Abstract
The advancement of genetic techniques has greatly boosted taxonomic studies in recent years. Within the genus Mycobacterium, 42 new species have been detected since 1990, most of which were grown from clinical samples. Along with species for which relatively large numbers of strains have been reported, some of the new species of mycobacteria have been detected rarely or even only once. From the phenotypic point of view, among the new taxa, chromogens exceed nonchromogens while the numbers of slowly and rapidly growing species are equivalent. Whereas conventional identification tests were usually inconclusive, an important role was played by lipid analyses and in particular by high-performance liquid chromatography. Genotypic investigations based on sequencing of 16S rRNA gene have certainly made the most important contribution. The investigation of genetic relatedness led to the redistribution of the species previously included in the classically known categories of slow and rapid growers into new groupings. Within slow growers, the intermediate branch related to Mycobacterium simiae and the cluster of organisms related to Mycobacterium terrae have been differentiated; among rapid growers, the group of thermotolerant mycobacteria has emerged. The majority of species are resistant to isoniazid and, to a lesser extent, to rifampin. Many of the new species of mycobacteria are potentially pathogenic, and there are numerous reports of their involvement in diseases. Apart from disseminated and localized diseases in immunocompromised patients, the most frequent infections in immunocompetent people involve the lungs, skin, and, in children, cervical lymph nodes. The awareness of such new mycobacteria, far from being a merely speculative exercise, is therefore important for clinicians and microbiologists.
INTRODUCTION
Two major periods may be distinguished in prokaryotic taxonomy, one characterized by the utilization of phenotypic studies and one characterized by a focus on genotypic characteristics. In mycobacterial taxonomy the first period lasted from the dawn of mycobacterial studies in the late 1880s to the end of the 1980s and the second started during the last decade of the 20th century and has continued to the present. While early genotypic studies confirmed the validity of previously defined phenotype-based taxa, subsequently, with the emergence of the higher discriminative power of these techniques, the splitting of some species and the ex novo definition of others resulted.
The rationale of genotypic taxonomy is linked to the detection, within the genome, of highly conserved regions harboring hypervariable sequences in which species-specific deletions, insertions, or replacements of single nucleotides are present. The gene encoding the 16S rRNA has been for many years, and still is, the primary target of molecular taxonomic studies, with several other genomic regions playing a minor role. Although the role of genetics has been preeminent in the recent advancement of mycobacterial taxonomy, an important contribution has also been made by chemotaxonomic investigations. This approach achieved excellent results with mycobacteria, thanks to the presence in their cell wall of an unusually heavy lipid burden which includes unique molecules such as mycolic acids. As a consequence of these two developments, the number of mycobacterial species has greatly increased in the last decade.
Important baseline articles concerning the nontuberculous mycobacteria exist. They include the milestone review by Wolinsky (175), properly updated 13 years later by Wayne and Sramek (169). Interestingly, 1992, the year the latter review was published, definitively sanctioned the success of the genotypic approach to mycobacterial taxonomy. The present paper, which aims to continue on the track of the above-mentioned authoritative reviews, focuses on the mycobacterial species described in the 1990s, with particular emphasis on those in which molecular and lipid analyses played an important role.
16S rRNA Gene
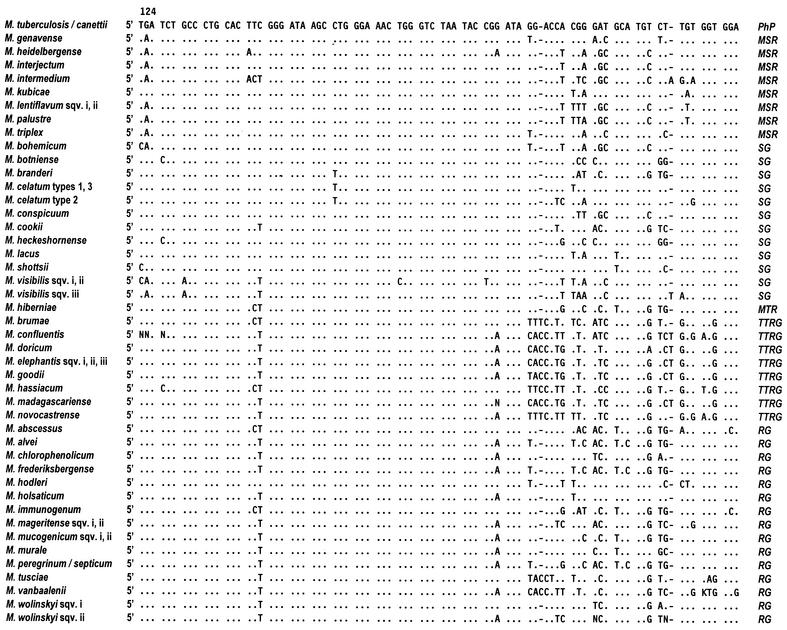
The 16S rRNA is an approximately 1,500-nucleotides sequence encoded by the 16S ribosomal DNA (rDNA). The latter is a highly conserved gene in which regions common to all living beings exist while nucleotide variations are concentrated in specific areas. In the mycobacterial 16S rDNA, the nucleotide stretches which are the most interesting are the ones shared by all the members of the genus Mycobacterium (5) and also the hypervariable regions, characterized by species-specific variability. The 16S rRNA-based genetic investigation of mycobacterial taxonomy and phylogeny focuses on two hypervariable sequences, known as region A and B, which correspond to the Escherichia coli positions around 130 to 210 and 430 to 500 respectively.
Like every single-stranded nucleotide sequence, the 16S rRNA folds up into a secondary structure (Fig. 1) characterized by loops (or helices). Helices 8, 9, 10, and 11 fall within region A, while helix 18 is in region B. The sequences of such helices present, in addition to nucleotide substitutions scattered all over the hypervariable regions, additional regions of variability that, being shared by clusters of species within the genus Mycobacterium, are of particular interest, mainly from the phylogenetic point of view (Fig. 2). Helix 10 may be extended by an insertion of one cytosine at the E. coli homologous position 184. Mycobacteria with such an insertion belong to a separate branch within the rapid growers and are known as “thermotolerant rapid growers,” even though only some of them are able to grow at temperatures over 37°C. More extensive is the insertion at position 455 in helix 18. The number of nucleotides involved ranges from 8 in M. scrofulaceum to 12 in the majority of slow growers and even 14 in M. terrae and a few other species, which therefore appear phylogenetically related. A short helix (i.e., without any insertion) is thought to be the original condition.
FIG. 1.
Secondary-structure model of the 16S rDNA (double lines indicate variable or hypervariable; gray lines indicate highly conserved; V1 to V9 indicate major variable regions). Reprinted from reference 8 with permission from the author.
FIG. 2.
Sequences of 16S rDNA hypervariable regions A and B of new mycobacteria. Positions, as derived from the E. coli sequence, are indicated. Only nucleotides that differ from M. tuberculosis are shown; dashes indicate deletions. sqv., sequevar. PhP, phylogenetic position; MSR, M. simiae related; SG, slow growers; MTR, M. terrae related; TTRG, thermotolerant rapid growers; RG, rapid growers.
Following the detection of the helix 18 polymorphism, it was found that the insertion-free type appeared to be characteristic of rapidly growing species (132) while the variably elongated helix was found in slow growers. M. simiae at first appeared to be the only exception, being characterized by slow growth and a short helix 18, but it has been subsequently joined by an increasing number of slow growers with the same molecular characteristic.
For identification purposes, the sequence of region A is usually sufficient. Region B may be considered confirmatory since in several cases its sequence is shared by a number of species. While slow growers contain one copy of the 16S rRNA gene (4), rapid growers, except for M. chelonae and M. abscessus, have two copies.
No cutoff exists for the minimal number of nucleotide differences indicating distinct taxa. The 5- to 15-base diversity in the 16S rRNA gene, proposed for other microorganisms (44), is not applicable to the genus Mycobacterium, whose members are more closely related to each other. Closely related species may differ only by a few bases or, in the presence of clear phenotypic differences, not differ at all. In contrast, several unequivocally defined species exist presenting genetic heterogeneity that, for various sequevars of M. avium complex (MAC), may involve differences of up to 7 bp.
Genetic sequencing, which is cumbersome (75) and often subject to interpretation errors when done manually, is nowadays made easier and reproducible due to the improved chemistry (e.g., sequenase and dye terminators) and sequencing methods and to the availability of fully automatic instrumentation. Furthermore, despite what is still believed, it has also become very inexpensive.
65-kDa Heat Shock Protein Gene
The 65-kDa heat shock protein gene (hsp65) is also highly conserved among mycobacterial species. However, it also presents hypervariable regions (positions 624 to 664 and 683 to 725 of the M. tuberculosis gene), whose sequences may be used for identification purposes (114). hsp65 is better known by taxonomists, however, for the PCR restriction enzyme pattern analysis (PRA) of a 441-bp sequence often referred to as the Telenti fragment (137). PRA involves visualizing fragment patterns obtained by cutting the PCR-amplified sequence with proper restriction enzymes (usually BstEII and HaeIII) (137). The digestion products, separated by agarose gel electrophoresis, appear as bands whose patterns are usually species specific. However, species with overlapping patterns and multiple patterns within a single species do exist.
Other Molecular Targets of Diagnostic Interest
Less commonly used taxonomic applications of PRA involve the 16S rRNA gene (31, 171) and the superoxide dismutase gene (123). An emerging target of taxonomic interest is the region between the genes encoding the 16S and the 23S rRNAs, which is commonly known as the internal transcribed spacer (ITS). Recently, PRA of the ITS was shown to be a useful tool (117) suitable for identifying most mycobacterial species and determining to which one of five clusters of closely related species the others belong. ITS sequencing is also increasingly used (115). This region, like most others which are less highly conserved than the 16S rDNA, is characterized by a higher rate of polymorphism, which allows, in some cases, a more precise species definition. In other cases, however, the presence of intraspecies variability may result in confusion.
Mycolic Acid Analysis
Mycolic acids are β-hydroxy fatty acids with a long side chain at position α that are a major component of the conspicuous lipid content of the mycobacterial cell wall. They differ in the number of carbon atoms, ranging from 60 to 90, and in the presence of different functional groups. The mycolic acid pattern of the cell wall generally varies with the species. Thus, mycolic acid analyses can be a useful tool for mycobacterial identification. Two direct approaches may be used for the analysis of mycolic acids: thin-layer chromatography (TLC) and high-performance liquid chromatography (HPLC). A third method, gas-liquid chromatography (GLC), in which mycolic acids are investigated in the form of their cleavage products, is also used.
TLC allows the differentiation of esters of seven known mycolic acid types: type i or α-mycolates, type ii or α′-mycolates, type iii or methoxy-mycolates, type iv or keto-mycolates, type v or epoxy-mycolates, type vi or wax esters, and type vii or ω-1-methoxy-mycolates. Type i mycolate is present in all mycobacterial species detected so far. Type vii is present in only a few species of nonpigmented rapidly growing mycobacteria (M. senegalense and M. alvei and several strains of M. porcinum, M. fortuitum, and M. peregrinum) (86). The other types are variously distributed among different species; most mycobacteria contain no more than two or three mycolic acid types. Given the limited number of mycolic acid types, many patterns are shared by more than one species.
Once mycolic acids have been liberated from whole cells, they are extracted as mycolates by means of methyl esterification (12). They are then spotted on silica gel sheets (stationary phase), where they are separated by the capillary progression of elution solvents (mobile phase). The identification of separated compounds, which look like spots, is determined by their position and is generally performed by comparison with spots from reference strains run in parallel with the sample.
In HPLC analysis, the mycolic acids are separated on the basis of their polarity and the carbon chain length, with the more polar and shorter eluting first. In contrast to TLC, the identification of eluted compounds is unimportant; the arrangement of major peaks in the chromatogram, the position on the basis of retention times, and the height in comparison with other peaks are the only pieces of information needed. Each species is characterized by a pattern with a particular number, position, and height of peaks. The visual comparison of HPLC chromatograms with ones of known species is still the most reliable identification procedure.
To obtain an HPLC chromatogram, the mycolic acids are liberated from whole cells by means of saponification and extracted with chloroform. They are then derivatized to UV-adsorbing bromophenacyl esters (17, 20). The extract is injected in the column, where it is eluted by the mobile phase, a gradient of methanol and methylene chloride. The eluted fractions are quantified by UV absorption, using a detector set to 254 to 260 nm. More recently, improved sensitivity (16 to 26 times greater) has been achieved by replacing the UV absorption detection technique with one based on fluorescence absorption. Although a different derivatization technique is required, which utilizes fluorescent labeling compounds (coumarins), the chromatograms are fully comparable with the ones obtained by standard UV-HPLC.
With GLC, not only mycolic acids but the whole lipid component of the cell wall is analyzed (53). At operating column temperatures, ranging from 275 to 300°C, the pyrolysis of mycolic acids occurs with consequent splitting into saturated methyl esters 22, 24, or 26 carbon atoms long (C22:0, C24:0, and C26:0). Along with the above mycolic acid cleavage products, saturated and unsaturated fatty acids are eluted, including exadecanoic (C16:0), octadecenoic (C18:1) octadecanoic (C18:0), and tuberculostearic (10-methyloctadecanoic.) acids and secondary alcohols, stemming from wax esters. The distribution of the GLC products varies qualitatively and quantitatively in different species. Different concentrations of tuberculostearic acid are present in different mycobacteria, with the exception of M. gordonae. The most frequently detected compounds include saturated and unsaturated 16- and 18-carbon fatty acids and, among mycolic acid cleavage products, the C24:0 methyl ester. For the identification of single species, the most important compounds include methyl-branched fatty acids, alcohols, and C22:0, C24:0, and C26:0 mycolic acid cleavage products. Many species contain common peaks of various heights; characteristic peaks, even those of poor height, are however more important for identification. As with TLC, species do exist with indistinguishable profiles.
The fatty acids, once extracted into methyl esters from whole cells by means of methanolysis, are subjected to trifluoroacetylation, which improves alcohol detection. They are then injected into the gas chromatograph, where they are driven to the temperature-controlled column by the carrier gas. The quantity of the various eluted products is determined by means of a flame ionization detector and the identification, achieved on the basis of retention times, may be confirmed by mass spectrometry.
Classification
Many classifications have been proposed for mycobacteria in the last 50 years. The classification based on major phenotypic features (growth rate and pigmentation type) proposed by Runyon (118) still remains popular. For the reader's ease, the species discussed here are arranged, in alphabetical order, within a simplified Runyon scheme. The only exception are the environmental mycobacteria, which are dealt with separately. As usual, the distinction between rapid and slow growth is based on the ability of strains to develop clearly visible colonies in less or more than 7 days, respectively.
PIGMENTED SLOW GROWERS
M. bohemicum
Three isolates of a new mycobacterium, grown in different sputum samples from a patient with pulmonary disease due to M. tuberculosis, were recognized in 1998 and considered to represent a new species, M. bohemicum (108). An identical mycobacterium, at that time unidentified, had been isolated 1 year earlier from a cervical lymph node of a child (148).
Phenotypic features.
M. bohemicum is a rod-shaped acid-fast bacillus which grows slowly at temperatures ranging from 25 to 40°C. The colonies are small (1 to 2 mm in diameter), smooth, and scotochromogenic, while the majority of biochemical tests are negative (Table 1). Its lipid component is characterized by an uncommon TLC pattern (Table 2), shared only by M. hassiacum and M. mucogenicum. Both GLC (Table 3) and HPLC profiles overlap with, or are hardly differentiable from, those of MAC and M. scrofulaceum (148). Interestingly, one strain, out of a group isolated from the environment, presented an HPLC chromatotype characterized by a single cluster, clearly different from all other strains isolated so far (140).
TABLE 1.
Major biochemical and cultural features of slowly growing mycobacteriaa
| Species | Arylsulfatase (3 days) | Semiquantitative catalase (mm) | 68°C catalase | Nitrate reduction | Tween 80 hydrolysis | Urea hydrolysis | Pigment |
|---|---|---|---|---|---|---|---|
| M. bohemicum | − | <45 | + | − | − | +/− | + |
| M. botniense | −b | <45 | − | − | − | − | + |
| M. branderi | +c | <45 | +c | − | − | − | − |
| M. canettii | ND | ND | − | + | − | ND | − |
| M. celatum | + | <45 | + | − | − | − | + |
| M. conspicuum | + | <45c | + | − | + | − | + |
| M. cookii | + | ND | ND | − | − | − | + |
| M. doricum | − | <45 | +c | + | − | + | + |
| M. genavense | − | >45 | + | − | − | + | − |
| M. heckeshornense | − | <45 | + | − | − | − | + |
| M. heidelbergense | − | ND | + | − | + | + | − |
| M. hiberniae | −d | >45c | +c | + | v | − | +e |
| M. interjectum | − | <45c | + | − | − | + | + |
| M. intermedium | + | >45c | + | − | + | + | + |
| M. kubicae | − | >45 | ND | + | − | − | + |
| M. lacus | ± | <45 | ND | + | ± | + | − |
| M. lentiflavum | − | <45 | ± | − | − | − | + |
| M. palustre | −d | <45 | +c | +/− | + | + | + |
| M. shottsii | − | ND | v | − | − | + | − |
| M. triplex | −b | >45 | + | + | − | + | − |
| M. tusciae | −b | <45 | + | + | + | + | + |
+, positive; −, negative; +/−, predominantly positive; ±, weak; ND, not done; v, variable.
Test positive at 14 days.
Unpublished data.
Test variable at 10 days.
Pink pigment.
TABLE 2.
Distribution of different mycolate types among various mycobacterial speciesa
| Species | Mycolic acid typeb
|
Species, with identical distribution | ||||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | VII | ||
| M. abscessus | + | + | − | − | − | − | − | M. chelonae |
| M. alvei | + | − | − | − | − | − | + | None |
| M. bohemicum | + | − | +c | +c | − | + | − | M. komossense |
| M. branderi | + | − | − | + | − | + | − | Very common |
| M. brumae | + | − | − | − | − | − | − | M. fallax, M. triviale |
| M. canettii | + | − | + | + | − | − | − | Common, including M. tuberculosis |
| M. celatum | + | − | − | + | − | + | − | Very common |
| M. chlorophenolicum | + | − | − | + | − | + | − | Very common |
| M. confluentis | + | − | − | + | − | − | − | M. bovis BCG |
| M. conspicuum | + | − | − | + | − | + | − | Very common |
| M. cookii | + | + | − | − | − | + | − | None |
| M. doricum | + | − | + | − | − | + | − | None |
| M. elephantis | + | − | − | + | − | + | − | Very common |
| M. frederiksbergense | + | − | − | + | − | + | − | Very common |
| M. genavense | + | + | − | + | − | − | − | M. heidelbergense, M. intermedium, M. lentiflavum, M. malmoense, M. simiae |
| M. hassiacum | + | − | − | − | − | + | − | M. mucogenicum |
| M. heckeshornense | + | − | − | + | − | + | − | Very common |
| M. heidelbergense | + | + | − | + | − | − | − | M. genavense, M. intermedium, M. lentiflavum, M. malmoense, M. simiae |
| M. hiberniae | + | − | − | + | − | + | − | Very common |
| M. hodleri | + | − | − | + | − | + | − | Very common |
| M. holsaticum | + | − | + | + | − | − | − | Common, including M. tuberculosis |
| M. interjectum | + | − | − | + | + | + | − | None |
| M. intermedium | + | + | − | + | − | − | − | M. genavense, M. heidelbergense, M. lentiflavum, M. malmoense, M. simiae |
| M. kubicae | + | − | + | + | − | − | − | Common, including M. tuberculosis |
| M. lentiflavum | + | + | − | + | − | − | − | M. genavense, M. heidelbergense, M. intermedium, M. malmoense, M. simiae |
| M. madagascariense | + | − | − | + | − | + | − | Very common |
| M. mageritense | + | + | − | − | + | − | − | M. chitae, M. farcinogenes, M. fortuitum, M. peregrinum, M. porcinum, M. senegalense, M. smegmatis |
| M. mucogenicumd | + | − | − | − | − | + | − | M. hassiacum |
| M. murale | + | − | − | + | − | + | − | Very common |
| M. novocastrense | + | − | − | + | − | + | − | Very common |
| M. peregrinum | + | + | − | − | + | − | −e | M. chitae, M. farcinogenes, M. fortuitum, M. mageritense, M. porcinum, M. senegalense, M. smegmatis |
| M. tusciae | + | − | − | + | − | + | − | Very common |
No information is available for M. botniense, M. goodii, M. immunogenum, M. lacus, M. palustre, M. septicum, M. shottsii, M. triplex, M. vanbaalenii, and M. wolinskyi.
I, α-mycolate; II, α′-mycolate; III, methoxy-mycolate; IV, keto-mycolate; V, epoxy-mycolate; VI, wax esters; VII, ω1-methoxy-mycolate.
Data from reference 140.
Data from reference 96.
Present in most strains (86).
TABLE 3.
GLC patterns of methyl-branched fatty acids, alcohols, and mycolic acid cleavage products in different mycobacterial speciesa,b
| Species | Methyl-branched fatty acidsc,d | Alcohols | Mycolic acid cleavage products |
|---|---|---|---|
| M. abscessus | 24:0 | ||
| M. alvei | x-Me 16:0 | 22:0 | |
| x-Me 18:1 | 24:0 | ||
| M. bohemicum | 2-Me 20:0 | 2OH 18:0 | 22:0 |
| 2,4-diMe 22:0 | 2OH 20:0 | 24:0 | |
| 2,4,6-triMe 22:0 | |||
| 2,4,6-triMe 24:0 | |||
| M. botniense | 2,4,6,x-tetraMe 20:0 2,4,6,x,x-pentaMe 22:0 | 2OH 20:0 2OH 22:0 | 24:0 26:0 |
| M. branderi | 2OH 20:0 | 24:0 | |
| 26:0 | |||
| M. brumae | 20:0 | ||
| 22:0 | |||
| M. canettii | |||
| M. celatum | 2OH 20:0 | 24:0 | |
| 2OH 22:0 | 26:0 | ||
| M. chlorophenolicum | 2OH 18:0 | ||
| 2OH 20:0 | |||
| M. conspicuum | 2-Me 12:0 | 2OH 18:0 | |
| 2OH 20:0 | |||
| M. cookii | 24:0 | ||
| M. doricum | 2OH 18:0 | ||
| 2OH 20:0 | |||
| M. frederiksbergense | 2OH 18:0 | 22:0 | |
| 2OH 20:0 | |||
| M. genavense | 24:0 | ||
| 26:0 | |||
| M. goodii | 24:0 | ||
| M. hassiacum | 9-Me 16:0 | 2OH 18:0 | |
| 10-Me 16:0 | 2OH 20:0 | ||
| M. heckeshornense | 10-Me 16:0 | 2OH 20:0 | 26:0 |
| 2OH 22:0 | |||
| M. heidelbergense | |||
| M. hiberniae | 2OH 18:0 | 22:0 | |
| 2OH 20:0 | 24:0 | ||
| M. hodleri | 10-Me 16:0 | 2OH 18:0 | 22:0 |
| 2OH 20:0 | 24:0 | ||
| M. holsaticum | 2OH 20:0 | ||
| M. interjectum | |||
| M. intermedium | 2-Me 12:0 | ||
| 2-Me 14:0 | |||
| M. lentiflavum | |||
| M. madagascariense | 2,4-diMe 20:0 | 2OH 18:0 | 22:0 |
| 2OH 20:0 | |||
| M. mucogenicumc | 10-Me 16:0 | 2OH 18:0 | 22:0 |
| 24:0 | |||
| M. murale | 2OH 20:0 | ||
| M. palustref | 2-Me 10:0 | 22:0 | |
| 2-Me,16-phe 16:0 | 24:0 | ||
| 2-Me 19:0 | 26:0 | ||
| 2-Me 20:0 | |||
| 2,9-diMe 20:0 | |||
| M. peregrinum | 24:0 | ||
| 26:0 | |||
| M. triplexg | 24:0 | ||
| 26:0 | |||
| M. tusciae | 8-Me 16:0 | 2OH 18:0 | |
| 2OH 20:0 | |||
| M. vanbaalenii | 10-Me 18:0 | ||
| M. wolinskyi | 24:0 |
No information is available for M. confluentis, M. elephantis, M. immunogenum, M. kubicae, M. lacus, M. mageritense, M. novocastrense, M. septicum, and M. shottsii.
