and Galpha13. Science. 1998;280:2109–2111. doi: 10.1126/science.280.5372.2109. [DOI] [PubMed] [Google Scholar]
- Li S, Wilkinson MF. Site-directed mutagenesis: a two-step method using PCR and DpnI. Biotechniques. 1997;23:588–590. doi: 10.2144/97234bm05. [DOI] [PubMed] [Google Scholar]
- Ma H, Gamper M, Parent C, Firtel RA. The Dictyostelium MAP kinase kinase DdMEK1 regulates chemotaxis and is essential for chemoattractant-mediated activation of guanylyl cyclase. EMBO J. 1997;16:4317–4332. doi: 10.1093/emboj/16.14.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein DJ, Titus MA, De Lozanne M, Spudich JA. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. ,. [DOI] [PubMed] [Google Scholar]
- Meili R, Ellsworth C, Lee S, Reddy TB, Ma H, Firtel RA. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nellen W, Datta S, Reymond C, Sivertsen A, Mann S, Crowley T, Firtel RA. Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 1987;28:67–100. doi: 10.1016/s0091-679x(08)61637-4. [DOI] [PubMed] [Google Scholar]
- Okaichi K, Cubitt AB, Pitt GS, Firtel RA. Amino acid substitutions in the Dictyostelium Gα subunit Gα2 produce dominant negative phenotypes and inhibit the activation of adenylyl cyclase, guanylyl cyclase, and phospholipase C. Mol Biol Cell. 1992;3:735–747. doi: 10.1091/mbc.3.7.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95:81–91. doi: 10.1016/s0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- Penn RB, Pronin AN, Benovic JL. Regulation of G protein-coupled receptor kinases. Trends Cardiovasc Med. 2000;10:81–89. doi: 10.1016/s1050-1738(00)00053-0. [DOI] [PubMed] [Google Scholar]
- Peracino B, Borleis J, Jin T, Westphal M, Schwartz JM, Wu LJ, Bracco E, Gerisch G, Devreotes P, Bozzaro S. G protein β subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J Cell Biol. 1998;141:1529–1537. doi: 10.1083/jcb.141.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posner BA, Mukhopadhyay S, Tesmer JJ, Gilman AG, Ross EM. Modulation of the affinity and selectivity of RGS protein interaction with Gα subunits by a conserved asparagine/serine residue. Biochemistry. 1999;38:7773–7779. doi: 10.1021/bi9906367. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Labortory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Siderovski DP, Hessel A, Chung S, Mak TW, Tyers M. A new family of regulators of G-protein-coupled receptors? Curr Biol. 1996;6:211–212. doi: 10.1016/s0960-9822(02)00454-2. [DOI] [PubMed] [Google Scholar]
- Sierra DA, Popov S, Wilkie TM. Regulators of G-protein signaling in receptor complexes. Trends Cardiovasc Med. 2000;10:263–268. doi: 10.1016/s1050-1738(00)00072-4. [DOI] [PubMed] [Google Scholar]
- Soll DR, Voss E. Two- and three-dimensional computer systems for analyzing how animal cells crawl. In: Soll DR, Wessels D, editors. Motion Analysis of Living Cells. New York: Wiley-Liss; 1998. pp. 25–52. [Google Scholar]
- Spink KE, Polakis P, Weis WI. Structural basis of the Axin-adenomatous polyposis coli interaction. EMBO J. 2000;19:2270–2279. doi: 10.1093/emboj/19.10.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutoh K. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid. 1993;30:150–154. doi: 10.1006/plas.1993.1042. [DOI] [PubMed] [Google Scholar]
- Tesmer JJ, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4-activated G(iα1): stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- van Es S, Devreotes PN. Molecular basis of localized responses during chemotaxis in amoebae and leukocytes. Cell Mol Life Sci. 1999;55:1341–1351. doi: 10.1007/s000180050374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan RA, Devreotes PN. Ligand-induced phosphorylation of the cAMP receptor from Dictyostelium discoideum. J Biol Chem. 1988;263:14538–14543. [PubMed] [Google Scholar]
- Watts DJ, Ashworth JM. Growth of myxameobae of the cellular slime mold Dictyostelium discoideum in axenic culture. Biochem J. 1970;119:171–174. doi: 10.1042/bj1190171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedegaertner PB. Lipid modifications and membrane targeting of Gα. Biol Signals Recept. 1998;7:125–135. doi: 10.1159/000014538. [DOI] [PubMed] [Google Scholar]
- Weijer CJ. Morphogenetic cell movement in Dictyostelium. Semin Cell Dev Biol. 1999;10:609–619. doi: 10.1006/scdb.1999.0344. [DOI] [PubMed] [Google Scholar]
- Wessels D, Voss E, Von Bergen N, Burns R, Stites J, Soll DR. A computer-assisted system for reconstructing and interpreting the dynamic three-dimensional relationships of the outer surface, nucleus and pseudopods of crawling cells. Cell Motil Cytoskeleton. 1998;41:225–246. doi: 10.1002/(SICI)1097-0169(1998)41:3<225::AID-CM4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Wessels DJ, Zhang H, Reynolds J, Daniels K, Heid P, Lu SJ, Kuspa A, Shaulsky G, Loomis WF, Soll DR. The internal phosphodiesterase RegA is essential for the suppression of lateral pseudopods during Dictyostelium chemotaxis. Mol Biol Cell. 2000;11:2803–2820. doi: 10.1091/mbc.11.8.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland T, Bahtijari N, Zhou XB, Kleuss C, Simon MI. Polarity exchange at the interface of regulators of G protein signaling with G protein α-subunits. J Biol Chem. 2000;I:28500–28506. doi: 10.1074/jbc.M004187200. ,. [DOI] [PubMed] [Google Scholar]
- Zhong H, Neubig RR. Regulator of G protein signaling proteins: novel multifunctional drug targets. J Pharmacol Exp Ther. 2001;297:837–845. [PubMed] [Google Scholar]