Numbers to the left of the colon indicate the number of carbon atoms; numbers to the right of the colon indicate the number of double bonds.
n-Me, methyl branch at position n.
In all the species, 10-Me 18:0 (tuberculostearic acid) is also present.
Data from reference 96.
Unpublished data.
Data from reference 133.
Three strains have been investigated for antimicrobial susceptibility; the pattern of two of them, determined in liquid medium (145, 148), is clearly less resistant than that of the strain on which the sp. nov. description is based (Table 4). It should, however, be noted that the latter strain was tested on solid media, which are characterized by yielding consistently higher, and less reliable, minimal inhibitory concentrations (MIC) (69).
TABLE 4.
| Species | No. of strains tested | Ethambutol | Isoniazid | Rifam pin | Streptomycin |
|---|---|---|---|---|---|
| M. bohemicumc | 3 | R | R | v | I |
| M. branderi | 9 | S | R | R | S |
| M. canettii | 2 | S | S | S | v |
| M. celatum | 24 | v | R | R | S |
| M. conspicuum | 2 | S | R | I | I |
| M. cookii | 17 | S | S | S | S |
| M. doricum | 1 | S | S | S | S |
| M. genavense | 8 | R | R | S | S |
| M. heckeshornense | 1 | S | R | v | S |
| M. heidelbergensec | 2 | v | v | v | v |
| M. hiberniae | 13 | S | R | R | R |
| M. interjectumc | 4 | R | R | S | v |
| M. intermedium | 1 | S | R | S | R |
| M. kubicae | 15 | S | R | R | R |
| M. lacus | 1 | S | S | S | I |
| M. lentiflavumc | 3 | R | R | R | R |
| M. palustrec | 1 | S | R | S | S |
| M. shottsii | 21 | S | R | S | S |
| M. triplex | 10 | S | R | R | R |
| M. tusciae | 1 | I | ND | S | S |
Genotypic features.
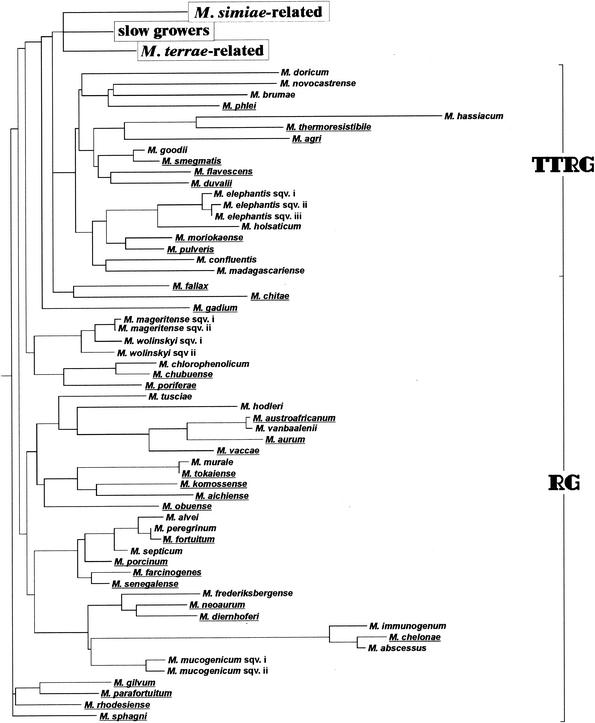
M. bohemicum occupies a phylogenetic position within the large cluster, including the majority of slowly growing mycobacteria (Fig. 3). Among environmental strains, several differences may be present in the otherwise homogeneous pattern of the species; 3 to 4 single-base mismatches have been detected in the 16S rDNA and one 14-nucleotide insertion is contained within the ITS region (140).
FIG. 3.
Phylogenetic tree of slow growers, including new mycobacteria along with previously defined species (underlined). M. visibilis and M. lepraemurium, of which only a partial 16S sequence is available, are not included. sqv., sequevar; MSR, M. simiae related; MTR, M. terrae related; SG, slow growers.
Clinical and epidemiological features.
In contrast to the strain isolated from sputum, there is evident clinical significance for the bacteria isolated from the two nonimmunocompromised children with lymphadenopathy reported so far (145, 148), as well as in a further unpublished similar case of mine.
A large group of M. bohemicum strains have been recently investigated (140); one of them was isolated from a generalized cutaneous infection in an elderly woman, three were isolated from the sputa of three different patients, one was isolated from the mesenteric lymph nodes of a goat, and five were isolated from stream water. The origin of one additional clinical isolate is unknown. Coisolation of M. bohemicum and M. palustre, both regarded as clinically insignificant, has also been reported (139). From such a survey, unusually rich for a species recognized less than 4 years ago, two major observations emerge: M. bohemicum should be regarded as a potential pathogen, and natural water is one of its reservoirs. The virulence of the species also seems confirmed: following intravenous infection of syngeneic gamma interferon-deficient and BALB/c mice with M. bohemicum, also the latter, nonimmunocompromised animals developed splenomegaly and granulomatous liver lesions (36).
Type strain: DSM 44277T. EMBL 16S rDNA sequence accession number: U84502.
M. celatum
M. celatum was first described in 1993 (19), when 24 strains, 22 of which had been isolated independently, were differentiated from MAC and, most importantly, from the phenotypically closely related species M. xenopi. With the exception of five, whose source is unknown, the strains were all of clinical origin, predominantly from the respiratory tract (sputa and bronchial washings) but also from blood, stools, and a bone biopsy specimen. Of 11 patients whose human immunodeficiency virus (HIV) status was known, 7 were seropositive.
Phenotypic features.
M. celatum is acid fast and predominantly rod shaped. It grows in 20 to 35 days, between 33 and 42°C, forming small, smooth, colonies described as unpigmented, but the majority of the strains are pale yellow and scotochromogenic. Polymorphic colonies have also been reported (39). The most important distinctive biochemical test seems to be the positive arylsulfatase activity (Table 1).
In TLC, M. celatum has a mycolate set indistinguishable from those of a number of both slowly and rapidly growing species (Table 2); in contrast, the GLC pattern is uncommon (Table 3). The close similarity of the M. celatum and M. xenopi HPLC profiles (151) resulted, in the years preceding the sp. nov. description, in the inclusion of M. celatum in a group named “M. xenopi-like.” However, a careful assessment of chromatograms allows the easy differentiation of the two species due to the earlier retention times of the peaks of the second cluster characterizing M. celatum.
A unique feature of M. celatum is the consistent resistance to rifampin, with very high MICs (Table 4) (151).
Genotypic features.
The PRA analysis of the hsp65 gene divided the strains investigated in the sp. nov. description into two groups, characterized by seven and six bands. This grouping was confirmed by the patterns obtained by multilocus enzyme electrophoresis and by the sequencing of the 16S rDNA (19). A 10-base mismatch between the two different sequevars (named type 1 and type 2) was detected. A third variant (type 3) was subsequently described on the basis of eight strains from AIDS patients. The 16S rDNA sequence of type 3 differs by 7 bases from that of type 1 and by 17 bases from that of type 2 (16). The mismatches are outside the hypervariable regions A and B.
Picardeau et al. (104) have recently described a new insertion sequence, IS1407, in M. celatum. Three or four copies of IS1407 are present in identical genomic positions in M. celatum types 1 and 3, while it is absent in type 2. Furthermore, the same authors uncovered the presence of a single pulsed-field gel electrophoresis (PFGE) pattern among all type 1 and type 3 isolates and polymorphic patterns among type 2 isolates (104). The fact that M. celatum type 2 lacks IS1407, differs by 10 and 17 bp in 16S rDNA sequences from types 1 and 3, and has very different PFGE genotype patterns model seems to support the notion that it is (or may be) a different species (104). Interestingly two strains of M. celatum have been detected that contain two copies of the 16S rRNA gene; they have different sequences, of type 1 and type 3, respectively (109, 158).
The presence of only a single nucleotide mismatch with the short stretch of M. tuberculosis complex 16S rRNA targeted by the GenProbe DNA probe (AccuProbe Mycobacterium tuberculosis complex; GenProbe, San Diego, Calif.) is responsible for the cross-hybridization with M. celatum type 1 (18). This cross-reactivity, usually moderate, is detectable only when the stringency of the hybridization conditions is lower than recommended (AccuProbe Mycobacterium tuberculosis complex, package insert). On the basis of the sequence identity of types 1 and 3 in hypervariable region A, a cross-reactivity, although not documented so far, can be assumed for type 3 too. Phylogenetically, M. celatum has genetic markers of slow growers, most closely related to M. branderi and M. xenopi, with which it forms a branch that diverged early in the evolution of the slowlygrowing species (Fig. 3).
Clinical and epidemiological features.
Several independent clinical strains, subsequently recognized as being identical to M. celatum (four of type 1 and one of type 2), had been phenotypically investigated in Finland before the sp. nov. description; they were isolated between 1972 and 1990 without being assigned to any species (11).
Numerous clinically significant isolations have been reported in AIDS patients, presenting as disseminated infection in 10 patients (7, 37, 48, 105, 151, 178), limited to the lungs in 5 cases (48), and exclusively extrapulmonary in one case of penile infection (29). In contrast, in nonimmunocompromised patients, only one case of fatal pulmonary disease in an elderly woman (21) and one of childhood lymphadenopathy (G. Haase, H. Skopnik, S. Bätge, and E. C. Böttger, Letter, Lancet 344:1020-1021, 1994) have been reported.
Although many clinical isolates of M. celatum are not clinically significant, the pathogenicity of the species seems, higher than that of the majority of nontuberculous mycobacteria, particularly in AIDS patients. Such a hypothesis also seems confirmed by the experimental inoculation of M. celatum into BALB/c and syngeneic gamma interferon-deficient mice. It gave rise to splenomegaly and granulomatous liver lesions in both immunocompetent and immunodeficient animals, demonstrating the high virulence of the strain (36).
Isolation from the environment has not been reported so far.
Type strains: ATCC 51131T (type 1), ATCC 51130T (type 2) and NCTC 12882T (type 3). EMBL 16S rDNA sequence accession numbers: L08169 (type 1), L08170 (type 2), and Z46664 (type 3).
M. conspicuum
Only the two strains, each represented by multiple isolates, on which the sp. nov. description was based in 1995 have been reported so far for M. conspicuum (129). Both were responsible for disseminated infections in heavily immunocompromised young patients, one with AIDS and the other with an unidentified cellular immunodeficiency.
Phenotypic features.
The mycobacterial cells are coccobacillary and acid fast. Growth on solid media is slow and requires, at temperatures from 22 to 31°C, at least 3 weeks. Growth at 37°C occurs only in liquid media. A pale yellow scotochromogenic pigment is present, and the majority of biochemical traits are negative (Table 1).
The TLC mycolic acid pattern is shared with other slow growers including MAC and M. terrae (Table 2). The GLC fatty acid set is unique (Table 3). Similarity, again to M. terrae, is detected by HPLC, with both profiles presenting two clusters of peaks, with the early cluster being major and the late cluster being minor. The profiles are nevertheless distinguishable since the peaks of M. conspicuum present slightly later retention times (129).
Although the species is not very resistant in vitro (Table 4), both infected patients remained culture positive until death, despite treatment with multiple antibiotics.
Genotypic features.
The 16S rDNA of M. conspicuum is, from the genetic point of view, characteristic of a slow grower; M. conspicuum occupies a phylogenetic position close to M. asiaticum (Fig. 2 and 3).
Clinical and epidemiological features.
Both patients from whom M. conspicuum was isolated had severe immune impairment. Nevertheless, the virulence of the species does not seem lower than that of other opportunistic agents like MAC and M. genavense, especially since the disseminated mycobacterial infection was probably a major cause of death in both subjects. The virulence in vivo of M. conspicuum seems to be confirmed by the development of splenomegaly and granulomatous liver lesions not only in syngeneic gamma interferon-deficient mice but also in BALB/c mice following intravenous infection (36).
Although only two strains of M. conspicuum have been detected so far, there is the possibility of underisolation due to the low growth temperature required for the isolation of M. conspicuum.
Type strain: DSM: 44136T. EMBL 16S rDNA sequence accession number: X88922.
M. doricum
M. doricum was described in 2001 (153) on the basis of a single isolate from the cerebrospinal fluid of a severely immunocompromised AIDS patient.
Phenotypic features.
M. doricum is scotochromogenic and forms smooth yellow colonies in about 2 weeks at temperatures ranging from 25 to 37°C. Only urea and nitrates, among the most frequently used biochemical tests, are positive (Table 1).
The mycolic acid composition is not common among scotochromogens (Table 2), while the GLC pattern is similar to that of M. chlorophenolicum (Table 3). The HPLC profile is unique, characterized by an early major cluster of peaks followed by a minor one (153).
Antimicrobial susceptibility testing, performed in liquid medium, revealed good activity of first-line antituberculous drugs (Table 4).
Genotypic features.
The nucleotide sequence of the 16S rRNA gene is characterized by a short helix 18 and a single cytosine insertion in helix 10; the combination of such features is considered the genetic signature of thermotolerant rapid growers, the group to which M. doricum presents the closest phylogenetic relatedness (Fig. 2 and 4). Among the slow growers not genetically related to M. simiae, M. doricum is at present the only species to present a short helix 18.
FIG. 4.
Phylogenetic tree of rapid growers, including new mycobacteria along with previously defined species (underlined). sqv., sequevar; TTRG, thermotolerant rapid growers.
Clinical and epidemiological features.
Although the patient infected with M. doricum suffered from headache and stiff neck, the correlation of such symptoms with mycobacterial isolation is not certain because Cryptococcus neoformans was isolated from blood cultures concurrently. The clinical significance of the strain, although possible, remains questionable. No other isolation of this species has been reported so far.
Type strain: DSM 44339T, CIP 106867T. EMBL 16S rDNA sequence accession number: AF264700.
M. heckeshornense
M. heckeshornense was described in 2000 (116) after one strain had been repeatedly isolated, during a 5-year period, from the sputum of a young, nonimmunocompromised woman with pulmonary cavitation and infiltrates. After resection of the lung, M. heckeshornense was also isolated from a biopsy sample.
Phenotypic features.
The growth of M. heckeshornense is scanty on Middlebrook agar medium and even poorer on egg medium. Growth is obtained after at least 4-weeks at 30 to 45°C. Many phenotypic features of M. heckeshornense are hardly distinguishable from those of M. xenopi: the colonies are small, scotochromogenic, and smooth, and the major biochemical tests are negative (Table 1).
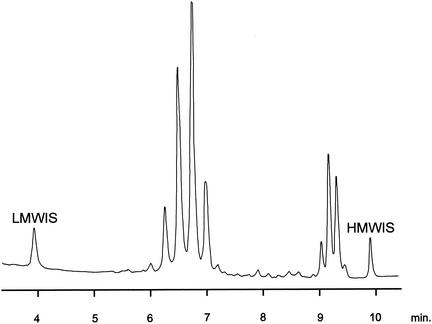
The combination of mycolic acids by TLC is very common among mycobacteria (Table 2), while the fatty acid profile revealed by GLC are close to those of Mycobacterium botniense (Table 3), a recently described new species related to M. xenopi (141). The HPLC pattern (Fig. 5) is practically overlapping with those of M. xenopi and M. botniense. Therefore, there is a high risk of misidentification as M. xenopi.
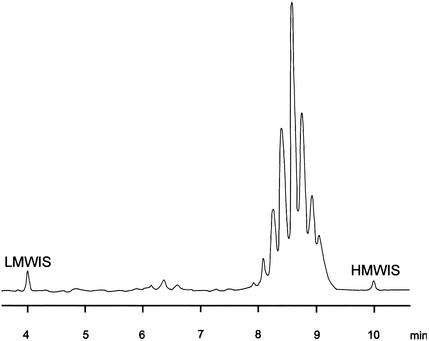
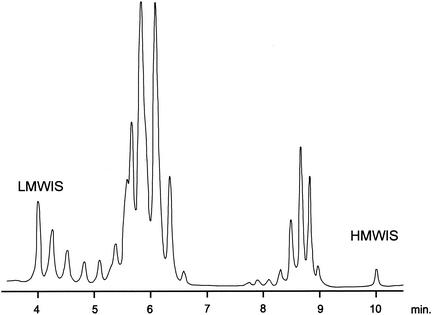
FIG. 5.
Mycolic acid pattern of M. heckeshornense obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
M. heckeshornense is susceptible to first-line antituberculous drugs except isoniazid (Table 4).
Genotypic features.
M. xenopi is the species most closely related in the phylogenetic tree constructed on the basis of the 16S rDNA (Fig. 3). The relatedness is confirmed, although with higher sequence divergence, by comparison of the ITS regions. One strain resembling M. xenopi and containing two different copies of the 16S rDNA, corresponding to M. xenopi and M. heckeshornense respectively, has been reported (158).
Clinical and epidemiological features.
The criteria required to demonstrate the pathogenicity of a nontuberculous mycobacterium (165) seem clearly fulfilled from in case that originated the sp. nov. description. Other isolations, although not documented in the literature, are inferred from the presence in the GenBank database of two identical 16S rDNA sequences. The first belongs to a strain from a patient with pulmonary disorders, and the second sequence came from a strain tentatively named “Mycobacterium sydneyiensis” (sic) (113).
Type strain: DSM 44428T. GenBank 16S rDNA sequence accession number: AF174290.
M. interjectum
M. interjectum was recognized as a new species in 1993 (127), when it was isolated twice from a lymph node of a child with lymphadenopathy. The first isolate was grown from a fragment of a partially resected lymph node and the second, three months later, when a fistula developed and a total surgical resection was performed.
Phenotypic features.
M. interjectum is a slowly growing (at 31 to 37°C) acid-fast coccobacillus that forms yellowish scotochromogenic colonies mostly resembling, in biochemical and cultural features, M. scrofulaceum (Table 1). A culture presenting both yellow and white colonies of M. interjectum at the same time has also been reported without any interconvertibility among the two chromatic variants detected (147).
The TLC pattern of mycolic acids is similar to those of MAC and M. scrofulaceum (Table 2). The pattern of fatty acids revealed by GLC is also close to the latter species (Table 3). Interestingly, two different HPLC profiles of mycolic acids may be obtained for M. interjectum; the first and most frequent grossly resembles those of MAC and M. scrofulaceum (143), while the other, which is unique, has a large cluster of peaks emerging during the second half of the elution (147). Available data concerning antimicrobial susceptibility show the species to be quite resistant to all antituberculous drugs (Table 4).
Genotypic features.
The identity of the whole hypervariable region B of the 16S rDNA demonstrates the close genetic relatedness of M. interjectum to M. simiae and other “intermediate” species (Fig. 2 and 3).
Clinical and epidemiological features.
Cervical lymphadenitis in childhood is the most frequent pathology attributable to M. interjectum. In addiction to the case which allowed the recognition of the species, four further similar cases have been reported (30, 84, 119). Two more cases of chronic lung disease (38, 84) and a repeated isolation from urine (143) should also be added. Only two isolations from AIDS patients have been reported (51, 147). The clinical significance of these isolations is, however, questionable. Well documented, although unpublished, is a case of pulmonary cavitary disease diagnosed following double bronchoscopic isolation of M. interjectum; in this case the patient was successfully treated for 5 months with clarithromycin.
No doubt exists about the potential pathogenicity of the species, as confirmed also by the investigation of virulence in vivo, carried out by experimental infection of BALB/c and syngeneic gamma interferon-deficient mice, which gave rise to splenomegaly and granulomatous liver lesions in both animals (36).
Type strain: DSM 44064T. EMBL 16S rDNA sequence accession number: X70961.
M. intermedium
The sp. nov. description dates from 1993 (90) and is related to three isolations from the sputum of a nonimmunocompromised patient with chronic bronchitis. The patient was treated and showed a significant improvement.
Phenotypic features.
The development of recognizable colonies on egg-based media requires at least 2 weeks at 25 to 41°C. Although the species has been described as photochromogenic, its yellow pigmentation seems enhanced rather than induced by light exposure. The coccobacillary cell morphology and the complex of cultural and biochemical traits (positive arylsulfatase, urease, and Tween 80 hydrolysis [Table 1]) result in this species being hard to distinguish from M. asiaticum.
The mycolate investigation, carried out using TLC, reveals a pattern overlapping that of a large group of species including M. simiae, M. malmoense, and M. genavense (Table 2). The GLC profile of fatty acids, pyrolysis products, and alcohols is easily distinguishable from those of other slow growers (Table 3). The unique HPLC pattern of mycolic acid bromophenacyl esters is characterized by a single, large cluster of peaks emerging in the second part of elution (147).
The only strain detected so far is resistant to isoniazid and streptomycin (Table 4).
Genotypic features.
The nucleotide sequence of hypervariable region B, in the 16S rDNA, overlapping those of M. simiae and related slow growers, is characterized by a short helix 18. This feature, emphasized also by the name (the Latin word “intermedius,” likewise “interjectus” indicating another mycobacterial species, means “halfway”), is responsible for the phylogenetic position of the species, intermediate between rapid and slow growers, close to M. simiae (Fig. 2 and 3). M. intermedium is the most ancient species to diverge from the slow growers, typically characterized by a long helix 18.
Clinical and epidemiological features.
No further isolation has been reported so far from patients or from the environment. The clinical significance of M. intermedium appears questionable. The repeated isolation and the presence of acid-fast bacilli in smears from sputum are not sufficient to demonstrate its pathogenic role in the only case reported so far. Furthermore, experimental infection with M. intermedium showed that it was pathogenic for gamma interferon-deficient mice but not for immunocompetent BALB/c mice (36).
Type strain: DSM 44049T. EMBL 16S rDNA sequence accession number: X67847.
M. kubicae
The new species M. kubicae, described in 2000, emerged from the investigation of a cluster of 15 strains that had been collected over several years at the Centers for Disease Control and Prevention (CDC) (41).
Phenotypic features.
The cells are strongly acid fast and are rod shaped and frequently bent. The colonies, which are yellow, smooth, and domed, are scotochromogenic and grow slowly between 28 and 37°C. Among the biochemical features, only semiquantitative catalase, clearly over 45 mm, stands out from other, mostly negative, tests (Table 1).
The TLC investigation of mycolic acids of M. kubicae reveals a common pattern among mycobacteria, including M. tuberculosis, while nothing is known about the GLC fatty acid composition. The well-documented HPLC pattern (41) is characterized by a single cluster of peaks grossly resembling the profile of M. asiaticum, hence the M. asiaticum-like appellation previously indicating such strains.
Genotypic features.
Because of the sequence identity of the 16S rDNA hypervariable region B, M. kubicae clusters with the steadily increasing group of M. simiae-related slow growers (Fig. 2 and 3).
Clinical and epidemiological features.
The only clinical information concerning 11 of 15 strains on which the description of M. kubicae is based is that the source was respiratory (sputum or bronchial aspirate). Thus no hypothesis may be made, at present, about its potential pathogenicity.
Type strains: ATCC 700732T, CIP 106428T. GenBank 16S rDNA sequence accession number: AF133902.
M. lentiflavum
M. lentiflavum was described in 1996 (130) on the basis of 22 isolates, 11 of which were independent (4 from gastric fluid, 4 from sputum, 2 from urine, and 1 from a biopsy specimen) while the others were associated with a contaminated bronchoscope.
Phenotypic features.
M. lentiflavum is a scotochromogenic slow grower (3 to 4 weeks is needed, on average, at 22 to 37°C) whose most common biochemical tests are negative (Table 1). The cells are acid fast and coccobacillary, while the colonies appear pale yellow, tiny and smooth.
The TLC pattern of mycolic acids reveals the presence of α-, α′-, and keto-mycolates, a pattern identical to those of M. simiae, M. malmoense, M. genavense, and M. intermedium (Table 2). The GLC profile does not present discriminative compounds (Table 3), while the HPLC chromatotype presents a three-clustered pattern very similar to the one of M. simiae; the two species are distinguishable but not easily (152).
The little available information about the susceptibility to antimycobacterial drugs suggests a high-level resistance to the majority of them (Table 4).
Genotypic features.
The genetic sequence of M. lentiflavum presents, within the hypervariable region B of 16S rDNA, 100% sequence homology to M. simiae and related mycobacteria, with which it phylogenetically clusters (Fig. 2 and 3). Two sequevars differing in two nucleotides within the 5′ end of 16S rDNA are present (http://www.ridom.de/static/download/dign_mycoba.pdf).
The PRA of hsp65 revealed, among the 22 isolates investigated at the time of the sp. nov. description, three different patterns, none of which overlapped with those of any previously evaluated mycobacterial species (130).
Clinical and epidemiological features.
Although the majority of M. lentiflavum isolates appear to lack clinical significance (47), several pathologies related to this species have been reported. Among the strains involved in the sp. nov. description, the one isolated from a vertebral disk biopsy specimen (130) was most probably the agent of spondylodiscitis from which the patient suffered and whose symptoms, following treatment with multiple antituberculous agents, markedly improved. Other pathologies were reported later: four cases of cervical lymphadenitis (22, 57, 144), one case of cavitary pulmonary disease (144), one disseminated infection in a woman undergoing steroid therapy (67a), and, in AIDS patients, again one disseminated infection (98) and a hepatic abscess (144).
No environmental isolation has been reported so far; however, PCR products similar to those of M. lentiflavum have been obtained from soil (91), and six strains have been detected presenting hsp65 restriction profiles typical of M. lentiflavum, although differing results were obtained from 16S rDNA and ITS sequences (81).
Experimental investigation revealed a low-level virulence for M. lentiflavum, which was not able to cause persistent infection in BALB/c or gamma interferon-deficient mice (36).
Type strain: ATCC 51985. EMBL 16S rDNA sequence accession number: X80769.
M. palustre
The description of M. palustre, in 2002 (139), is based on eight environmental strains from four Finnish streams, three clinical isolates (two from sputum and one from a child's cervical lymph node), and two veterinary isolates from porcine submandibular lymph nodes.
Phenotypic features.
M. palustre grows in 3 to 4 weeks at temperatures ranging from 22 to 42°C (optimum, 37°C) and forms smooth, yellow, scotochromogenic colonies. Tween 80 hydrolysis and urease are positive (Table 1).
No TLC mycolic acid investigation has been performed. The GLC fatty acid pattern is unique among mycobacteria (Table 2). The HPLC profile of M. palustre presents peaks arranged in a single cluster; such peaks, although corresponding to the ones of M. kubicae (41), differ from them in height (139).
Susceptibility testing, performed only on the isolate of human origin, revealed that all antituberculous drugs except isoniazid had low MICs, as did quinolones and macrolides (152).
Genotypic features.
The sharing, within the 16S rDNA sequence of the hypervariable region B with M. simiae-related slowly growing mycobacteria determines the clustering of M. palustre with the latter group, close to M. kubicae (Fig. 2 and 3). All strains are characterized by a unique nucleotide sequence in the ITS region (139).
M. palustre strains yield different results when tested with two commercially available DNA probes. With AccuProbe, they hybridize with MAC-specific probe but not with the ones specific for M. avium and M. intracellulare (152). With INNO LiPA Mycobacteria (Innogenetics) they hybridize only with the genus Mycobacterium-specific probe, being negative with the others including those specific for the M. avium-intracellulare-scrofulaceum grouping and for the single species M. avium and M. intracellulare (150, 152).
Clinical and epidemiological features.
The involvement in a case of childhood lymphadenopathy places M. palustre among potentially pathogenic species. There remains uncertainty concerning the significance of lymphatic infections in animals, given that M. palustre was grown from apparently healthy slaughtered pigs. Natural waters may well represent the environmental reservoir of the new species
Type strains: ATCC BAA-377T, DSM 44572T. EMBL 16S rDNA sequence accession number: AJ308603.
M. tusciae
M. tusciae was described in 1999 (149) on the basis of one clinical and two environmental isolates, the first from a child's cervical lymph node and the others from potable water.
Phenotypic features.
The cells of M. tusciae are rod shaped and acid fast; the growth is visible, on egg medium, in about 1 month at temperatures ranging from 25 to 32°C. Growth at 37°C is achieved only in liquid media or on Middlebrook agar. The colonies are rough and scotochromogenic and are characterized by a very elevated center surrounded by a flat and uneven fringe. All major biochemical tests are positive (Table 1).
The mycolic acid composition of the M. tusciae cell wall is one of the most common among slow-growers. It is shared, among others, by MAC, M. xenopi, and M. scrofulaceum (Table 2). The problems associated with distinguishing M. tusciae from M. scrofulaceum are present also in GLC analysis (Table 3), while the HPLC pattern, characterized by an early major cluster of peaks, differs from those of any species reported so far (149).
The species is characterized in vitro by moderate susceptibility to antituberculous drugs (Table 4).
Genotypic features.
The 16S rDNA sequence has the distinctive features of thermotolerant rapid growers, i.e., a short helix 18 and a single cytosine insertion in helix 10. Nevertheless, in the phylogenetic tree, M. tusciae, although clustering with rapid growers, lies on a branch far from thermotolerant rapid growers and even farther from M. simiae-related organisms, with which it shares growth rate and a short helix 18 (Fig. 2 and 4).
Clinical and epidemiological features.
Two strains of M. tusciae were isolated from potable water. Since the mouth is the usual route for agents of such childhood pathologies, the water may well represent a source for lymphonodal cervical infections. The isolation from a sterile site supports the clinical relevance of M. tusciae, especially considering that the slight immunosuppression of the child from whom it was grown (as a result of treatment with low-dose steroids) does not seem to have played a major role in this case.
Type strain: DSM 44338T. EMBL 16S rDNA sequence accession number: AF058299.
NONPIGMENTED SLOW GROWERS
M. branderi
In 1995, nine strains, repeatedly isolated from the sputum of separate subjects including patients with pulmonary cavitary disease insensitive to drug treatment, were the basis for the description of the new species M. branderi (78).
Phenotypic features.
The species is nonchromogenic, grows slowly, and forms smooth, often umbonate colonies after incubation at temperatures ranging from 25 to 45°C. Biochemical tests, except for arylsulfatase, are negative (Table 1).
The mycolic acid composition (TLC) is very common among slowly growing mycobacteria (MAC, M. scrofulaceum, etc., [Table 2]), while the profile of GLC fatty acids and the HPLC pattern are not distinguishable from those of M. celatum (Table 3) (17).
Like M. celatum, M. branderi is resistant to rifamycins and to isoniazid (Table 4).
Genotypic features.
The 16S rRNA gene contains a long helix 18, made even longer by a one-nucleotide insertion. The most closely related species is M. celatum (Fig. 2 and 3).
Clinical and epidemiological features.
The potential pathogenicity of M. branderi, hypothesized at the time of sp. nov. description, was confirmed by a subsequent report (174) describing the isolation of this species from the hand of a woman with ulcerative tenosynovitis. The isolation of M. branderi from bronchoalveolar lavage fluid of an elderly patient with pneumonic infiltrates, reported in the same paper, was, in contrast, considered not significant since a MAC isolate had also been grown from the sputum of the subject. Infection with M. branderi gives rise to splenomegaly and granulomatous liver lesions in both BALB/c and syngeneic gamma interferon-deficient mice, thus demonstrating virulence for immunocompetent animals as well (36).
Type strain: ATTC 51789T. EMBL 16S rDNA sequence accession number: X82234.
“M. canettii”
The smooth Canetti strain of M. tuberculosis has long been considered a curiosity and only recently (28) was shown to differ from common rough strains by the presence of large amounts of lipooligosaccharides. Apart from the historical strain isolated in 1969 from a patient about whom no information is available, the first human isolate of the smooth variant of M. tuberculosis dates back to 1993, when it was cultured from a cervical lymph node of a Somali child (159).
Phenotypic features.
Although all the commonly investigated phenotypic tests (Table 1), including niacin accumulation, and the antimicrobial susceptibility pattern are compatible with M. tuberculosis (Table 4), its growth is much faster since as the colonies are clearly visible within 6 days. Serial subcultures of the strains reveal a consistent conversion of colonies from smooth to rough with a 1:500 rate and without any reversion from rough to smooth. The above colony conversion is accompanied by the loss of lipooligosaccharides.
No difference in the composition of cell wall mycolic acids (TLC) (Table 2) and fatty acids (GLC) (Table 3) exists between M. canettii and M. tuberculosis. The HPLC pattern too, unreported so far, is indistinguishable from that of M. tuberculosis (Fig. 6).
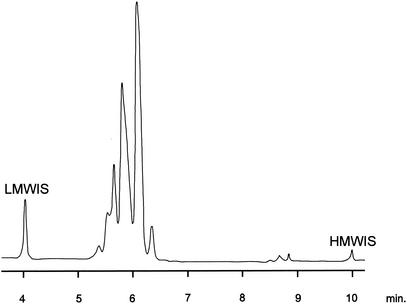
FIG. 6.
Mycolic acid pattern of M. canettii obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
Genotypic features.
Molecular comparisons show that the sequence of the 16S rDNA of the smooth variant, in the regions known to harbor genus- and species-specific variations, is identical to that of M. tuberculosis (102). Among polymorphic genetic markers, a previously unreported pattern emerges for IS6110 and IS1081. The spacer oligonucleotide typing (spoligotyping) pattern is characterized by two spacer sequences only, a highly unusual pattern previously encountered, although with different spacers, in only four M. microti isolates. Almost all the above molecular markers (159) detected in the recently isolated M. tuberculosis smooth variant are shared by the historical Canetti strain. Recently, the PRA of hsp65, using the restriction enzyme HhaI, was shown to clearly distinguish M. canettii strains, characterized by three bands, from other members of M. tuberculosis complex, which consistently have four bands (49).
Clinical and epidemiological features.
An additional isolation has been reported in Switzerland from an AIDS patient with mesenteric tuberculosis who had worked for more than 20 years in Africa (102). Experimentally infected guinea pigs revealed tuberculous lesions and severe loss of body fat at autopsy. Such lesions were much more disseminated in animals inoculated with the smooth variant than in controls infected with M. tuberculosis H37Rv, demonstrating a greater virulence of the former. The shift from smooth to rough colonies is higher in vivo than in vitro, suggesting that the rough variant is better adapted to the in vivo conditions.
On the basis of the experimental data, the Canetti variant seems to be the most divergent member of the M. tuberculosis complex; nevertheless, it is still uncertain whether it represents a new species. van Soolingen et al. (159) suggest that the M. tuberculosis complex should be regarded as a single species, including several subspecies, and propose M. tuberculosis subsp. Canetti as name of the novel taxon.
The lack of other reports concerning isolates of M. canettii may well be due to misdiagnosis attributable to the extremely unusual colony morphology as well as its rarity. Animals are considered the most probable reservoir at present.
Type strain: CIPT 140010059T. EMBL 16S rDNA sequence accession numbers: AJ007315 (hypervariable region A), AJ007316 (hypervariable region B).
M. genavense
Although M. genavense was not the first new species to be recognized in the era of molecular taxonomy, it is certainly the one that has attracted the most widespread awareness and that accounts for most of the literature. The species was described in 1993 (9) on the basis of a conspicuous group of strains (more than 30), the first of which had been reported, as unidentified, 3 years earlier (65). Extensive clinical information concerning the AIDS patients from whom the majority of the above-mentioned strains had been isolated has been discussed in a previous paper (10).
Phenotypic features.
The most striking phenotypic feature of M. genavense is the extremely slow growth, obtained almost exclusively in liquid media. The large majority of strains were isolated with the radiometric Bactec 460TB instrumentation (Becton Dickinson); other liquid media were subsequently found suitable to support the growth. Scanty growth may be obtained after prolonged incubation on solid media such as Middlebrook 7H11 enriched with 2 μg of mycobactin J per ml or on Middlebrook 7H10 supplemented with 10% human blood (87).
The cell morphology is coccobacillary, and the colonies require 1 to 3 months to become visible. They are initially transparent but may become creamy and white with age. A wide range of temperatures, from 25 to 42°C, with preference for the higher ones, support growth. Because of the difficulty in obtaining enough biomass, very little is known about the phenotypic features of this species. Catalase (both semiquantitative and 68°C) and urease are positive, while niacin, nitrate, Tween 80 hydrolysis, and arylsulfatase are negative (Table 1).
Important features for the presumptive identification of M. genavense are the clear enhancement of growth achieved in acidified (pH 6.2) broth and the inhibition of growth, like M. tuberculosis (155) but unlike other nontuberculous mycobacteria, by p-nitro-α-acetylamino-β-hydroxypropiophenone (131).
Knowledge concerning M. genavense lipid patterns is limited to HPLC analysis, which reveals a close similarity to M. simiae. The patterns of the two species are characterized by three almost overlapping late clusters of peaks (27).
Little is known about the antimicrobial susceptibility of M. genavense (Table 4). A pattern overlapping that of MAC has been hypothesized (101), but several clinical and microbiological data seem to suggest a lower resistance level, with only isoniazid and ethambutol being clearly inactive (9, 155).
Genotypic features.
From the molecular point of view, M. genavense is characterized, within the 16S rDNA, by a sequence in hypervariable region B identical to the one of M. simiae and related mycobacteria (Fig. 2 and 3).
Clinical and epidemiological features.
In the years preceding the introduction of highly effective antiretroviral treatments, M. genavense was the second most frequently isolated species, after M. avium, responsible for disseminated infections in AIDS patients. Most of these patients present with fever, diarrhea, and weight loss (101). M. genavense was isolated mainly from blood but also from various biopsy specimens (lymph node, spleen, and duodenal mucosa). Cases published before 1998 have been reviewed by Tortoli et al. (146). More recently, 20 additional isolations from AIDS patients have been reported, including cutaneous (S. Fournier and V. Vincent, Letter, Ann. Intern. Med. 128:409, 1998) and genital (160) infections.
Only four M. genavense infections have been reported in HIV-seronegative patients. Two were disseminated, in an immunosuppressed woman (6) and in a leukemic patient (79), while the other two were represented by lymphadenopathy in a child (82) and a soft tissue infection in an immunodeficient, HIV-negative patient (80a). In two cases, M. genavense DNA was detected using PCR in asymptomatic subjects (33). Noteworthy is the finding of M. genavense in hospital tap water; this is a possible link to the disseminated infections in HIV-infected patients (64).
M. genavense is the most common mycobacterial infection in birds (106), which to date are the only identified potential reservoir. Les frequent are infections in other animals, including a disseminated infection in an immunodeficient cat (67).
The many examples of clinical documentation, unusual for nontuberculous mycobacteria, show clearly that M. genavense is pathogenic, at least for HIV-infected persons and for birds. It may be responsible for localized or disseminated lesions, and death is not an exceptional outcome.
A parallel intravenous injection of M. genavense into immunocompetent and gamma interferon-deficient mice produced persistent infection of the liver and spleen only in the latter animals, suggesting that the virulence of the species is not sufficiently high for it to survive and spread in the presence of gamma interferon-activated macrophages (35). However, its persistence in gamma interferon-deficient mice closely resembles the disseminated infection of M. genavense in immunodeficient patients.
Type strain: ATCC 51234T. EMBL 16S rDNA sequence accession number: X60070.
M. heidelbergense
M. heidelbergense was described in 1997 (55) based on two isolates grown 5 years earlier (56) from lymph nodes of an immunocompetent child with lymphadenitis and recurrent fistula formation.
Phenotypic features.
Growth on egg medium is problematic. Colonies are small, nonpigmented, and smooth. Growth occurs at 30 to 37°C, not before the week 4 of incubation; it may, however, be observed earlier on agar and liquid Middlebrook media. Tween 80 hydrolysis and urease are positive (Table 1).
The mycolic acid pattern is identical to those of M. simiae, M. malmoense, and M. intermedium (Table 2). The GLC (Table 3) and HPLC patterns practically overlap with those of M. malmoense (55).
Unusually for a nontuberculous mycobacterium, the type strain of M. heidelbergense is susceptible to isoniazid in addition to other major antituberculous drugs (Table 4).
Genotypic features.
The similarity, at the phenotypic level, to M. malmoense is not confirmed when the 16S rDNA is observed. M. heidelbergense presents complete homology of hypervariable region B to M. simiae and consequently a close phylogenetic relatedness to the latter species (Fig. 2 and 3). The PRA of hsp65 also is suitable to clearly differentiate M. heidelbergense from M. malmoense.
Clinical and epidemiological features.
Like other strains isolated from lymph nodes, which usually require surgical, seldom repeated, intervention, M. heidelbergense must be considered pathogenic, and confirmation of its clinical significance comes from an isolate from a patient with a tumor-mimicking pulmonary infection (103). In contrast to the strain from which the species was described, the susceptibility pattern of the latter was characterized by resistance to all drugs tested but ciprofloxacin, a discrepancy that cannot be attributed to the different testing techniques which were used. Six other independent M. heidelbergense isolations, probably not clinically significant, have been reported: four from sputum, one from gastric fluid, and one from urine (55).
Intravenous inoculation of M. heidelbergense into immunocompetent (BALB/c) and gamma interferon-deficient mice gave rise to splenomegaly and granulomatous liver lesions only in the latter, demonstrating that gamma interferon is decisive for the outcome of infections with this mycobacterium (36).
Type strain: ATCC 51253T. EMBL 16S rDNA sequence accession number: X70960.
M. lacus
The only strain isolated so far was obtained from the synovial tissue excised from the elbow of a woman with bursitis of posttraumatic origin, and the new species was described in 2002 (156).
Phenotypic features.
The strain, which is microscopically characterized by large bacilli with prominent beading, grows in 2 to 3 weeks at 25 to 42°C. The colonies are small, nonpigmented and dry. Among classical biochemical tests, only nitrate reduction and urease are positive (Table 1).
The lipidic pattern, which was not investigated by TLC and GLC, is characterized in HPLC by a single cluster of peaks practically overlapping that of M. gastri (156).
M. lacus is susceptible, in addition to antitubercular drugs, to various molecules including aminoglycosides, quinolones, and macrolides (Table 4).
Genotypic features.
The sequences of the 16S rDNA and the ITS region are unique (Fig. 2). M. malmoense and M. marinum are the most closely related species (Fig. 3). The pattern obtained by PRA of hsp65 is distinct from every one reported previously.
Clinical and epidemiological features.
The isolation from a tissue presenting caseating granulomas unquestionably supports the pathogenic role of the organism. Since the infection was probably contracted during of a trauma while the patient was swimming in a lake, the environment may well represent the natural reservoir of M. lacus.
Type strains: ATCC BAA-323T, DSM 44577T. GenBank 16S rDNA and ITS sequence accession number: AF406783.
M. shottsii
Among epizootic mycobacteria isolated from striped bass in the Chesapeake Bay (110), 21 strains were isolated from granulomatous lesions, mainly of the spleen, and had identical phenotypic and genotypic features. A first report, published in 2001 (110), was supported 2 years later when the new species M. shottsii was described (111).
Phenotypic features.
Acid-fast coccobacilli form dysgonic, flat, rough, unpigmented colonies after 4 to 6 weeks of incubation at 23°C, while no growth is obtained at temperatures over 30°C; less frequently, smooth colonies develop. Almost all the most frequently investigated biochemical traits are negative, apart from urease and niacin accumulation (Table 1). The latter is an extremely rare feature among nontuberculous mycobacteria, except for M. simiae, in which the production of niacin may be present, although only inconsistently.
HPLC analysis, the only lipid investigation performed so far, revealed a single cluster of peaks, grossly resembling the one characterizing the M. tuberculosis complex but easily distinguishable from it by the clearly lower retention time of each peak (111).
M. shottsii is susceptible to major antituberculous drugs, with isoniazid being the only exception (Table 4).
Genotypic features.
The 16S rDNA presents the structure typical of slow growers (Fig. 2 and 3).
Clinical and epidemiological features.
The presence, in all the infected fish, of granulomatous lesions clearly demonstrates the pathogenicity of the species for striped bass; nothing is known about the potential pathogenicity for other fish and humans. Seawater seems a plausible candidate as the natural reservoir.
Type strains: ATCC 700981T, NCTC 13215T. GenBank 16S rDNA sequence accession number: AY005147.
M. triplex
Ten independent strains from various states in the United States, referred to CDC between 1990 and 1994, were assigned to the new species M. triplex in 1996 (42). Six strains had been isolated from sputum, two from lymph nodes, and one from cerebrospinal fluid; the origin of the other remains unknown. The strains, initially suspected of belonging to the MAC group, failed to hybridize with specific commercial DNA probes and were temporarily assigned to the SAV (simiae-avium) group.
Phenotypic features.
The species, whose cells are short and acid fast, grows slowly, forming smooth unpigmented colonies after 2 or 3 weeks at 37°C. All major biochemical tests except Tween 80 hydrolysis are positive (Table 1).
Little is known about the lipid structure of the cell wall. The only technique investigated, HPLC, revealed a pattern characterized by three clusters of peaks, hardly distinguishable from that of M. simiae (42).
As with the majority of M. simiae-related species, M. triplex is characterized by high MICs of all antituberculous drugs (Table 4).
Genotypic features.
M. triplex has an identical hypervariable region B (Fig. 2 and 3) with M. simiae and related species.
Clinical and epidemiological features.
No clinical information is available concerning the patients from whom the strains investigated at CDC had been isolated. In contrast, a case of disseminated M. triplex infection in an AIDS patient, reported in Italy (25), is well documented. The isolation from pericardial and peritoneal fluid of a young girl receiving a liver transplant (66) and from an immunocompetent patient with a pulmonary infection (89) has also been reported. The hypothesis of the potential pathogenic role of M. triplex emerging from these reports is strengthened by the presence of two strains recovered from lymph nodes within the CDC cluster.
Five other strains have been reported, but their presence in the species M. triplex is questionable. Of the three which were clinically significant, two were responsible for lymphadenopathy in children (61, 152) and one caused chronic pulmonary disease (133). These strains are characterized, in comparison with M. triplex, by a one-nucleotide mismatch in 16S rDNA hypervariable region A. Two of them, the only ones investigated by HPLC, have practically overlapping profiles. Disagreement for other major phenotypic features such as nitrate reduction and pigmentation, however, shows that there are differences between each of the above strains.
Type strain: ATCC 70071T. EMBL 16S rDNA sequence accession number: U57632.
PIGMENTED RAPID GROWERS
M. elephantis
M. elephantis was described in 2000 (125) on the basis of a single isolate from a pulmonary abscess of an elephant that died due to chronic respiratory disease. Very recently, in Canada, 11 independent human isolates have been reported (157): 10 from sputum and 1 from an axillary lymph node.
Phenotypic features.
M. elephantis, whose cells are coccobacillary and weakly acid fast, grows rapidly between 25 and 45°C. It was initially considered not chromogenic, although the appearance of yellowish pigmentations on aging was reported. Subsequently, two types of colonies have been detected in the type strain, one of which is clearly scotochromogenic (157). Among biochemical tests, semiquantitative catalase, nitrate reduction, Tween 80 hydrolysis, and urease are positive (Table 5).
TABLE 5.
Major biochemical and cultural features of rapidly growing mycobacteriaa
| Species | Arylsulfatase (3 days) | Semiquantitative catalase (mm) | 68°C catalase | Nitrate reduction | Tween 80 hydrolysis | Urea hydrolysis | Pigment | Growth at ≥42°C | 5% NaCl tolerance | Growth on MacConkey agar |
|---|---|---|---|---|---|---|---|---|---|---|
| M. abscessus | + | >45 | v | − | − | + | − | − | + | + |
| M. alvei | + | ND | + | + | + | + | − | − | − | − |
| M. brumae | − | ND | + | + | + | + | − | − | − | − |
| M. chlorophenolicum | + | ND | ND | − | ND | ND | + | − | +b | ND |
| M. confluentis | − | >45 | + | + | − | + | − | −c | − | −d |
| M. elephantis | − | >45 | + | + | + | + | + | + | + | − |
| M. frederiksbergense | ND | ND | ND | + | + | − | + | − | ND | − |
| M. goodii | − | <45 | − | + | ND | ND | − | + | + | + |
| M. hassiacum | − | >45 | + | − | − | + | + | + | + | −d |
| M. hodleri | ND | ND | ND | − | + | + | + | − | ND | ND |
| M. holsaticum | − | ND | − | + | v | + | − | − | + | − |
| M. immunogenum | + | ND | ND | − | ND | ND | − | − | − | + |
| M. madagascariense | + | ND | ND | − | + | + | + | − | − | − |
| M. mageritense | + | ND | − | + | − | + | − | + | + | + |
| M. mucogenicum | + | <45 | − | v | + | + | − | − | − | + |
| M. murale | + | ND | ± | − | + | + | + | − | − | − |
| M. novocastrense | +d | >45 | +d | + | + | +d | + | + | + | +/− |
| M. peregrinum | + | >45 | + | + | v | + | − | − | + | − |
| M. septicum | NDe | ND | ND | + | ND | ND | − | − | + | + |
| M. vanbaalenii | + | ND | ND | + | + | + | + | − | ND | ND |
| M. wolinskyi | − | <45 | + | + | ND | ND | − | − | + | + |
+, positive; −, negative; +/−, predominantly positive; v, variable; ±, weak; ND, not done.
3% NaCl was tested.
Positive at 41°C.
Unpublished data.
Test positive at 14 days.
The only lipid study performed so far is HPLC. The HPLC pattern shows an early group of five peaks followed by a larger cluster (157). The type of mycolic acids present in the cell wall and the GLC pattern are unknown.
While the strain isolated from the elephant was resistant to ciprofloxacin, all the human strains detected subsequently were susceptible to a large number of drugs including quinolones (Table 6). This discrepancy is probably due to the use, in the first case, of a clearly out-of-date method on solid medium (resistance ratio).
TABLE 6.
| Species | No. of strains tested | Ethambutol | Isoniazid | Rifampin | Streptomycin | Amikacin | Cephalothin | Ciprofloxacin | Clarithromycin | Tobramycin |
|---|---|---|---|---|---|---|---|---|---|---|
| M. abscessusc | 99 | R | R | R | R | S | R | R | S | I |
| M. alvei | 6 | S | R | R | R | ND | ND | ND | ND | ND |
| M. brumae | 11 | S | R | R | R | ND | ND | ND | ND | ND |
| M. confluentis | 1 | S | S | ND | S | ND | ND | ND | ND | ND |
| M. elephantisd | 5 | S | S | v | S | S | ND | S | v | ND |
| M. goodii | 8 | S | R | R | ND | S | v | I | v | I |
| M. hassiacume | 2 | S | R | R | S | S | ND | S | S | S |
| M. holsaticum | 9 | S | R | R | S | ND | ND | ND | ND | ND |
| M. immunogenum | 12 | ND | ND | ND | ND | S | R | v | S | R |
| M. mageritense | 11 | R | R | ND | ND | S | R | S | R | R |
| M. mucogenicumf | 84 | ND | ND | ND | ND | v | S | S | S | ND |
| M. murale | 2 | S | S | S | S | S | ND | S | S | ND |
| M. novocastrense | 1 | S | ND | R | S | ND | ND | S | ND | ND |
| M. peregrinumc | 8 | R | R | R | R | S | R | S | S | S |
| M. septicum | 1 | ND | ND | ND | R | S | ND | S | ND | S |
| M. wolinskyi | 3 | ND | R | R | ND | S | v | I | v | R |
No information is available for M. chlorophenolicum, M. frederiksbergense, M. hodleri, M. madagascariense, and M. vanbaalenii.
R, resistant; S susceptible; I, moderately susceptible; v, variable; ND, not done.
Data of sp. nov. descriptions integrated with information from the literature (157) and unpublished data.
Data of sp. nov. descriptions integrated with information from the literature (154).
Data from reference 166.
Genotypic features.
The sequence of the 16S rDNA is characteristic of the thermotolerant rapid growers; i. e., it has a cytosine insertion in helix 10 and a short helix 18. The helix 18, however, although short, is 2 nucleotides longer than in other rapid growers (Fig. 2 and 4). The sequencing of clinical isolates and of both morphological variants of the type strain showed mismatches in four bases between the two morphotypes. They differed by three bases and one base from the genetically homogeneous group of clinical isolates (157). Very recently a further sequevar has been reported (E. Tortoli et al., submitted for publication). The PRA of hsp65 gene reveals a unique and previously unpublished pattern for M. elephantis.
Clinical and epidemiological features.
While the clinical significance of 10 strains isolated, only once, from the sputum of elderly patients with preexisting nonmycobacterial disease is questionable, no doubt exists about the pathogenic role of the isolate grown from the granulomatous tissue of an axillary lymph node. The hypothesis is strengthened by the evident involvement of M. elephantis in the fatal pathology of the elephant from which the type strain was isolated.
Three strains, whose 16S rDNA sequence already was present in the public domain databases under the name MCRO17 (128), were recently recognized as belonging to M. elephantis (157). This clarification allowed to definitively identify as M. elephamtis two strains isolated in the 1990s from a bronchial aspirate and a sputum sample (142) and a third, unpublished strain, lacking clinical significance, however.
Type strain: DSM 44368T. EMBL 16S rDNA sequence accession number: AJ010747.
M. hassiacum
M. hassiacum was described in 1997 based on an isolate grown from urine (121).
Phenotypic features.
The species is characterized by striking growth conditions. It is able to grow in 2 to 6 days at temperatures ranging from 30 to 65°C. The colonies, which are smooth and scotochromogenic, are moist and slimy when grown at 37°C and dry when grown at 60°C. Growth also occurs on 5% NaCl and MacConkey agar without crystal violet (Table 5).
Mycolic acids detected by TLC include α-mycolates and wax esters, a pattern also shown by M. bohemicum and M. mucogenicum (Table 2). The GLC fatty acid set is unique (Table 3). The HPLC profile (154) grossly resembles that of M. flavescens.
As with a majority of rapidly growing species, susceptibility testing reveals resistance to isoniazid and rifampin (Table 6).
Genotypic features.
The 16S rDNA arrangement is peculiar: like thermotolerant rapid growers, M. hassiacum has an extended helix 10, due to the insertion of an extra cytosine, and a short helix 18 (Fig. 2). However, because of the large number of base mismatches, the use of different algorithms may result either in its phylogenetic clustering with thermotolerant rapid growers (Fig. 4) or in a position close to the slowly growing species M. xenopi.
Clinical and epidemiological features.
Only one additional isolation of M. hassiacum, again from urine, has been reported subsequently (154). The urinary origin, the occasional isolation, and the absence of important clinical findings seem to exclude any medical significance for both cases reported so far. The contamination of clinical samples by environmental strains is at present the most probable hypothesis.
Type strain: DSM 44199T. EMBL 16S rDNA sequence accession number: U49401.
M. novocastrense
The only strain of M. novocastrense isolated to date was reported in 1997 and was obtained from a slowly spreading skin granuloma on a child's hand (124).
Phenotypic features.
M. novocastrense is a rapid grower which forms smooth, butyrous, photochromogenic colonies at temperatures ranging from 25 to 43°C. Most biochemical tests are positive (Table 5).
The mycolic acid composition of the cell wall is rather common and does not allow the identification of the species (Table 2). The HPLC and GLC patterns are unique and can play a decisive role in the identification of the species (Fig. 7).
FIG. 7.
Mycolic acid pattern of M. novocastrense obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
The susceptibility pattern is characterized by resistance to rifampin only (Table 6).
Genotypic features.
The 16S rDNA sequence is characterized by a long helix 10 and by a short helix 18. Although the nucleotide inserted in helix 10 is thymine, M. novocastrense clusters with thermotolerant rapid growers, which are generally characterized by the insertion of one cytosine. Interestingly M. novocastrense shares full homogeneity of the hypervariable region B with other mycobacterial species, including M. alvei and M. septicum. However, there is no significant phylogenetic relatedness between M. novocastrense and these mycobacteria (Fig. 2 and 4).
Clinical and epidemiological features.
The isolation from a typical granulomatous lesion strongly supports the potential pathogenicity of the strain.
Type strain: DSM 44203T. EMBL 16S rDNA sequence accession number: U96747.
NONPIGMENTED RAPID GROWERS
M. abscessus
The previously named M. chelonae subsp. abscessus was recognized as new species in 1992 (80) following the detection of low DNA homology to M. chelonae sensu stricto (hybridization test results, 35%). M. abscessus is a nonchromogenic rapid grower whose phenotypic (Tables 2, 3, 5, and 6), genotypic, and clinical features (162) were well known before its elevation to a species. It is therefore outside the purpose of the present review.
Type strain: ATCC 19977T. GenBank 16S rDNA sequence accession number: X82235.
M. alvei
The description of M. alvei, in 1992, is based on six strains, four of which were environmental (from water and soil) and two were isolated from sputa (3).
Phenotypic features.
The organism, whose cells are short and strongly acid fast, grows rapidly at 25 to 30°C but needs 2 weeks at 37°C. It is nonchromogenic and is characterized by large, rough colonies. Among major biochemical tests, arylsulfatase, Tween 80 hydrolysis, nitrate reduction, and urease are positive (Table 5).
The mycolic acid pattern is unique; in addition to the ever-present α-mycolate, M. alvei contains ω1-methoxy-mycolates only. Conversely, the GLC distribution of fatty acids is very common among nonphotochromogenic rapid growers. The HPLC profile is characterized by an impressive, uninterrupted series of peaks taking practically the whole range of retention times typical of mycobacterial mycolic acids (17).
The species is resistant to all antituberculous drugs except ethambutol (Table 6).
Genotypic features.
The 16S sequence, not determined at the time of the sp. nov. description, was determined later. Hypervariable region B is identical to that of the closely related species M. septicum and M. peregrinum and to that of M. novocastrense, which, in contrast, clusters with the thermotolerant rapid growers (Fig. 2 and 4).
Clinical and epidemiological features.
The environmental origin of most of the strains and the unlikely involvement in the pathologies of the two respiratory strains suggest an environmental reservoir of the species and indicates that their presence in clinical samples is due to contamination.
Type strain: CIP 103464T. EMBL 16S rDNA sequence accession number: AF023664.
M. brumae
M. brumae was described in 1993 (85) based on 10 environmental isolates (eight from river water and two from soil) plus one from a sputum sample.
Phenotypic features.
The species grows within 4 to 5 days at temperatures ranging from 25 to 37°C; it forms large, rough, nonchromogenic colonies. Arylsulfatase only, among the main biochemical tests, is negative (Table 5).
A single mycolic acid (α-mycolate) is revealed by TLC, a feature shared only by M. fallax and M. triviale (Table 2). The GLC pattern is close to that of M. xenopi (Table 3). The HPLC profile is similar to those of M. fallax and M. triviale and is characterized by a single late cluster of peaks (17).
Genotypic features.
The 16S rDNA, not sequenced at the time of the sp. nov. description, was subsequently investigated and revealed that M. brumae, like M. novocastrense, has a long helix 10 with a one-thymine insertion. Although this insertion differs from the one (cytosine) characterizing thermotolerant rapid growers, both species cluster with the latter group in the phylogenetic tree (Fig. 2 and 4).
Clinical and epidemiological features.
Present knowledge indicates that M. brumae probably belongs to the nonpathogenic environmental rapid growers.
Type strain: CIP: 103465T. SmartGene 16S rDNA sequence accession number: RM1999.
M. confluentis
The new species M. confluentis was described in 1992 based on a single isolate obtained from the sputum of a healthy male (76).
Phenotypic features.
M. confluentis is characterized by acid-fast coccobacillary cells. The colonies, smooth and unpigmented, grow in 2 to 4 days at 22 to 41°C, become brownish after about 2 months, and subsequently turn black.
The only available strain gave positive results for nitrate and urea only among the most widely used biochemical tests (Table 5) and was susceptible to all antituberculous drugs (Table 6).
Information about the lipid component of the cell wall is poor since no investigations have been reported using either TLC or GLC. The recently reported HPLC pattern (17) shows a conspicuous number of peaks arranged in a single, late-emerging cluster.
Genotypic features.
The unique sequence of the 16S rDNA is characterized by a short helix 18 and a cytosine insertion in helix 10, placing this species within the group of thermotolerant rapid growers (Fig. 2 and 4).
Clinical and epidemiological features.
None of the parameters needed to establish the clinical significance of the species are present in the only reported strain of M. confluentis. Intravenously inoculated M. confluentis is eradicated not only in immunocompetent mice (BALB/c) but also in gamma interferon-deficient mice, thus suggesting low or absent virulence (36).
Type strain: DSM 44017T. EMBL 16S rDNA sequence accession number: X63608.
M. goodii
Like M. wolinskyi, M. goodii was differentiated from M. smegmatis, with which it was previously confused, by a cooperative study group in 1999 (14). The description of the new species is based on 28 strains.
Phenotypic features.
M. goodii grows rapidly at temperatures ranging from 30 to 45°C and shares a majority of phenotypic features with M. smegmatis (Table 5). In most strains, however, the colonies, which are smooth to mucoid, develop yellowish pigmentation which intensifies with age.
The mycolic acid pattern of M. goodii has not been studied by TLC. GLC analysis is unable to differentiate the new species from M. smegmatis (Table 3); these two species are equally indistinguishable using HPLC, where the profiles are characterized by peaks with overlapping relative retention times and with close height proportions. Less problematic is the differentiation from M. mageritense and M. wolinskyi, in which the peaks of the first cluster are clearly lower (14).
M. goodii is susceptible to ethambutol, and chloramphenicol; the susceptibility to tobramycin is characterized by MICs of >1 μg/ml for over 80% of strains, a feature useful also for the differentiation from M. smegmatis (Table 6).
Genotypic features.
M. goodii shares the sequence of hypervariable region B within the 16S rDNA with the other M. smegmatis-related species. Hypervariable region A is unique and has, like that of M. smegmatis sensu stricto, a cytosine insertion in helix 10. Phylogenetically, M. goodii is therefore included in the thermotolerant rapid growers (Fig. 2 and 4).
The PRA of hsp65, using AciI, is suitable for differentiation of M. goodii, which has a single pattern, from most but not from all other M. smegmatis-related organisms. Several (16%) M. smegmatis sensu stricto strains present a pattern identical to that of M. goodii. The 16S rRNA gene, investigated using PRA, allows unambiguous differentiation of M. goodii.
Clinical and epidemiological features.
The 28 strains investigated were isolated from humans with traumatic osteomyelitis following iatrogenic infections or with respiratory infections (lipoid pneumonia). In many cases, patients yielded multiple positive cultures. The chronic lung disease produced by M. goodii, which accounts for almost 25% of pathology due to such organisms, is invariably characterized by lipoid pneumonia, in contrast to disease caused by M. fortuitum. One case of bursitis due to M. goodii has also been recently reported (46). The pathogenicity of M. goodii therefore appears unquestionable.
Type strain: ATCC 700504T. EMBL 16S rDNA sequence accession number: Y12872.
M. holsaticum
M. holsaticum was described in 2002 based on nine clinical strains isolated from nine independent clinical samples (112).
Phenotypic features.
Cells are coccoid and acid fast and form visible colonies within 1 week. A wide temperature range is tolerated, from 20 to 40°C. However, while the colonies are dysgonic at 20°C they are smooth and moist at higher temperatures. The species is nonchromogenic, but yellow pigment may develop with age. The most significant conventional test results are the positivity of nitrate reduction and the growth on 5% NaCl Lowenstein-Jensen medium (Table 5).
TLC reveals a common mycolic acid combination, identical, among others, to that of M. tuberculosis (Table 2). The GLC pattern resembles that of M. murale (Table 3). The HPLC profile, not published so far, is unique (Fig. 8).
FIG. 8.
Mycolic acid pattern of M. holsaticum obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
Resistance to isoniazid and rifampin characterize the antimicrobial susceptibility of M. holsaticum (Table 6).
Genotypic features.
The 16S rDNA is characterized, in hypervariable region A, by an extraordinary similarity to M. tuberculosis; three nucleotides only differ in this stretch. Region B, in contrast, presents the short helix 18 typical of rapid growers. Within the ITS sequence, a 10-bp discordance is present that splits the species in two sequevars. With the algorithm adopted here, M. holsaticum seems to be one of the thermotolerant rapid growers, which is surprising since it lacks any insertion (either cytosine or, less commonly, thymine) in helix 10 (Fig. 2 and 4).
The hsp65 gene, investigated by both sequencing and PRA, gives unique results suitable for differentiating M. holsaticum from other slowly growing mycobacteria.
Like M. celatum, the very close sequence similarity to M. tuberculosis in the sequence targeted by AccuProbe Mycobacterium tuberculosis complex requires the use of the recommended stringency conditions to avoid a nonspecific hybridization with the latter probe.
Clinical and epidemiological features.
The lack of any clinical information about the patients from whom M. holsaticum was isolated and the specimen type (sputum in six cases and urine and gastric fluid in two others) do not support the pathogenicity of M. holsaticum.
Type strain: DSM 44478T, CCUG 46266T. EMBL 16S rDNA sequence accession number: AJ310467.
M. immunogenum
The description of the new taxon M. immunogenum, in 2001, was based on 112 isolates that, on hsp65 PRA, had revealed a pattern apparently halfway between those of M. chelonae and M. abscessus (171, 172).
Phenotypic features.
M. immunogenum are acid-fast curved bacilli. The colonies are nonchromogenic and predominantly rough and grow in less than 1 week at 30 to 35°C.
The HPLC pattern is the only lipid aspect studied so far. The profile of M. immunogenum is not distinguishable from that of M. abscessus, while minor differences in the height of the peaks of the second cluster allow the differentiation from M. chelonae (171).
Arylsulfatase only is positive among the most frequently used biochemical tests (Table 5).
The resistance to cephalothin and the high MIC of tobramycin are important for the differential diagnosis from M. mucogenicum and M. chelonae, respectively (Table 6).
Genotypic features.
The 16S rDNA nucleotide sequence of M. immunogenum overlaps, in hypervariable region B, the one shared by M. fortuitum and M. chelonae, while in region A, it differs from that of any other rapid grower. Phylogenetically, M. immunogenum clusters with M. chelonae and M. abscessus in a branch separated from other species (Fig. 2 and 4). Essentially equivalent results are obtained by hsp65 gene sequencing.
Interestingly, M. immunogenum has two copies of the rDNA operon, which differs from M. abscessus and M. chelonae, the only rapid growers known to have a single copy.
PFGE Separation of the restriction enzyme-digested bacterial DNA reveals 14 different patterns. When hsp65 PRA is performed using MspI and HhaI, the pattern of M. immunogenum is indistinguishable from that of M. chelonae. While using HinI and HaeIII, the pattern overlaps that of M. abscessus (171).
Clinical and epidemiological features.
The spectrum of diseases due to M. immunogenum includes cutaneous infections, keratitis, and catheter-related infections. Of the 112 strains investigated for the sp. nov. description, 98 had been grown from the environment of metal-working industries, where aerosolized oil is known to be associated with hypersensitivity pneumonitis (93). Of the others, 11 were clinical strains (from skin, eye, urine, synovial fluid, and bronchoalveolar lavage fluid) while the remaining strains were isolated from five nosocomial pseudo-outbreaks involving contaminated bronchoscopes. Three further environmental isolates associated with hypersensitivity pneumonitis (122) and two clinical strains, only one of which published (136), have been also documented.
The pathogenic role of M. immunogenum is demonstrated by numerous cases of diseases directly attributable to this species. Furthermore, the involvement of the species in hypersensitivity pneumonitis was hypothesized when high titers of antibody immunoglobulin G against M. immunogenum were detected in workers exposed to metal-working fluids (122).
Type strains: ATCC 700505T, CIP 106684T. EMBL16S rDNA sequence accession number: AJ011771.
M. mageritense
Five clinical strains, isolated from the sputum of different patients, were studied in 1997 (32) and found to be different from any other species recognized at that time.
Phenotypic features.
The cells of M. mageritense are strongly acid-fast long rods. The colonies, which are nonpigmented, smooth, and mucoid (but sometimes also rough), develop in 2 to 5 days at temperatures ranging from 22 to 45°C. Like many rapid growers M. mageritense can grow on both NaCl-containing Lowenstein-Jensen medium and MacConkey agar (Table 5).
TLC of mycolic acids present in the cell wall does not distinguish M. mageritense from M. smegmatis and M. fortuitum (Table 2). The GLC pattern has not been reported so far. The mycolic acid profile obtained by HPLC (14) follows the outline common to most rapid growers, which is characterized by two late clusters of peaks, but it shows extremely poor first cluster and a very narrow second one. One isolate characterized by two major peaks has also been reported (163).
The species is resistant to isoniazid, ethambutol, and clarithromycin and susceptible to ciprofloxacin (Table 6).
Because of the sharing of many phenotypic features, it seems likely that at least some of the strains so far assigned to the M. fortuitum third biovariant complex (164) actually belong to M. mageritense (163).
Genotypic features.
M. mageritense is characterized, in 16S rDNA region B, by a sequence different from that shared by other M. smegmatis-related organisms, which also include M. goodii and M. wolinskyi, but identical to the sequence of M. hodleri. Furthermore, it lacks the cytosine insertion in helix 10. A single nucleotide change (guanine replaced by adenine) is found in the helix 18, which is short, in a few strains. The closest phylogenetically related species is M. wolinskyi (Fig. 2 and 4). The 16S rRNA and the sodA genes sequences yielded a not substantially different phylogenetic relatedness.
hsp65 PRA results in an identical pattern for all isolates tested so far, with six of the seven tested restriction enzymes. The pattern is easily distinguishable from that of other mycobacteria.
Clinical and epidemiological features.
No clinical information about the Spanish patients from whom M. mageritense was isolated is available. Six further strains were isolated in the United States, and apart from the two isolates of pulmonary origin, they appeared pathogenic; two came from surgical wounds, one came from the blood culture of an immunosuppressed patient with catheter-related sepsis, and one came from a patient with severe sinusitis (163). No isolation has been reported so far from the environment.
Type strain: CIP 104973T. EMBL 16S rDNA sequence accession number: AJ011335.
M. mucogenicum
The group of M. chelonae-like organisms (MCLO) included mycobacteria considered, in the large majority of cases, not to be pathogenic or, at most, responsible for nosocomial pseudo-infections associated predominantly with water. Nevertheless, of 87 clinical (mostly respiratory) MCLOs reviewed by Wallace et al. in 1993 (166), about 20, all nonrespiratory, proved to be pathogenic. In 1995, a genetic investigation performed on three MCLO reference strains (126) revealed their diversity from any other mycobacterial species, and the status of new species, M. mucogenicum, was proposed.
Phenotypic features.
M. mucogenicum, whose cells are partially acid fast, grows in 2 to 4 days at 28 to 37°C and forms nonpigmented mucoid colonies.
The distinction of M. mucogenicum from M. chelonae by means of conventional tests, whose results are often variable, is problematic (Table 5). The most important differentiating feature seems to be the susceptibility of M. mucogenicum, but not of M. chelonae, to cephalothin (Table 6).
The determination of mycolic acids by TLC, performed after the sp. nov. description, reveals a pattern identical to those of M. xenopi and M. terrae (96) and also to those of M. bohemicum and M. hassiacum (Table 2). The GLC profile, initially considered identical to that of M. chelonae, turned out subsequently to possess two additional peaks (96) (Table 3). Finally, the HPLC pattern is characterized by two well-separated clusters of peaks (166).
Genotypic features.
The sequencing of the 16S rRNA gene revealed, in one of the strains examined, a 4-base mismatch within hypervariable region B (Fig. 2). This diversity was, however, not considered sufficient to split the group in two taxa. A unique pattern or two similar patterns were found by PRA of hsp65, depending to the restriction enzyme used.
Clinical and epidemiological features.
In contrast to the other MCLOs, M. mucogenicum is frequently involved in pathologies, mainly posttraumatic wounds and catheter-related sepsis. One case of fatal progressive granulomatous hepatitis (diagnosed by culture at autopsy) (50) and one case of bacteremia (100) have also been reported.
M. mucogenicum was grown from bottled mineral water (23) and was recovered from 41% of samples of ice and of public drinking water investigated in the United States (26).
Type strain: ATCC 49650T. EMBL 16S rDNA sequence accession numbers: X80771 (ATCC 49650), X80773 (ATCC 49649).
M. peregrinum
The previous M. fortuitum subsp. peregrinum was recognized as a new species in 1992 (80) following the detection of a low level of DNA homology to M. fortuitum sensu stricto (hybridization test results below 50%). M. peregrinum is a nonchromogenic rapid grower whose phenotypic (Tables 2, 3, 5, and 6), genotypic, and clinical features were well known before its elevation to species (162). It is therefore outside the scope of this review.
Type strain: ATCC 14467T. EMBL 16S rDNA sequence accession number: AF130308.
M. septicum
The description of the species M. septicum, in 2000, is based on a single strain isolated four times from the same 2-year-old patient with a metastatic hepatoblastoma. Three isolates came from blood cultures, and one was from a central venous catheter (120).
Phenotypic features.
The bacterial cells are pleomorphic, mostly coccobacillary, and acid fast. The colonies, unpigmented (beige) and rough, grow in less than 1 week at temperatures between 28 and 35°C.
Almost all of the most common biochemical tests are positive (Table 5). The strain is susceptible in vitro to aminoglycosides except streptomycin (Table 6) and to most of the substances suitable for rapidly growing mycobacteria testing, including ciprofloxacin, doxycycline, and imipenem.
The HPLC chromatotype is indistinguishable from that of M. fortuitum and M. peregrinum (120), while other lipid investigations such as GLC and TLC have not been done so far.
Genotypic features.
Surprisingly, since it is not stated in the article describing the new species, M. septicum is not differentiable from M. peregrinum on the basis of the sequences of 16S rDNA hypervariable regions A and B (Fig. 2). The two species are fully homologous in hypervariable region A, and region B is also shared by M. fortuitum, M. alvei, and M. novocastrense, M. septicum and M. peregrinum differ in only four nucleotides within a 26-bp stretch included in the last one-third of the gene. Phylogenetic relatedness emerges with M. peregrinum, M. fortuitum, and M. alvei but not with M. novocastrense (Fig. 4).
Ribotyping of restriction enzyme-digested (SalI) genomic DNA gave an identical pattern for the four isolates of M. septicum, which clearly differed from those of the closely related species M. fortuitum, M. peregrinum, and M. senegalense.
Clinical and epidemiological features.
The isolation site, along with clinical information concerning the only patient with M. septicum infection reported so far, clearly fulfill the criteria for the pathogenicity of the species.
Type strains: ATCC 700731T, DSM 44393T. GenBank 16S rDNA sequence accession number: AF111809.
M. wolinskyi
Like M. goodii, M. wolinskyi was recognized in 1999 (14) as part of a cooperative study focused on the heterogeneity present among the strains assigned, at that time, to the species M. smegmatis. Eight different strains were investigated.
Phenotypic features.
M. wolinskyi grows in 2 to 4 days, is nonchromogenic, and produces smooth to mucoid colonies at temperatures ranging from 30 to 45°C. Its biochemical traits resemble those of M. smegmatis (Table 5).
The high MIC of tobramycin (2 to 8 μg/ml in 100% of the strains) is of primary importance in distinguishing M. wolinskyi from other M. smegmatis-related rapid growers as far as susceptibility patterns are concerned (Table 6).
While the mycolic acids of the cell wall have not been studied using TLC, both the GLC (Table 3) and HPLC patterns closely resemble those of M. smegmatis. In particular, the HPLC peaks of M. wolinskyi show retention times overlapping the ones of M. smegmatis and M. mageritense, with the profile of the latter showing the closer resemblance. Nevertheless, M. wolinskyi can be differentiated because of the clearly greater height of the peaks of the first cluster (14).
Genotypic features.
M. wolinskyi, like M. mageritense but unlike the other species of the group of M. smegmatis-related rapid growers, lacks the cytosine insertion in helix 10 within hypervariable region A of the 16S rRNA gene. Region B, in contrast, is identical to that of M. goodii among M. smegmatis-related organisms and to that of M. frederiksbergense as well (Fig. 2). From the phylogenetic point of view, the most closely related species is M. mageritense (Fig. 4). For one of six M. wolinskyi isolates sequenced in the cooperative study, a 4-nucleotide difference emerged in hypervariable region A. The correct genetic identification of this variant is, however, possible by referring to a third hypervariable region (corresponding to positions 1001 to 1027 of E. coli), whose sequence is shared by all M. wolinskyi strains.
PRA of hsp65 using MspI or AciI is able to differentiate M. wolinskyi from every other M. smegmatis-related organism. However, while a unique pattern is obtained with MspI, AciI splits the strains into two subgroups. Also, the restriction fragment length polymorphism (RFLP) analysis of the 16S rRNA gene can be used to differentiate M. wolinskyi.
Clinical and epidemiological features.
All the strains forming the very homogeneous cluster investigated for the description of the new species M. wolinskyi were isolated, in many cases repeatedly, from surgical infections and traumatic cellulitis and were almost always associated with osteomyelitis. Therefore, there is no doubt about the pathogenicity of the species.
Type strain: ATCC 700010T. EMBL 16S rDNA sequence accession number: Y12871.
SOLELY ENVIRONMENTAL SPECIES
M. botniense
Two strains grown from the water of the same Finnish stream represent the basis for the description, in 2000, of the new species M. botniense (141).
Phenotypic features.
The colonies are thin, smooth, and yellow and are characterized by better growth on egg medium than on Middlebrook's agar. Several features of M. botniense resemble those of M. xenopi. It grows very slowly (5 to 8 weeks) and prefers high incubation temperatures (from 36 to 50°C). The major biochemical tests are negative with the exception of arylsulfatase (Table 5). The strains are resistant to all first-line antituberculous drugs (Table 6).
TLC investigation of mycolic acids has not yet been reported. The GLC pattern is identical to that of another recently described mycobacterium, M. heckeshornense (Table 3) (116). The HPLC profile is similar to that of M. xenopi: their two clusters of peaks have overlapping retention times although with slightly different height proportions (Fig. 9).
FIG. 9.
Mycolic acid pattern of M. botniense obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
Genotypic features.
The 16S rDNA sequence of the strains shows closest similarity to M. xenopi (Fig. 2 and 3), from which it differs by only 22 nucleotides. The relatedness to M. xenopi is also confirmed in the sequence of the ITS region.
Epidemiological features.
M. botniense has been isolated so far exclusively from the environment. However, the extremely high risk of misidentification as M. xenopi may well be responsible for the lack of reports from clinical samples.
Type strain: ATCC 700701T. EMBL 16S rDNA and ITS sequence accession number: AJ012756.
M. chlorophenolicum
One strain previously assigned to the genus Rhodococcus was moved to the genus Mycobacterium in 1994 because of the detection of typical mycobacterial mycolic acids. This strain was further investigated, along with two other similar isolates (60).
Phenotypic features.
The most frequent cell morphology is coccoid due to fragmentation of unbranched rods which develop in culture. Colonies are yellow to orange and slightly mucoid and usually grow within 1 week at temperatures ranging from 18 to 37°C. This species has very unusual nutritional characteristics including the ability to degrade chlorinated phenols, guaiacols, and syringols.
Mycolic acids detected by TLC include α-mycolates, keto-mycolates, and wax esters, which is a very common pattern (Table 2). In contrast, the GLC fatty acid profile is uncommon, resembling only that of M. doricum (Table 3). The HPLC pattern (17) is characterized by major early and minor late clusters of peaks.
Genotypic features.
The phylogeny based on 16S rDNA sequence places M. chlorophenolicum with the rapid growers (Fig. 2 and 4) (13).
Epidemiological features.
All strains were environmental and were isolated from various chlorophenol-contaminated Finnish soils. The possibility of growth of M. chlorophenolicum from clinical samples appears negligible.
Type strains: ATCC 49826T, CIP 104189T; DSM 43826T. EMBL 16S rDNA sequence accession number: X79094.
M. cookii
Seventeen environmental strains from sphagnum and water in New Zealand represent the organisms used for the description of the new species M. cookii in 1990 (73).
Phenotypic features.
M. cookii is rod shaped and acid fast; the colonies are smooth and yellow-orange and grow slowly at 22 to 31°C. Arylsulfatase only is positive among the major biochemical tests (Table 5).
The TLC mycolic acid pattern is unique and includes α- and α′-mycolates and wax esters (Table 2). The GLC profile is unusual mainly because of the almost complete lack of tuberculostearic acid in most of the strains (Table 3). The HPLC pattern shows three clusters of peaks whose height steadily decreases with increasing elution times (17).
Susceptibility testing reveals the efficacy of all first-line antituberculous drugs (Table 6).
Genotypic features.
Although considered intermediate between rapid and slow growers on the basis of the partial 16S rDNA sequence (73), M. cookii is characterized by a long helix 18, made even longer by a 2-nucleotide insertion, and unquestionably clusters with slow growers (Fig. 2 and 3).
Epidemiological features.
M. cookii is clearly saprophytic, as confirmed by inoculation in laboratory animals (mouse, guinea pig, and rabbit). It has been suggested to be responsible for false-positive results with M. bovis tuberculin in cattle (73).
Type strain: ATCC 49103T. EMBL 16S rDNA sequence accession number: 53896.
M. frederiksbergense
The species M. frederiksbergense is based on one strain isolated from coal tar-contaminated soil in Denmark and was described in 2001 (170).
Phenotypic features.
M. frederiksbergense is scotochromogenic, characterized by cadmium-yellow smooth colonies and by growth in 5 to 7 days at 30°C (range, 15 to 37°C). Interestingly, it is able to mineralize phenanthrene, fluoranthrene, and pyrene. Among biochemical tests, Tween 80 hydrolysis and nitrate are positive (Table 5).
The mycolic acid pattern detected by TLC is very common among mycobacteria (Table 2). GLC reveals a unique profile (Table 3). The HPLC mycolic acid profile is unusual and includes a crowded first cluster of very high peaks clearly separated from a less important second cluster (170).
Genotypic features.
The sequence of 16S rDNA is characterized by a unique hypervariable region A and by a region B identical to those of M. smegmatis and M. wolinskyi (Fig. 2).
Epidemiological features.
The species is clearly environmental and saprophytic. No other isolation has been reported so far.
Type strains: DSM 44346T, NRRL B24126T. EMBL accession number of 16S rDNA sequence: AJ276274.
M. hiberniae
The species M. hiberniae was described in 1993 from 13 environmental strains isolated from sphagnum, moss, and soil in Ireland (71).
Phenotypic features.
The morphology is coccobacillary, and colonies are pink and grow slowly at 22 to 37°C, becoming progressively rougher. Nitrate reduction is the only biochemical reaction consistently positive (Table 1).
M. hiberniae is resistant to almost all first-line antituberculous drugs (Table 4).
The mycolic acid composition is shared by many other mycobacteria (Table 2), while the GLC profile has a unique pattern (Table 3). The HPLC profile includes an early and a late cluster of peaks, with prominent peaks in the first one (17).
Genotypic features.
The 16S rDNA sequence of M. hiberniae is characterized by a long helix 18, 2 nucleotides longer than most of the other slow growers. This is a feature characteristic of M. terrae and M. nonchromogenicum, with which it forms a separate branch among slow growers (Fig. 2 and 4).
Epidemiological features.
The habitat and the lack of pathogenicity for experimentally inoculated animals (mouse, guinea pig, and rabbit) suggest the inclusion of M. hiberniae among saprophytic environmental species. The reported cross-reaction with M. bovis tuberculin may give rise to a false-positive skin test in cattle.
Type strain: ATCC 49874T. EMBL 16S rDNA sequence accession number: X67096.
M. hodleri
M. hodleri was described in 1996; the only strain reported so far was isolated from soil polluted with fluoranthrene (77).
Phenotypic features.
The strain is scotochromogenic and grows rapidly between 18 and 37°C; the colonies are usually smooth, but rough variants may also be present.
Tween 80 hydrolysis and urease tests are positive (Table 5).
No information is available about the antimicrobial susceptibility.
TLC reveals a mycolic acid composition very common among both rapid and slow growers (Table 2); in contrast, the GLC pattern is easily distinguishable from that of other rapid growers. The HPLC profile of mycolic acids, unpublished so far, is characterized by two clearly separated clusters of peaks (Fig. 10).
FIG. 10.
Mycolic acid pattern of M. hodleri obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
Genotypic features.
Even though M. hodleri has an identical sequence in hypervariable region B of 16S rDNA to that of M. mageritense, the two species occupy a clearly distinct position among rapid growers on the phylogenetic tree (Fig. 2 and 4).
Epidemiological features.
The peculiar metabolic activities, mainly the possibility of degrading several polycyclic aromatic hydrocarbons, unquestionably confirms the exclusively environmental nature of the species.
Type strain: DSM 44183T. EMBL 16S rDNA sequence accession number: X93184.
M. madagascariense
M. madagascariense was first recognized in 1992 from four strains isolated from sphagnum in Madagascar (72).
Phenotypic features.
M. madagascariense grows rapidly at temperatures ranging from 22 to 31°C (the optimum is at 22°C) and is scotochromogenic. The colonies are yellow-orange and smooth. The species shows biochemical tests positive for arylsulfatase, Tween 80 hydrolysis, and urease (Table 5).
The mycolic acid pattern, as detected by TLC, is common within rapid growers (Table 2). The GLC pattern reveals similarity to M. aurum, M. neoaurum, and M. gadium (Table 3). The HPLC pattern is characterized by two widely separated peak clusters (17).
Genotypic features.
The phylogeny based on 16S rDNA places M. madagascariense, which has a cytosine insertion in helix 10 and a short helix 18, within the group of thermotolerant rapid growers, close to M. confluentis (Fig. 2 and 4) (76).
Epidemiological features.
The exclusively environmental nature of the species seems unquestionable. Negative pathogenicity testing on laboratory animals (rabbit, guinea pig, and mouse) confirm the clinical unimportance of the species.
Type strain: ATCC: 49865T. SSU 16S rRNA sequence accession number: X55600.
M. murale
The new species M. murale was described in 1999 based on five strains isolated from a water-damaged wall (161).
Phenotypic features.
M. murale grows rapidly at temperatures as low as 10°C and up to 37°C (optimum, 30°C). The colonies are smooth and scotochromogenic (saffron yellow). Cells are coccobacillary and acid fast. The majority of biochemical tests (arylsulfatase, Tween 80 hydrolysis, and urea) are positive (Table 5), and the strains are susceptible to first-line antituberculous drugs (Table 6).
The TLC pattern of mycolic acids is very common (Table 2); M. holsaticum, M. aurum, and M. hodleri are the most closely related species on the basis of the GLC results (Table 3). HPLC investigation of cell wall mycolic acids, not reported to date, presents two major clusters of peaks (Fig. 11).
FIG. 11.
Mycolic acid pattern of M. murale obtained by HPLC analysis. LMWIS, low-molecular-weight internal standard; HMWIS, high-molecular-weight internal standard.
Genotypic features.
The 16S rDNA is characterized by a short helix 18 (Fig. 2 and 4). Ribotyping with two different restriction enzymes gave an almost identical pattern for all the isolates of M. murale, distinct from those of even most closely related species of rapidly growing mycobacteria.
Epidemiological features.
At present, the environment seems the sole reservoir, and no indication of potential pathogenicity exists.
Type strain: DSM 44340T. GenBank 16S rDNA sequence accession number: Y08857.
M. vanbaalenii
Mycobacterium sp. strain PYR-1, isolated from oil-contaminated sediment (62), was recognized as new species in 2002 (74). Because of its ability to degrade environmentally hazardous chemicals like high-molecular-weight polycyclic aromatic hydrocarbons, M. vanbaalenii has potential use in environmental bioremediation.
Phenotypic features.
The growth is rapid and occurs, even on minimal media, at 24 to 37°C, with formation of smooth saffron yellow colonies. All major biochemical tests but niacin are positive (Table 5).
The TLC pattern of mycolic acids is unknown, while the GLC pattern is close to but distinguishable from that of M. austroafricanum (Table 3). The HPLC profile of cell wall mycolic acids is characterized by a three-clustered descending profile (74).
Genotypic features.
Despite one cytosine insertion in helix 10 (167), M. vanbaalenii clusters far from thermotolerant rapid growers, close to M. austroafricanum and M. aurum (Fig. 2 and 4). The distinction from M. austroafricanum is, however, confirmed by the low level (below 40%) of DNA-DNA hybridization among the two species and by the different restriction patterns obtained by PFGE analysis.
Epidemiological features.
The only isolate obtained so far is environmental.
Type strain: DSM 7251T, NRRL B-24157T. GenBank 16S rDNA sequence accession number: X84977.
“VIRTUAL” MYCOBACTERIA
Genetic molecular techniques often allow the detection and classification, on the basis of their nucleic acid specific sequences, of organisms that are impossible to grow in culture and, in some cases, even to observe. My definition of such organisms, “virtual” mycobacteria, refers to the very limited possibility of knowledge they offer at present, with their phenotype remaining wrapped in mystery. The grounds of the taxonomic status of such organisms are still questionable.
“M. visibilis”
Mycobacterial DNA was obtained, by means of PCR amplification, from cutaneous lesions of three cats observed during the last 20 years (88). In 2002, on the basis of the homologies of their DNA, they were hypothesized as belonging to the same species, M. visibilis (1), whose name should, however, be corrected to M. visibile according to the rules of Latin grammar (E. Tortoli, submitted for publication).
Phenotypic features.
M. visibilis presents microscopically as filamentous acid-fast bacilli, at times in clumps, which, unusually for mycobacteria, are visible in tissues stained with hematoxylin and eosin. Only in one case was scanty growth achieved, but the strain rapidly lost viability.
Genotypic features.
The three strains have only 99.4% similarity to each other in the first 550 bp (the only segment sequenced so far) of the 16S rRNA gene, and the most closely related species appears to be M. leprae. PRA of dnaJ gene reveals a pattern with no matches to any patterns reported so far.
Clinical and epidemiological features.
The strains investigated were pathogenic and were responsible for “feline multisystemic granulomatous mycobacteriosis,” whose most evident signs are extensive ulcerative skin lesions. No hypothesis is possible about the reservoir of the mycobacterium.
GenBank 16S rDNA sequence accession numbers: AY061984, AY061985, AY061986.
NEW SUBSPECIES
M. avium subsp. avium
After DNA-DNA hybridization studies (177) had demonstrated that the previously named species M. avium and M. paratuberculosis and a cluster of strains designated “wood pigeon mycobacteria” showed high genetic relatedness, numerical taxonomy and PFGE of the above taxa demonstrated that they represented subspecies of a single species (138), for which the name M. avium was proposed.
Phenotypic, genotypic, and clinical features of the previous species M. avium, many of which were known before it was declassed to M. avium subsp. avium, were thoroughly investigated in subsequent years because of the frequent involvement of this bacterium in opportunistic infections in AIDS patients (68). It is therefore outside the purpose of the present review.
Type strain: ATCC 25291T.
M. avium subsp. paratuberculosis
M. avium subsp. paratuberculosis (138), previously M. paratuberculosis, is mycobactin dependent. It is an obligate pathogen of ruminants, in which it causes paratuberculosis, and it is suspected to be involved in Crohn's disease in humans. The phenotypic (53a), genotypic, and clinical features of M. paratuberculosis were well known before it was declassed to M. avium subsp. paratuberculosis (24); it is therefore outside the purpose of present review.
Type strain: ATCC 19698T.
M. avium subsp. silvaticum
In contrast to other subspecies of M. avium, M. avium subsp. silvaticum does not overlap a previously known species (138).
Phenotypic features.
The phenotypic features are almost identical to those of M. avium subsp. avium, from which it differs by its inability to grow on egg medium and for stimulation of growth at pH 5.5.
Genotypic features.
An insertion sequence element, repeated 10 to 12 times, has been detected within the genome of M. avium subsp. silvaticum (94). This element, IS902, has homology to IS900 (94) of M. avium subsp. paratuberculosis.
Clinical and epidemiological features.
M. avium subsp. silvaticum is an obligate pathogen, causing tuberculosis in birds, that may be the cause of chronic enteritis in calves. The specific element IS902 has also been detected in long-term cultures of samples from Crohn's disease patients and ulcerative colitis patients, as well as from noninflammatory bowel disease controls (95). The ecological niche represented by birds seems the most likely natural reservoir.
Type strain: CIP 103317T.
M. avium subsp. hominissuis
Very recently, while the present review was being prepared, the new subspecies M. avium subsp. hominissuis was proposed. It is defined by the presence of a signature nucleotide within the ITS, by the IS1245 RFLP pattern, and by growth temperature tolerance as well. Refer to the original paper (92) for details.
M. bovis subsp. caprae
A total of 121 strains isolated from lymph nodes or pulmonary lesions of 119 goats, 1 sheep, and 1 pig, although belonging to the M. tuberculosis complex, had phenotypic and genotypic features which differentiate them from other members of the complex. They were thought, in 1999, to belong to the subspecies caprae within the species M. tuberculosis (2). Three years later, on the basis of biochemical and genetic characteristic, the species designation of the new taxon was replaced by M. bovis subsp. caprae (99).
Phenotypic features.
The strains grow in 3 to 4 weeks on solid media at 36°C, and the addition of pyruvate evidently stimulates growth. Among biochemical tests, the most significant are positive pyrazinamidase and negative niacin and nitrate results.
The TLC pattern of mycolic acids has not been investigated; the GLC analysis reveals a profile shared by other members of M. tuberculosis complex.
Like M. tuberculosis sensu stricto, M. bovis subsp. caprae is susceptible to all first-line antitubercular drugs, including pyrazinamide.
Genotypic features.
The sequences of 16S rRNA gene, and of the ITS region as well, are identical to those shared by other members of the M. tuberculosis complex; as expected, the M. bovis subsp. caprae sequence hybridizes with AccuProbe M. tuberculosis complex. The M. tuberculosis complex-specific sequences IS6110 (63), IS1081 (83), and mpb70 (173) are present in all the strains. The analysis of gene polymorphisms carried out by different techniques on the pncA, katG, oxyR, and gyrA genes revealed a combination of polymorphism unique within the M. tuberculosis complex; such a separation is confirmed also by spoligotyping.
The detection of M. bovis-specific mutations in oxyR and girB loci of M. bovis subsp. caprae and the absence of the mtp40 sequence were the reason for the attribution to the species M. bovis; the point mutation in the pncA gene, which causes the susceptibility of the subspecies to pyrazinamide, is the only obvious genetic difference from M. bovis (99).
The isolation of M. bovis subsp. caprae has suggested that it represents a variant, considered ancestral on the basis of its special combination of polymorphisms, more adapted to goats than is M. bovis (2).
Clinical and epidemiological features.
Apart from goats, M. bovis subsp. caprae was initially isolated from a sheep, a pig (2), and three humans (54) who had been in close contact with goats. Isolation from humans, cattle, and red deer, but not from goats, has been reported in Austria (107), and the DNA fingerprinting was suggestive of transmission in and among these species.
M. bovis subsp. caprae is an addition to the agents of human tuberculosis contracted from animals. Pulmonary disease was found in all the patients studied so far; in one of them it was lethal because of miliary dissemination after transplantation, while in another, resistance to isoniazid and rifampin developed (107).
Type strain: CIP 105776T. EMBL 16S rDNA sequence accession number: AJ131120.
UNIDENTIFIED MYCOBACTERIA
Despite the great progress of mycobacterial taxonomy in the last decade, a number of yet undescribed taxa have still not emerged. Many are in fact the new mycobacterial sequences deposited in major genetic databases not corresponding to any of species published in the literature.
In a recent study (142), 72 mycobacteria isolated from humans, the environment, or animals were investigated using conventional tests, HPLC, and 16S rDNA genetic sequencing. They represented 53 taxonomic entities, none of which corresponded to any officially recognized species. They were almost homogeneously distributed among major systematic mycobacterial clusters: thermotolerant rapid growers, M. terrae related, slow growers, M. simiae related, and rapid growers. The presence of a fairly large percentage of mycobacteria still not identifiable represents an important reservoir of “new species” that shows no signs of diminishing.
CONCLUSIONS
Isolation Source and Phenotypic Characters
Forty-two new mycobacterial species were officially recognized in the last 14 years (Table 7). Most of them (23 species [55%]) were isolated from clinical samples only, 10 were isolated from both the environment and clinical samples, and 9 were isolated from the environment only. Since the environment is universally considered the main reservoir of nontuberculous mycobacteria, it seems plausible that the isolation exclusively from biological samples of over half the new species is biased by the limited number of environmental investigations.
TABLE 7.
Major phenotypic and epidemiological features of new mycobacterial species
| Sample origin | No. of slow growers
|
No. of rapid growers
|
Total no. | ||
|---|---|---|---|---|---|
| Chromogens | Nonchromogens | Chromogens | Nonchromogens | ||
| Clinical | 8a | 6 | 3 | 6 | 23 |
| Clinical and environmental | 3 | 1 | 6 | 10 | |
| Environmental | 3 | 6 | 9 | ||
| Subtotals | 14 | 7 | 9 | 12 | |
| Grand total | 42 | ||||
The species M. shottsi, a pathogen for striped bass, is included.
Chromogens largely predominate among slow growers (14 of 21), while rapid growers are mainly nonchromogens (12 of 21). If M. intermedium, whose assignment as a photochromogen is questionable, is disregarded, M. novocastrense remains the only new photochromogenic species detected. Nine essentially environmental species are all scotochromogenic and are predominantly rapid growers (six of nine).
Species for which two morphological colony types have been reported or ones in which various pigmentation tones (from white to yellow) may be present are rare. Two HPLC chromotypes have been described for M. bohemicum and M. interjectum. Additionally, M. interjectum exhibits both a pigmented and an unpigmented variant.
Phylogenetic Investigation
The extra information obtained by a genetic approach to taxonomic studies allows the reconstruction of the phylogenetic tree of related systematic entities. Highly conserved regions present in particular genomic regions are the ideal target for such studies since the number, type, and position of mutations appearing through evolution represent a sensitive metric of phylogenetic distances.
Some perplexity arises, however, at least for nontaxonomists, when phylogenetic trees based on different highly conserved regions grossly differ or when different clusterings emerge from the analysis of the same genetic sequences performed with various mathematic algorithms.
The phylogenetic trees reported here (Fig. 3 and 4) were developed by submitting to the BLAST program (version 2.0; European Molecular Biology Laboratory [http://dove.embl-heidelberg.de/Blast2/]) the 16S rDNA sequences present in publicly available databases for all the established species of the genus Mycobacterium. The 5′ and 3′ ends were cut to an identical position, at E. coli bp 131 to bp 1332, respectively, thus obtaining a stretch of about 1,200 bases, i.e., the longest available for all the sequences.
There are five major phylogenetic groups within the genus Mycobacterium on the basis of 16S rDNA sequences. The most homogeneous is certainly the one related to M. simiae. It includes eight new species characterized by an identical sequence, presenting a short helix 18, in hypervariable region B (Fig. 2). One species only, M. simiae, was known within this group before 1990.
Among the slow growers, which include a large number of the classically known mycobacteria, are nine new species (Table 8). The distinctive feature of the group is the long helix 18, and each species differs from the others in both hypervariable regions of the 16S rDNA (Fig. 2).
TABLE 8.
Relationship between genotypic and phenotypic features of new mycobacterial species
| Genotypic group | No. of slow growers
|
No. of rapid growers
|
Total no. | ||
|---|---|---|---|---|---|
| Chromogens | Nonchromogens | Chromogens | Nonchromogens | ||
| M. simiae related | 5 | 3 | 0 | 0 | 8 |
| Slow growers | 6 | 3 | 1 | 0 | 10 |
| M. terrae related | 1 | 0 | 0 | 0 | 1 |
| Rapid growers | 1 | 0 | 5 | 9 | 15 |
| Thermotolerant rapid growers | 1 | 0 | 4 | 3 | 8 |
One new species only, M. hiberniae, clusters with M. terrae and M. nonchromogenicum. It is characterized by the presence of two nucleotide insertions in the long helix 18 (Fig. 2).
The group of rapid growers includes the largest number of new species, 15. Four sequences of hypervariable region B are shared by several taxa: two sequences are shared by three species, and two are shared by two species (Fig. 2).
Finally, eight new species cluster with the thermotolerant rapid growers. All of them have a 1-nucleotide insertion in helix 10. This nucleotide, which is typically cytosine, is replaced in two cases by thymine (Fig. 2). Interestingly, two species of this group have sequence homology in hypervariable region B to mycobacteria clustering within the slow growers.
Molecular Microheterogeneity within 16S rDNA
While the large majority of taxa are characterized by absolute homogeneity at the level of the 16S rRNA gene, several species exist which are characterized by limited nucleotide variations. This is the case for M. celatum, M. bohemicum, and M. elephantis, for which three sequevars are known, and M. immunogenum, M. interjectum, M. lentiflavum, M. mageritense, and M. wolinskyi, for which two sequences are known. The molecular microheterogeneity only rarely involves nucleotides within hypervariable regions A and B. It is therefore possible that such microheterogeneity is much more frequent than is thought at present, given that the sequencing is often limited to the most variable trait, the first (5′) 500 bp of the gene or even to hypervariable region A only.
Phenotype-Genotype Relations
In the large majority of cases, the new mycobacteria characterized by evident phenotypic similarity (biochemical tests and lipid structure) to other species also share a close phylogenetic relatedness with them. The most striking geno-phenotypic resemblances are among M. heckeshornense and M. botniense with M. xenopi, among M. lentiflavum and M. triplex with M. simiae, between M. kubicae and M. palustre, between M. wolinskyi and M. mageritense, and between M. goodii and M. smegmatis. It is, in contrast, less common to find a mycobacterium where the phenotype can be confused for a different, genetically unrelated, species, as is the case for M. heidelbergense and M. malmoense.
One also finds discrepancies when species are included within groups on the basis of genetic features. This is the case for M. tusciae, which despite a short helix 18, grows slowly, and for M. doricum, which grows slowly and at temperature no higher than 37°C despite the presence of the genetic markers of thermotolerant rapid growers.
Limitations of Susceptibility Testing
A feature almost always reported in papers describing new species is their antimicrobial susceptibility in vitro. Unfortunately, collection of this important experimental information is almost always thwarted by technical problems. (i) In many cases, only the four first-line drugs used in the treatment of tuberculosis are tested. (ii) The proportion method, a technique developed and validated for M. tuberculosis only, is almost invariably used. (iii) The drug concentrations fixed for M. tuberculosis testing are generally used, although they are largely below the ones recommended for the nontuberculous mycobacteria (Table 9) (97). (iv) Only occasionally are the MICs determined. It is more common that a qualitative investigation, furthermore on poorly reliable egg-based medium, is performed.
TABLE 9.
Recommendations for quantitative susceptibility testing of rapidly growing mycobacteriaa
| Drug | Twofold concn (μg/ml) | Susceptibility breakpoint (μg/ml) | Resistance breakpoint (μg/ml) | Notes |
|---|---|---|---|---|
| Amikacin | 1-128 | ≤16 | ≥64 | To be reported only in case of resistance to tobramycin |
| Cefoxitin | 2-256 | ≤16 | ≥128 | |
| Ciprofloxacin | 0.125-16 | ≤1 | ≥4 | |
| Clarithromycin | 0.06-64 | ≤2 | ≥8 | |
| Doxycycline | 0.25-32 | ≤1 | ≥16 | |
| Imipenem | 1-64 | ≤4 | ≥16 | Not to be reported for M. chelonae or M. abscessus |
| Sulfamethoxazole | 1-64 | ≤32 | ≥64 | MIC is 80% inhibition of growth |
| Tobramycin | 1-32 | ≤4 | ≥16 | To be tested for M. chelonae only |
Modified from reference 97.
The consequence of the aforementioned drawbacks is that the reliability of emerging drug susceptibility data is very poor. Multidrug resistance of nontuberculous mycobacteria is often, but sometime improperly, evoked without being objectively proven. Apart from resistance, what factors may emerge when drugs like isoniazid or pyrazinamide, which are highly effective against M. tuberculosis but unquestionably not efficacious for nontuberculous mycobacteria, are tested? What happens when breakpoints are used that are clearly not suitable for organisms that, unlike M. tuberculosis, are not characterized by a wide gap between drug MICs for resistant and susceptible strains? Furthermore, lack of standardization means that the susceptibility results obtained from different laboratories are not comparable. It is well known, for instance, that the MICs are consistently higher when measured on solid than in liquid media.
When such limitations are disregarded, the highly heterogeneous data reported for new mycobacteria reviewed here show that isoniazid, as expected, is the least effective drug with almost 76% of the species being resistant. Species resistant to ethambutol and rifampin make up 57%, while resistance to streptomycin is found in 40%. While the resistances to isoniazid and streptomycin are almost identically distributed among slowly and rapidly growing species, the former are much more resistant to ethambutol and susceptible to rifampin than are the latter.
However, when proper quantitative methods are used, consistent data, important for orientation of therapy, emerge and often give rise to identification characteristics (14, 171).
Significant or Not Significant? That's the Problem
A major problem when dealing with nontuberculous mycobacteria is the determination of their medical relevance. While, in the absence of disease, the clinical insignificance of the isolation is granted, it is very difficult to determine clinical significance when a pathogenic manifestation is present. A substantial consensus exists, represented by the American Thoracic Society recommendations (Table 10), that establish minimal criteria for acknowledgement of the pathogenicity of the finding (165). Further important information is provided by the results of microscopy and the number of colonies grown in culture. For low-virulence organisms like nontuberculous mycobacteria, the bacterial load represents a major factor in deciding pathogenicity.
TABLE 10.
Recommended diagnostic criteria for pulmonary disease caused by nontuberculous mycobacteriaa
| For patients with pulmonary cavitary lung disease |
| Presence of two or more sputum specimens that are acid-fast bacillus smear positive and/or result in moderate to heavy growth of nontuberculous mycobacteria on culture |
| Exclusion of other reasonable causes for the disease process |
| For patients with noncavitary lung disease |
| Presence of two or more sputum specimens that are acid-fast bacillus smear positive and/or result in moderate to heavy growth of nontuberculous mycobacteria on culture |
| Failure of the sputum culture to clear with bronchial toilet or within 2 wk of institution of specific mycobacterial drug therapy |
| Exclusion of other reasonable causes for the disease process |
| For patients with cavitary or noncavitary lung disease whose sputum evaluation is nondiagnostic or another disease cannot be excluded |
| A transbronchial or open-lung biopsy that yields the organism and shows mycobacterial histopathologic features (granulomatous infiltration, with or without acid-fast bacilli); no other criteria needed |
| A transbronchial or open-lung biopsy that fails to yields the organism but shows mycobacterial histopathologic features in the absence of a history of other granulomatous or mycobacterial disease plus: |
| Presence of two or more positive cultures of sputum |
| Exclusion of other reasonable causes for granulomatous disease |
Data from reference 165.
In the sp. nov. descriptions, little emphasis, if any, is traditionally placed on clinical information. However, this gap is usually filled in by case reports that generally follow the recognition of a new species. The information concerning the response to treatment is, however, often missing, and only very rarely is successful therapy reported. The widespread use of improper regimens (those which are, in effect, directed against M. tuberculosis) may well be one of the reasons.
Most Frequent Pathologies
Among newly detected slow growers, 100% of nonchromogenic and 50% of chromogenic species are potentially pathogenic. Among rapid growers, the pathogenicity is shared by eight taxa, of which only one species is pigmented.
Except for lung diseases, for which both mycobacterial growth groups may be responsible, different pathologies are caused by slowly and rapidly growing species. Childhood cervical lymphadenitis and AIDS-related disseminated diseases are frequently caused by slow growers while posttraumatic infections are frequently due to rapid growers.
When the potentially pathogenic species for which several cases have been reported are considered, a preferential association with specific diseases emerges. Among slow growers, M. bohemicum and M. interjectum are almost exclusively involved in cervical lymphadenitis while M. celatum and M. genavense are most frequently responsible for opportunistic infections in AIDS patients. Large studies are available for the rapid growers M. abscessus, M. goodii, M. mucogenicum, and M. wolinskyi. These are very frequently associated with posttraumatic and postsurgical wounds including osteomyelitis. M. abscessus is also frequently responsible for chronic lung disease (52) and disseminated cutaneous infections in immunosuppressed people (45). In adjunct to pathologies directly attributable to M. immunogenum, this species is commonly isolated from aerosols in industries using metal-working abrasives known to be associated with hypersensitivity pneumonitis.
M. scrofulaceum, Organism in Decline?
Well documented, and still unexplained by epidemiologists, is the sudden decrease of childhood lymphadenopathies caused by M. scrofulaceum (176). On the other hand, eight of the clinically significant new species were isolated just from cervical lymphadenitis, and, interestingly five of them are, like M. scrofulaceum, scotochromogenic slow growers. Is there a link among such events? A certain answer is not possible at present, but it seems likely that at least some of the strains isolated from lymph nodes, in the years when the great majority of cases of mycobacterial lymphadenitis of childhood were attributed to M. scrofulaceum, were actually simply yellow slowly growing mycobacteria not unquestionably identifiable by the technologies available at that time and presenting the closest resemblance to M. scrofulaceum.
Suggested Minimal Standards for the Description of New Taxa
On the basis of the tests adopted for the large majority of new species descriptions reviewed here, several tentative suggestions for increasing standardization emerge. Despite their declining importance, several conventional tests still have a role. The most common and, ipso facto, recommended cultural tests include growth temperatures, growth rate, pigmentation type, colony morphology, plus for rapidly growing organisms, NaCl tolerance, and MacConkey growth. Among biochemical tests, 3-day arylsulfatase, nitrate reduction, Tween 80 hydrolysis, urea hydrolysis, 68°C catalase, and semiquantitative catalase are the most widely used. Lipid analysis cannot be disregarded, and none of the available techniques, TLC, GLC, and HPLC, should be neglected. The lack of information for one or two of them would deprive many diagnostic centers of an important identification criterion, especially since most of the laboratories use only one lipid approach. Finally, not in question is the primary role of genetics; the sequencing of the entire 16S rRNA gene or at least of the first 500 bp is critical.
Following the detection of the higher guanine-plus-cytosine content (61 to 71 mol%) characterizing the mycobacterial genome in comparison with other microorganisms (135), this parameter became one of the most frequently investigated in the new species. Although this is a still frequently provided piece of information, it appears to be of limited value and can be omitted. Very rarely is DNA-DNA homology determined for the sp. nov. description these days. This technique represents the most important approach to a quantitative definition of species and is the sole technique reflecting the whole genomic complexity. The rationale for DNA-DNA hybridization is that within the same species, the DNA relatedness should be at least 70% and the divergence should be below 5% (168). Unfortunately, this technique has become less and less popular in recent years, mainly because it is complex, expensive, and labor intensive; its revival seems unlikely.
The taxonomic relevance of new species recognized on the basis of a single strain, with no additional isolates, remains uncertain. Furthermore, with a single isolate, the weight attached to a phenotypic, and even a genotypic, characteristic may mislead the investigator since no certainty exists that such features really reflect the typical profile of the species. Therefore, it seems urgent that a decision about the minimal number of strains needed for the description of new species should be made. Although the reports concerning new mycobacteria, even if represented by single strains, are indisputably useful, it seems advisable that their official recognition as new species should remain pending the detection of further independent isolates. How many? Two, five, ten? This should be left to a consensus among taxonomists or, to be more exact, to mycobacteriological taxonomists, in consideration of the particular genetic relatedness existing among such organisms.
A personal remark about the names of new mycobacteria: stemming of the spreading habit of using the name of living people seems advisable. An exception may be made for names of scientists whose role in the advancement of mycobacteriology is preeminent (M. goodii, M. kubicae, M. wolinskyi), but it seems of questionable taste in remaining cases.
What Is the Impact of Expanded Taxonomy on Routine Mycobacteriology Laboratory Practice?
The explosive expansion of mycobacterial taxonomy in the 1990s further supports the recommendation of concentrating higher-level mycobacterial diagnostics in reference laboratories. Commercially available DNA probes are available that allow nonspecialized laboratories to correctly identify the most frequently encountered species. The identification of strains that are not easy identified should, however, be discouraged in centers other than reference centers. At the same time, the implementation of modern identification systems by reference centers is required.
Both genetic and chemotaxonomic approaches have their own role; both of them, however, have limitations. Furthermore, no single test methodology can provide 100% accurate results (70). TLC and GLC cannot be used alone, mainly because they are hindered by the large number of species with shared patterns. With HPLC too, which is more discriminative, there are also problems in differentiating species with similar profiles; this is an emerging problem, in particular among rapid growers, with the increase in the number of species. PRA, which is suitable for the identification of most species, has not yet been tested for many of new taxa. Furthermore, its interpretation may at times be made problematic by species with identical patterns or with multiple patterns.
Genetic sequencing at last! Although the combination of multiple approaches is needed, the sequencing of the 5′ end (about 500 bp) of 16S rDNA is unquestionably the only method that, even when used alone, provides the most reliable identifications. Nevertheless, limitations do exist, mainly due to the limited portion of the genome surveyed by 16S rDNA analysis and to the highly conserved nature of the gene that makes species variations less evident then in investigations concerning DNA-DNA homology. To the well-known species pairs with an identical 5′ end, M. gastri-M. kansasii and M. marinum-M. ulcerans, others exist among newly described taxa. Overlapping exists for M. septicum and M. peregrinum, for M. murale and M. tokaiense, and for M. novocastrense and one of the sequevars of M. flavescens. Furthermore, several species contain multiple sequevars are present (see “Molecular microheterogeneity within 16S rDNA”). The most critical issue is represented, however, by the lack of quality control of sequence entries in major public sequence databases. As a consequence, their use in similarity searches often results in inaccurate identifications, with the best match often favoring ragged sequences and faulty or outdated entries. A very important source of data of the highest quality has been built out in the RIDOM Mycobacteria project (http://www.ridom.de/mycobacteria/) (D. Harmsen, J. Rothganger, C. Singer, J. Albert, and M. Frosch, Letter, Lancet 353:291, 1999), which offers an excellent free platform for analyzing mycobacterial 16S sequences (158). Moreover, it contains reviewed data and descriptions on the phenotype of all mycobacteria.
Final Remarks
The recently described mycobacterial species represent only the emergence of a natural diversity previously unrecognized because of the inadequacy of available identification methods. Many newly described mycobacteria had probably already been isolated in the last decades but had been considered variants of, at that time, officially recognized species, with natural variability being most frequently invoked to justify discrepant test results.
It is commonly thought that nontuberculous mycobacteria, and newly described species in particular, are restricted to developed countries. Again, a shortage of technology may well explain the missed isolation of such problematic microorganisms in developing countries, especially since, when suitable means have been employed, nontuberculous mycobacteria have been detected in these countries too (34, 40).
Physicians do not regard taxonomy highly since these data are often considered distant from clinical practice. Such an attitude appears to be groundless if we consider that mycobacterial species differ in virulence and sometimes present characteristic antimicrobial patterns. Correct identification may therefore be of use in the decision about whether a therapeutic regimen may be appropriate and, in some cases, in the determination of the drugs to use.
Acknowledgments
I thank Vivian Jonas (Gen-Probe, San Diego, Calif.) for valuable suggestions and thorough revision of the manuscript; Marina Luquin and Pedro-Luis Valero (University of Barcelona, Barcelona, Spain) for punctual revision of tables concerning TLC and GLC analyses; Nalin Rastogi (Pasteur Institute of Guadeloupe, Pointe a Pitre, France) for providing several M. canettii strains for HPLC analysis; Leslie Hall (Mayo Clinic, Rochester, Minn.) for providing information about the genetic sequence of M. mageritense; Elvira Richter (National Reference Center for Mycobacteria, Borstel, Germany) and Christine Turenne (National Reference Center for Mycobacteriology, Winnipeg, Canada) for providing, before publication, manuscripts concerning the description of M. holsaticum and M. lacus, respectively. E. Richter also donated two strains of M. holsaticum for HPLC analysis.
REFERENCES
- 1.Appleyard, G. D., and E. G. Clark. 2002. Histologic and genotypic characterization of a novel Mycobacterium species found in three cats. J. Clin. Microbiol. 40:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranaz, A., E. Liebana, E. Gomez Mampaso, J. C. Galan, D. Cousins, A. Ortega, J. Blazquez, F. Baquero, A. Mateos, G. Suarez, and I. Dominguez. 1999. Mycobacterium tuberculosis subsp caprae subsp nov.: a taxonomic study of a new member of the Mycobacterium tuberculosis complex isolated from goats in Spain. Int. J. Syst. Bacteriol. 49:1263-1273. [DOI] [PubMed] [Google Scholar]
- 3.Ausina, V., M. Luquín, M. García-Barceló, M. A. Lanéelle, V. Lévy-Frébault, F. Belda, and G. Prats. 1992. Mycobacterium alvei sp. nov. Int. J. Syst. Bacteriol. 42:529-535. [DOI] [PubMed] [Google Scholar]
- 4.Bercovier, H., O. Kafri, and S. Sela. 1986. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem. Biophys. Res. Commun. 136:1136-1141. [DOI] [PubMed] [Google Scholar]
- 5.Böddinghaus, B., T. Rogall, T. Flohr, H. Blöcker, and E. C. Böttger. 1990. Detection and identification of mycobacteria by amplification of rRNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogdan, C., P. Kern, E. Richter, A. Tannapfel, S. Rüsch-Gerdes, T. Kirchner, and W. Solbach. 1997. Systemic infection with Mycobacterium genavense following immunosuppressive therapy in a patient who was seronegative for human immunodeficiency virus. Clin. Infect. Dis. 24:1245-1247. [DOI] [PubMed] [Google Scholar]
- 7.Bonomo, R. A., J. M. Briggs, W. Gross, M. Hassan, R. C. Graham, W. R. Butler, and R. A. Salata. 1999. Mycobacterium celatum infection in a patient with AIDS. Clin. Infect. Dis. 26:243-245. [DOI] [PubMed] [Google Scholar]
- 8.Böttger, E. C. 1996. Approaches for identification of microorganisms. Despite longer experience with fatty acid profiles, DNA-based analysis offers several advantages. ASM News 62:247-250. [Google Scholar]
- 9.Böttger, E. C., B. Hirschel, and M. B. Coyle. 1993. Mycobacterium genavense sp. nov. Int. J. Syst. Bacteriol. 43:841-843. [DOI] [PubMed] [Google Scholar]
- 10.Böttger, E. C., A. Teske, P. Kirschner, S. Bost, H. R. Chang, V. Beer, and B. Hirschel. 1992. Disseminated “Mycobacterium genavense” infection in patients with AIDS. Lancet 340:76-80. [DOI] [PubMed] [Google Scholar]
- 11.Brander, E., E. Jantzen, R. Huttunen, A. Julkunen, and M. L. Katila. 1992. Characterization of a distinct group of slowly growing mycobacteria by biochemical tests and lipid analyses. J. Clin. Microbiol. 30:1972-1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan, P. J., M. Heifets, and B. P. Ullom. 1982. Thin-layer chromatography of lipid antigens as a means of identifying nontuberculous mycobacteria. J. Clin. Microbiol. 15:447-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Briglia, M., R. I. L. Eggen, D. J. van Elsas, and W. M. De Vos. 1994. Phylogenetic evidence for transfer of pentachlorophenol-mineralizing Rhodococcus chlorophenolicus PCP-I-T to the genus Mycobacterium. Int. J. Syst. Bacteriol. 44:494-498. [DOI] [PubMed] [Google Scholar]
- 14.Brown, B. A., B. Springer, V. A. Steingrube, R. W. Wilson, G. E. Pfyffer, M. J. Garcia, M. C. Menendez, B. Rodriguez-Salgado, K. C. Jost, Jr., S. H. Chiu, G. O. Onyi, E. C. Böttger, and R. J. Wallace, Jr. 1999. Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov. rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49:1493-1511. [DOI] [PubMed] [Google Scholar]
- 15.Brown, B. A., J. M. Swenson, and R. J. Wallace, Jr. 1992. Broth microdilution MIC test for rapidly growing mycobacteria, p. 5.11.1-5.11.10. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C.
- 16.Bull, T. J., D. C. Shanson, L. C. Archard, M. D. Yates, M. E. Hamid, and D. E. Minnikin. 1995. A new group (type 3) of Mycobacterium celatum isolated from AIDS patients in the London area. Int. J. Syst. Bacteriol. 45:861-862. [DOI] [PubMed] [Google Scholar]
- 17.Butler, W. R., and L. S. Guthertz. 2001. Mycolic acid analysis by high-performance liquid chromatography for identification of Mycobacterium species. Clin. Microbiol. Rev. 14:704-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler, W. R., S. P. O'Connor, M. A. Yakrus, and W. M. Gross. 1994. Cross-reactivity of genetic probe for detection of Mycobacterium tuberculosis with newly described species Mycobacterium celatum. J. Clin. Microbiol. 32:536-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler, W. R., S. P. O'Connor, M. A. Yakrus, R. W. Smithwick, B. B. Plikaytis, C. W. Moss, M. M. Floyd, C. L. Woodley, J. O. Kilburn, F. S. Vadney, and W. M. Gross. 1993. Mycobacterium celatum sp. nov. Int. J. Syst. Bacteriol. 43:539-548. [DOI] [PubMed] [Google Scholar]
- 20.Butler, W. R., L. Thibert, and J. O. Kilburn. 1992. Identification of Mycobacterium avium complex strains and some similar species by high-performance liquid chromatography. J. Clin. Microbiol. 30:2698-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bux-Gewehr, I., H. P. Hagen, S. Rüsch-Gerdes, and G. E. Feurle. 1998. Fatal pulmonary infection with Mycobacterium celatum in an apparently immunocompetent patient. J. Clin. Microbiol. 36:587-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cabria, F., M. V. Torres, J. I. Garcia-Cia, M. N. Dominguez-Garrido, J. Esteban, and M. S. Jimenez. 2002. Cervical lymphadenitis caused by Mycobacterium lentiflavum. Pediatr. Infect. Dis. J. 21:574-575. [DOI] [PubMed] [Google Scholar]
- 23.Caroli, G., E. Levrè, A. Baggiani, E. Tortoli, and C. Baglioni. 1998. Mycobacterium mucogenicum found in a bottled mineral water, p. 63-64. In M. Casal (ed.), Clinical mycobacteriology. Prous Science, Barcelona, Spain.
- 24.Chiodini, R. J. 1989. Crohn's disease and the mycobacterioses: a review and comparison of two disease entities. Clin. Microbiol. Rev. 2:90-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cingolani, A., M. Sanguinetti, A. Antinori, L. M. Larocca, F. Ardito, B. Posteraro, G. Federico, G. Fadda, and L. Ortona. 2000. Brief report: disseminated mycobacteriosis caused by drug-resistant Mycobacterium triplex in a human immunodeficiency virus-infected patient during highly active antiretroviral therapy. Clin. Infect. Dis. 31:177-179. [DOI] [PubMed] [Google Scholar]
- 26.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyle, M. B., L. D. C. Carlson, C. K. Wallis, R. B. Leonard, V. A. Raisys, J. O. Kilburn, M. Samadpour, and E. C. Böttger. 1992. Laboratory aspects of “Mycobacterium genavense,” a proposed species isolated from AIDS patients. J. Clin. Microbiol. 30:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daffé, M., M. McNeil, and P. J. Brennan. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30:373-388. [DOI] [PubMed] [Google Scholar]
- 29.Dahl, D. M., D. Klein, and A. Morgentaler. 1996. Penile mass caused by the newly described organism Mycobacterium celatum. Urology 47:266-268. [DOI] [PubMed] [Google Scholar]
- 30.De Baere, T. T., M. Moerman, L. Rigouts, C. Dhooge, H. Vermeersch, G. Verschraegen, and M. Vaneechoutte. 2001. Mycobacterium interjectum as causative agent of cervical lymphadenitis. J. Clin. Microbiol. 39:725-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domenech, P., M. C. Menendez, and M. J. García. 1994. Restriction fragment length polymorphisms of 16S rRNA genes in the differentiation of fast-growing mycobacterial species. FEMS Microbiol. Lett. 116:19-24. [DOI] [PubMed] [Google Scholar]
- 32.Domenech, P., M. S. Jiménez, M. C. Menendez, T. J. Bull, S. Samper, A. Manrique, and M. J. Garcia. 1997. Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47:535-540. [DOI] [PubMed] [Google Scholar]
- 33.Dumonceau, J., A. van Gossum, M. Adler, J. P. van Vooren, P. A. Fonteyne, H. De Beenhouwer, and F. Portaels. 1997. Detection of fastidious mycobacteria in human intestines by the polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 16:358-363. [DOI] [PubMed] [Google Scholar]
- 34.Eaton, T., J. O. Falkinham III, T. O. Aisu, and T. M. Daniel. 1995. Isolation and characteristics of Mycobacterium avium complex from water and soil samples in Uganda. Tubercle Lung Dis. 76:570-574. [DOI] [PubMed] [Google Scholar]
- 35.Ehlers, S., and E. Richter. 2000. Gamma interferon is essential for clearing Mycobacterium genavense infection. Infect. Immun. 68:3720-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehlers, S., and E. Richter. 2001. Differential requirement for interferon-gamma to restrict the growth of or eliminate some recently identified species of nontuberculous mycobacteria in vitro. Clin. Exp. Immunol. 124:229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emler, S., P. Praplan, P. Rohner, R. Auckenthaler, and B. Hirschel. 1996. Disseminierte Infektion mit Mycobacterium celatum. Schweiz. Med. Wochenschr. 126:1062-1065. [PubMed] [Google Scholar]
- 38.Emler, S., T. Rochat, P. Rohner, C. Perrot, R. Auckenthaler, L. Perrin, and B. Hirschel. 1994. Chronic destructive lung disease associated with a novel mycobacterium. Am. J. Respir. Crit. Care Med. 150:261-265. [DOI] [PubMed] [Google Scholar]
- 39.Fattorini, L., L. Baldassarri, Y. J. Li, M. G. Ammendolia, Y. Fan, S. Recchia, E. Iona, and G. Orefici. 2000. Virulence and drug susceptibility of Mycobacterium celatum. Microbiology 146:2733-2742. [DOI] [PubMed] [Google Scholar]
- 40.Fine, P. E., S. Floyd, J. L. Stanford, P. Nkhosa, A. Kasunga, S. Chaguluka, D. K. Warndorff, P. A. Jenkins, M. Yates, and J. M. Ponnighaus. 2001. Environmental mycobacteria in northern Malawi: implications for the epidemiology of tuberculosis and leprosy. Epidemiol. Infect. 126:379-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Floyd, M. M., W. M. Gross, D. A. Bonato, V. A. Silcox, R. W. Smithwick, B. Metchock, J. T. Crawford, and W. R. Butler. 2000. Mycobacterium kubicae sp. nov., a slowly growing, scotochromogenic mycobacterium. Int. J. Syst. Evol. Microbiol. 50:1811-1816. [DOI] [PubMed] [Google Scholar]
- 42.Floyd, M. M., L. S. Guthertz, V. A. Silcox, P. S. Duffey, Y. Jang, E. P. Desmond, J. T. Crawford, and W. R. Butler. 1996. Characterization of an SAV organism and proposal of Mycobacterium triplex sp. nov. J. Clin. Microbiol. 34:2963-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reference deleted.
- 44.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 45.Fraser, V., and R. J. Wallace, Jr. 1996. Nontuberculous mycobacteria, p. 1224-1237. In C. G. Mayhal (ed.), Hospital epidemiology and infection control. The Williams & Wilkins Co., Baltimore, Md.
- 46.Friedman, N. D., and D. J. Sexton. 2001. Bursitis due to Mycobacterium goodii, a recently described, rapidly growing mycobacterium. J. Clin. Microbiol. 39:404-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galarraga, M. C., A. Torreblanca, and M. S. Jimenez. 2002. Aislamiento de Mycobacterium lentiflavum en un caso sospechoso de cancer de pulmon. Enferm. Infecc. Microbiol. Clin. 20:93-94. [PubMed] [Google Scholar]
- 48.Gholizadeh, Y., A. Varnerot, C. Maslo, B. Salauze, H. Badaoui, V. Vincent, and A. Buré-Rossier. 1998. Mycobacterium celatum infection in two HIV-infected patients treated prophylactically with rifabutin. Eur. J. Clin. Microbiol. Infect. Dis. 17:278-281. [DOI] [PubMed] [Google Scholar]
- 49.Goh, K. S., E. Legrand, C. Sola, and N. Rastogi. 2001. Rapid differentiation of “Mycobacterium canettii” from other Mycobacterium tuberculosis complex organisms by PCR-restriction analysis of the hsp65 gene. J. Clin. Microbiol. 39:3705-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldblatt, M. R., and J. A. Ribes. 2002. Mycobacterium mucogenicum isolated from a patient with granulomatous hepatitis. Arch. Pathol. Lab Med. 126:73-75. [DOI] [PubMed] [Google Scholar]
- 51.Green, B. A., and B. Afessa. 2000. Isolation of Mycobacterium interjectum in an AIDS patient with diarrhea. AIDS 14:1282-1284. [DOI] [PubMed] [Google Scholar]
- 52.Griffith, D. E., W. M. Girard, and R. J. Wallace, Jr. 1993. Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am. Rev. Respir. Dis. 147:1271-1278. [DOI] [PubMed] [Google Scholar]
- 53.Guerrant, G. O., M. A. Lambert, and C. W. Moss. 1981. Gas-chromatographic analysis of mycolic acid cleavage products in mycobacteria. J. Clin. Microbiol. 13:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Gunnarsson, E., and F. H. Fodstad. 1979. Cultural and biochemical characteristics of Mycobacterium paratuberculosis isolated from goats in Norway. Acta Vet. Scand. 20:122-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gutiérrez, M., S. Samper, M. S. Jiménez, J. D. A. van Embden, J. F. García Marín, and C. Martín. 1997. Identification by spoligotyping of a caprine genotype in Mycobacterium bovis strains causing human tuberculosis. J. Clin. Microbiol. 35:3328-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haas, W. H., W. R. Butler, P. Kirschner, B. B. Plikaytis, M. B. Coyle, B. Amthor, A. G. Steigerwalt, D. J. Brenner, M. Salfinger, J. T. Crawford, E. C. Böttger, and H. J. Bremer. 1997. A new agent of mycobacterial lymphadenitis in children: Mycobacterium heidelbergense sp. nov. J. Clin. Microbiol. 35:3203-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haas, W. H., P. Kirschner, S. Ziesing, H. J. Bremer, and E. C. Böttger. 1993. Cervical lymphadenitis in a child caused by a previously unknown mycobacterium. J. Infect. Dis. 167:237-240. [DOI] [PubMed] [Google Scholar]
- 57.Haase, G., H. Kentrup, H. Skopnik, B. Springer, and E. C. Böttger. 1997. Mycobacterium lentiflavum: an etiologic agent of cervical lymphadenitis. Clin. Infect. Dis. 25:1245-1246. [DOI] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Reference deleted.
- 60.Häggblom, M. M., L. J. Nohynek, N. J. Palleroni, K. Kronqvist, E. L. Nurmiaho-Lassila, M. S. Salkinoja-Salonen, S. Klatte, and R. M. Kroppenstedt. 1994. Transfer of polychlorophenol-degrading Rhodococcus chlorophenolicus (Apajalahti et al. 1986) to the genus Mycobacterium as Mycobacterium chlorophenolicum comb. nov. Int. J. Syst. Bacteriol. 44:485-493. [DOI] [PubMed] [Google Scholar]
- 61.Hazra, R., M. M. Floyd, A. Sloutsky, and R. N. Husson. 2001. Novel mycobacterium related to Mycobacterium triplex as a cause of cervical lymphadenitis. J. Clin. Microbiol. 39:1227-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heitkamp, M. A., W. Franklin, and C. E. Cerniglia. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 54:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hermans, P. W. M., D. van Soolingen, J. W. Dale, A. R. J. Schuitema, R. A. McAdam, D. Catty, and J. D. A. van Embden. 1990. Insertion element IS 986 from Mycobacterium tuberculosis: a useful tool for diagnosis and epidemiology of tuberculosis. J. Clin. Microbiol. 28:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hillebrand Haverkort, M. E., A. H. J. Kolk, L. F. F. Kox, J. J. A. M. TenVelden, and J. H. TenVeen. 1999. Generalized Mycobacterium genavense infection in HIV-infected patients: detection of the mycobacterium in hospital tap water. Scand. J. Infect. Dis. 31:63-68. [DOI] [PubMed] [Google Scholar]
- 65.Hirschel, B., H. R. Chang, N. Mach, P. F. Piguet, J. Cox, J. D. Piguet, M. T. Silva, L. Larsson, P. R. Klatser, J. E. R. Thole, L. Rigouts, and F. Portaels. 1990. Fatal infection with a novel unidentified mycobacterium in a man with the acquired immunodeficiency syndrome. N. Engl. J. Med. 323:109-113. [DOI] [PubMed] [Google Scholar]
- 66.Hoff, E., M. Sholtis, G. Procop, C. Sabella, J. Goldfarb, R. Wyllie, R. Cunningham, and L. H. G. Stockman. 2001. Mycobacterium triplex infection in a liver transplant patient. J. Clin. Microbiol. 39:2033-2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hughes, M. S., N. W. Ball, D. N. Love, P. J. Canfield, D. I. Wigney, D. Dawson, P. E. Davis, and R. Malik. 1999. Disseminated Mycobacterium genavense infection in a FIV-positive cat. J. Feline Med Surg. 1:23-29. [DOI] [PubMed] [Google Scholar]
- 67a.Ibàñez, R., R. Serrano-Heranz, M. Jiménez-Palop, C. Román, M. Corteguera, and S. Jiménez. 2002. Disseminated infection caused by slow-growing Mycobacterium lentiflavum. Eur. J. Clin. Microbiol. Infect. Dis. 21:691-692. [DOI] [PubMed] [Google Scholar]
- 68.Inderlied, C. B., C. A. Kemper, and L. E. M. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Inderlied, C. B., and K. A. Nash. 1996. Microbiology and in vitro susceptibility testing, p. 109-195. In J. A. Korvick, and C. A. Benson (ed.), Mycobacterium avium complex infection: progress in research and treatment. Marcel Dekker, Inc., New York, N.Y.
- 70.Janda, J. M., and S. L. Abbott. 2002. Bacterial identification for publication: when is enough enough? J. Clin. Microbiol. 40:1887-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kazda, J., R. Cooney, M. Monaghan, P. J. Quinn, E. Stackebrandt, M. Dorsch, M. Daffé, K. Müller, B. R. Cook, and Z. S. Tarnok. 1993. Mycobacterium hiberniae sp. nov. Int. J. Syst. Bacteriol. 43:352-357. [DOI] [PubMed] [Google Scholar]
- 72.Kazda, J., H. J. Müller, E. Stackebrandt, M. Daffé, K. Müller, and C. Pitulle. 1992. Mycobacterium madagascariense sp. nov. Int. J. Syst. Bacteriol. 42:524-528. [Google Scholar]
- 73.Kazda, J., E. Stackebrandt, J. Smida, D. E. Minnikin, M. Daffé, J. H. Parlett, and C. Pitulle. 1990. Mycobacterium cookii sp. nov. Int. J. Syst. Bacteriol. 40:217-223. [DOI] [PubMed] [Google Scholar]
- 74.Khan, A. A., S. J. Kim, D. D. Paine, and C. E. Cerniglia. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int. J. Syst. Evol. Microbiol. 52:1997-2002. [DOI] [PubMed] [Google Scholar]
- 75.Kirschner, P., A. Meier, and E. C. Böttger. 1993. Genotypic identification and detection of mycobacteria—facing novel and uncultured pathogens, p. 173-190. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 76.Kirschner, P., A. Teske, K. H. Schröder, R. M. Kroppenstedt, J. Wolters, and E. C. Böttger. 1992. Mycobacterium confluentis sp. nov. Int. J. Syst. Bacteriol. 42:257-262. [DOI] [PubMed] [Google Scholar]
- 77.Kleespies, M., R. M. Kroppenstedt, F. A. Rainey, L. E. Webb, and E. Stackebrandt. 1996. Mycobacterium hodleri sp. nov., a new member of the fast-growing mycobacteria capable of degrading polycyclic aromatic hydrocarbons. Int. J. Syst. Bacteriol. 46:683-687. [DOI] [PubMed] [Google Scholar]
- 78.Koukila-Kähkölä, P., B. Springer, E. C. Böttger, L. Paulin, E. Jantzen, and M. L. Katila. 1995. Mycobacterium branderi sp. nov., a new potential human pathogen. Int. J. Syst. Bacteriol. 45:549-553. [DOI] [PubMed] [Google Scholar]
- 79.Krebs, T., S. Zimmerli, T. Bodmer, and B. Lammle. 2000. Mycobacterium genavense infection in a patient with long-standing chronic lymphocytic leukaemia. J. Intern. Med. 248:343-348. [DOI] [PubMed] [Google Scholar]
- 80.Kusunoki, S., and T. Ezaki. 1992. Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42:240-245. [DOI] [PubMed] [Google Scholar]
- 80a.Leautez, S., D. Boutoille, P. Bemer-Melchior, T. Ponge, and F. Raffi. 2000. Localized Mycobacterium genavense soft tissue infection in an immunodeficient HIV-negative patient. Eur. J. Clin. Microbiol. Infect. Dis. 19:51-52. [DOI] [PubMed] [Google Scholar]
- 81.Leclerc, M. C., N. Haddad, R. Moreau, and M. F. Thorel. 2000. Molecular characterization of environmental Mycobacterium strains by PCR-restriction fragment length polymorphism of hsp65 and by sequencing of hsp65, and of 16S and ITS1 rDNA. Res. Microbiol. 151:629-638. [DOI] [PubMed] [Google Scholar]
- 82.Liberek, V., C. Soravia, B. Ninet, B. Hirschel, and C. A. Siegrist. 1996. Cervical lymphadenitis caused by Mycobacterium genavense in a healthy child. Pediatr. Infect. Dis. J. 15:269-270. [DOI] [PubMed] [Google Scholar]
- 83.Liébana, E., A. Aranaz, B. Francis, and D. Cousins. 1996. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 34:933-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lumb, R., A. Goodwin, R. Stapledon, A. Holland, and I. Bastian. 1997. Phenotypic and molecular characterization of three clinical isolates of Mycobacterium interjectum. J. Clin. Microbiol. 35:2782-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luquín, M., V. Ausina, V. Vincent Lévy-Frébault, M. A. Lanéelle, F. Belda, M. García-Barceló, G. Prats, and M. Daffé. 1993. Mycobacterium brumae sp. nov., a rapidly growing, nonphotochromogenic mycobacterium. Int. J. Syst. Bacteriol. 43:405-413. [Google Scholar]
- 86.Luquín, M., M. A. Lanéelle, V. Ausina, M. García-Barceló, F. Belda, C. Alonso, and G. Prats. 1991. Distribution of a novel mycolic acid in the genus Mycobacterium. Int. J. Syst. Bacteriol. 41:390-394. [DOI] [PubMed] [Google Scholar]
- 87.Maier, T., E. Desmond, J. McCallum, W. Gordon, K. Young, and Y. Jang. 1995. Isolation of Mycobacterium genavense from AIDS patients and cultivation of the organism on Middlebrook 7H10 agar supplemented with human blood. Med. Microbiol. Lett. 4:173-179. [Google Scholar]
- 88.Matthews, J. A., and H. D. Liggitt. 1983. Disseminated mycobacteriosis in a cat. J. Am. Vet. Med. Assoc. 183:701-702. [PubMed] [Google Scholar]
- 89.McMullan, R., J. Xu, M. Kelly, T. Stanley, J. E. Moore, B. C. Millar, M. Wylie, C. E. Goldsmith, and D. R. T. Shepherd. 2002. Mycobacterium triplex pulmonary infection in an immunocompetent patient. J. Infect. 44:263-264. [DOI] [PubMed] [Google Scholar]
- 90.Meier, A., P. Kirschner, K. H. Schröder, J. Wolters, R. M. Kroppenstedt, and E. C. Böttger. 1993. Mycobacterium intermedium sp. nov. Int. J. Syst. Bacteriol. 43:204-209. [DOI] [PubMed] [Google Scholar]
- 91.Mendum, T. A., B. Z. Chilima, and P. R. Hirsch. 2000. The PCR amplification of non-tuberculous mycobacterial 16S rRNA sequences from soil. FEMS Microbiol. Lett. 185:189-192. [DOI] [PubMed] [Google Scholar]
- 92.Mijs, W., P. de Haas, R. Rossau, L. T. Van der Laan, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and ‘M. avium subsp. hominissuis' for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 93.Moore, J. S., M. Christensen, R. W. Wilson, R. J. Wallace, Jr., Y. Zhang, D. R. Nash, and B. Shelton. 2000. Mycobacterial contamination of metalworking fluids: involvement of a possible new taxon of rapidly growing mycobacteria. AIHAJ 61:205-213. [DOI] [PubMed] [Google Scholar]
- 94.Moss, M. T., Z. Malik, M. L. V. Tizard, E. P. Green, J. D. Sanderson, and J. Hermon-Taylor. 1992. IS 902, an insertion element of the chronic enteritis causing Mycobacterium avium subsp. silvaticum. J. Gen. Microbiol. 138:139-145. [DOI] [PubMed] [Google Scholar]
- 95.Moss, M. T., J. D. Sanderson, M. L. V. Tizard, J. Hermon-Taylor, F. A. K. El-Zaatari, D. C. Markesich, and D. Y. Graham. 1992. Polymerase chain reaction detection of Mycobacterium paratuberculosis and Mycobacterium avium subsp. silvaticum in long term cultures from Crohn's disease and control tissues. Gut 33:1209-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Muñoz, M., E. Julian, M. García-Barceló, V. Ausina, and M. Luquín. 1997. Easy differentiation of Mycobacterium mucogenicum from other species of the Mycobacterium fortuitum complex by thin-layer and gas chromatography of fatty esters and alcohols. J. Chromatogr. Ser. B 689:341-347. [DOI] [PubMed] [Google Scholar]
- 97.NCCLS 2000. Antimycobacterial susceptibility testing for mycobacteria, Nocardia and other aerobic actinomycetes. Tentative standard M24-T2, 2nd ed. NCCLS, Wayne, Pa.
- 98.Ngo Niobe, S., C. M. Bebear, M. Clerc, J. L. Pellegrin, C. Bebear, and J. Maugein. 2001. Disseminated Mycobacterium lentiflavum infection in a human immunodeficiency virus-infected patient. J. Clin. Microbiol. 39:2030-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2002. Biochemical and genetic evidence for the transfer of Mycobacterium tuberculosis subsp. caprae Aranaz et al. 1999 to the species Mycobacterium bovis Karlson and Lessel 1970 (approved lists 1980) as Mycobacterium bovis subsp. caprae comb. nov. Int. J. Syst. Evol. Microbiol. 52:433-436. [DOI] [PubMed] [Google Scholar]
- 100.Paterson, D. L., N. Singh, T. Gayowski, and I. R. Marino. 1998. Mycobacterium mucogenicum bacteremia in a patient with cirrhosis. J. Clin. Gastroenterol. 27:346-347. [DOI] [PubMed] [Google Scholar]
- 101.Pechère, M., M. Opravil, A. Wald, J. P. Chave, M. Bessesen, A. Sievers, R. Hein, J. von Overbeck, R. A. Clark, E. Tortoli, S. Emler, P. Kirschner, V. Gabriel, E. C. Böttger, B. Hirschel, and the Swiss HIV Cohort Study. 1995. Clinical and epidemiological features of infection with Mycobacterium genavense. Arch. Intern. Med. 155:400-404. [DOI] [PubMed] [Google Scholar]
- 102.Pfyffer, G. E., R. Auckenthaler, J. D. A. van Embden, and D. van Soolingen. 1998. Mycobacterium canettii, the smooth variant of M. tuberculosis, isolated from a Swiss patient exposed in Africa. Emerg. Infect. Dis. 4:631-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pfyffer, G. E., W. Weder, A. Strassle, and E. W. Russi. 1998. Mycobacterium heidelbergense species nov. infection mimicking a lung tumor. Clin. Infect. Dis. 27:649-650. [DOI] [PubMed] [Google Scholar]
- 104.Picardeau, M., T. J. Bull, G. Prod'Hom, A. L. Pozniak, D. C. Shanson, and V. Vincent. 1997. Comparison of a new insertion element, IS 1407, with established molecular markers for the characterization of Mycobacterium celatum. Int. J. Syst. Bacteriol. 47:640-644. [DOI] [PubMed] [Google Scholar]
- 105.Piersimoni, C., E. Tortoli, F. de Lalla, D. Nista, D. Donato, S. Bornigia, and G. De Sio. 1997. Isolation of Mycobacterium celatum from patients infected with human immunodeficiency virus. Clin. Infect. Dis. 24:144-147. [DOI] [PubMed] [Google Scholar]
- 106.Portaels, F., L. Realini, L. Bauwens, B. Hirschel, W. M. Meyers, and W. De Meurichy. 1996. Mycobacteriosis caused by Mycobacterium genavense in birds kept in a zoo: 11-year survey. J. Clin. Microbiol. 94:319-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Prodinger, W. M., A. Eigentler, F. Allerberger, M. Schönbauer, and W. Glawischnig. 2002. Infection of red deer, cattle, and humans with Mycobacterium bovis subsp. caprae in Western Austria. J. Clin Microbiol. 40:2270-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reischl, U., S. Emler, Z. Horak, J. Kaustova, R. M. Kroppenstedt, N. Lehn, and L. Naumann. 1998. Mycobacterium bohemicum sp. nov., a new slow-growing scotochromogenic mycobacterium. Int. J. Syst. Bacteriol. 48:1349-1355. [DOI] [PubMed] [Google Scholar]
- 109.Reischl, U., K. Feldmann, L. Naumann, B. J. M. Gaugler, B. Ninet, B. Hirschel, and S. Emler. 1998. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of 16S rRNA gene. J. Clin. Microbiol. 36:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rhodes, M. W., H. Kator, S. Kotob, P. van Berkum, I. Kaattari, W. Vogelbein, M. M. Floyd, W. R. Butler, F. D. Quinn, C. Ottinger, and E. Shotts. 2001. A unique Mycobacterium species isolated from an epizootic of striped bass (Morone saxatilis). Emerg. Infect. Dis. 7:896-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rhodes, M. W., H. Kator, S. Kotob, P. van Berkum, I. Kaattari, W. Vogelbein, F. Quinn, M. M. Floyd, W. R. Butler, and C. A. Ottinger. 2003. Mycobacterium shottsii sp. nov., a slowly growing species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 53:421-424. [DOI] [PubMed] [Google Scholar]
- 112.Richter, E., S. Niemann, F. O. Gloeckner, G. E. Pfyffer, and S. Rüsch-Gerdes. 2002. Mycobacterium holsaticum sp. nov. Int. J. Syst. Evol. Microbiol. 52:1991-1996. [DOI] [PubMed] [Google Scholar]
- 113.Richter, E., S. Niemann, S. Rüsch-Gerdes, and D. Harmsen. 2001. Description of Mycobacterium heckeshornense sp. nov. J. Clin. Microbiol. 39:3023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ringuet, H., C. Akoua-Koffi, S. Honore, A. Varnerot, V. Vincent, P. Berche, J. L. Gaillard, and C. Pierre-Audigier. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852-857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Roth, A., M. Fisher, M. E. Hamid, S. Michalke, W. Ludwig, and H. Mauch. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Roth, A., U. Reischl, N. Schonfeld, L. Naumann, S. Emler, M. Fischer, H. Mauch, R. Loddenkemper, and R. M. Kroppenstedt. 2000. Mycobacterium heckeshornense sp. nov., a new pathogenic slowly growing Mycobacterium sp. causing cavitary lung disease in an immunocompetent patient. J. Clin. Microbiol. 38:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roth, A., U. Reischl, A. Streubel, L. Naumann, R. M. Kroppenstedt, M. Habicht, M. Fischer, and H. Mauch. 2000. Novel diagnostic algorithm for identification of mycobacteria using genus-specific amplification of the 16S-23S rRNA gene spacer and restriction endonucleases. J. Clin. Microbiol. 38:1094-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Runyon, E. H. 1959. Anonymous mycobacteria in pulmonary disease. Med. Clin. North Am. 43:273-290. [DOI] [PubMed] [Google Scholar]
- 119.Rustscheff, S., L. Maroti, M. Holberg-Petersen, M. Steinbakk, and S. E. Hoffner. 2000. Mycobacterium interjectum: a new pathogen in humans? Scand. J. Infect. Dis. 32:569-571. [DOI] [PubMed] [Google Scholar]
- 120.Schinsky, M. F., M. M. McNeil, A. M. Whitney, A. G. Steigerwalt, B. A. Lasker, M. M. Floyd, G. G. Hogg, D. J. Brenner, and J. M. Brown. 2000. Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteremia. Int. J. Syst. Evol. Microbiol. 50:575-581. [DOI] [PubMed] [Google Scholar]
- 121.Schröder, K. H., L. Neumann, R. M. Kroppenstedt, and U. Reischl. 1997. Mycobacterium hassiacum sp. nov., a new rapidly growing thermophilic mycobacterium. Int. J. Syst. Bacteriol. 47:86-91. [DOI] [PubMed] [Google Scholar]
- 122.Shelton, B. G., W. D. Flanders, and G. K. Morris. 1999. Mycobacterium sp. as a possible cause of hypersensitivity pneumonitis in machine workers. Emerg. Infect. Dis. 5:270-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Shivannavar, C. T., V. M. Katoch, V. D. Sharma, M. A. Patil, K. Katoch, V. P. Bharadwaj, R. K. Sharma, A. S. Bhatia, and B. M. Agrawal. 1996. Determination of mycobacterial phylogeny on the basis of immunological relatedness of superoxide dismutases. Int. J. Syst. Bacteriol. 46:1164-1169. [DOI] [PubMed] [Google Scholar]
- 124.Shojaei, H., M. Goodfellow, J. G. Magee, R. Freeman, F. K. Gould, and C. G. Brignall. 1997. Mycobacterium novocastrense sp. nov., a rapidly growing photochromogenic mycobacterium. Int. J. Syst. Bacteriol. 47:1205-1207. [DOI] [PubMed] [Google Scholar]
- 125.Shojaei, H., J. G. Magee, R. Freeman, M. Yates, N. U. Horadagoda, and M. Goodfellow. 2000. Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic mycobacterium isolated from an elephant. Int. J. Syst. Evol. Microbiol. 50:1817-1820. [DOI] [PubMed] [Google Scholar]
- 126.Springer, B., E. C. Böttger, P. Kirschner, and R. J. Wallace, Jr. 1995. Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45:262-267. [DOI] [PubMed] [Google Scholar]
- 127.Springer, B., P. Kirschner, G. Rost-Meyer, K. H. Schröder, R. M. Kroppenstedt, and E. C. Böttger. 1993. Mycobacterium interjectum, a new species isolated from a patient with chronic lymphadenitis. J. Clin. Microbiol. 31:3083-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Springer, B., L. Stockman, K. Teschner, G. D. Roberts, and E. C. Böttger. 1996. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J. Clin. Microbiol. 34:296-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Springer, B., E. Tortoli, I. Richter, R. Grünewald, S. Rüsch-Gerdes, K. Uschmann, F. Suter, M. D. Collins, R. M. Kroppenstedt, and E. C. Böttger. 1995. Mycobacterium conspicuum sp. nov., a new species isolated from patients with disseminated infections. J. Clin. Microbiol. 33:2805-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Springer, B., W. K. Wu, T. Bodmer, G. Haase, G. E. Pfyffer, M. Reiner, R. M. Kroppenstedt, K. H. Schröder, S. Emler, J. O. Kilburn, P. Kirschner, A. Telenti, M. B. Coyle, and E. C. Böttger. 1996. Isolation and characterization of an unique group of slowly growing mycobacteria: description of Mycobacterium lentiflavum sp. nov. J. Clin. Microbiol. 34:1100-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Stager, C. E., J. R. Davis, and S. H. Siddiqi. 1988. Identification of Mycobacterium tuberculosis in sputum by direct inoculation of the BACTEC NAP vial. Diagn. Microbiol. Infect. Dis. 10:67-73. [DOI] [PubMed] [Google Scholar]
- 132.Stahl, D. A., and J. W. Urbance. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172:116-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Suomalainen, S., P. Koukila-Kähkölä, E. Brander, M. L. Katila, A. Piilonen, L. Paulin, and K. Mattson. 2001. Pulmonary infection caused by an unusual, slowly growing nontuberculous mycobacterium. J. Clin. Microbiol. 39:2668-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Swenson, J. M., R. J. Wallace, Jr., V. A. Silcox, and C. Thornsberry. 1985. Antimicrobial susceptibility of five subgroups of Mycobacterium fortuitum and Mycobacterium chelonae. Antimicrob. Agents Chemother. 28:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Takeuchi, M., and K. Hatano. 1998. Gordona rhizosphera sp. nov., isolated from the mangrove rhizosphere. Int. J. Syst. Bacteriol. 48:607-612. [DOI] [PubMed] [Google Scholar]
- 136.Taylor, T. B., C. Patterson, Y. Hale, and W. W. Safranek. 1997. Routine use of PCR-restriction fragment length polymorphism analysis for identification of mycobacteria growing in liquid media. J. Clin. Microbiol. 35:79-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Thorel, M. F., M. Krichevsky, and V. Vincent Lévy-Frébault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov. Mycobacterium avium subsp. paratuberculosis subsp. nov. and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 139.Torkko, P., S. Suomalainen, E. Iiavanainen, E. Tortoli, M. Suutari, J. Seppänen, L. Paulin, and M. L. Katila. 2002. Mycobacterium palustre sp. nov., a potentially pathogenic slow-growing mycobacterium isolated from veterinary and clinical specimens, and Finnish stream water. Int. J. Syst. Evol. Microbiol. 52:1519-1525. [DOI] [PubMed] [Google Scholar]
- 140.Torkko, P., S. Suomalainen, E. Iivanainen, M. Suutari, L. Paulin, E. Rudback, E. Tortoli, V. Vincent, R. Mattila, and M. L. Katila. 2001. Characterization of Mycobacterium bohemicum isolated from human, veterinary, and environmental sources. J. Clin. Microbiol. 39:207-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Torkko, P., S. Suomalainen, E. Iivanainen, M. Suutari, E. Tortoli, L. Paulin, and M. L. Katila. 2000. Mycobacterium xenopi and related organisms isolated from stream waters in Finland and description of Mycobacterium botniense sp. nov. Int. J. Syst. Evol. Microbiol. 50:283-289. [DOI] [PubMed] [Google Scholar]
- 142.Tortoli, E., A. Bartoloni, E. C. Böttger, S. Emler, C. Garzelli, E. Magliano, A. Mantella, N. Rastogi, L. Rindi, C. Scarparo, and P. Urbano. 2001. Burden of unidentifiable mycobacteria in a reference laboratory. J. Clin. Microbiol. 39:4058-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tortoli, E., A. Bartoloni, C. Burrini, D. Colombrita, A. Mantella, G. Pinsi, M. T. Simonetti, G. Swierczynski, and E. C. Böttger. 1996. Characterization of an isolate of the newly described species Mycobacterium interjectum. Zentbl. Bakteriol. Int. J. Med. Microbiol. 283:286-294. [DOI] [PubMed] [Google Scholar]
- 144.Tortoli, E., A. Bartoloni, M. L. Erba, E. Levrè, N. Lombardi, A. Mantella, and L. Mecocci. 2002. Human infections due to Mycobacterium lentiflavum. J. Clin. Microbiol. 40:728-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tortoli, E., A. Bartoloni, V. Manfrin, A. Mantella, C. Scarparo, and E. Böttger. 2000. Cervical lymphadenitis due to Mycobacterium bohemicum. Clin. Infect. Dis. 30:210-211. [DOI] [PubMed] [Google Scholar]
- 146.Tortoli, E., F. Brunello, A. E. Cagni, D. Colombrita, D. Dionisio, L. Grisendi, V. Manfrin, M. Moroni, C. Passerini Tosi, G. Pinsi, C. Scarparo, and M. T. Simonetti. 1998. Mycobacterium genavense in AIDS patients: report of 24 cases in Italy and review of the literature. Eur. J. Epidemiol. 14:219-224. [DOI] [PubMed] [Google Scholar]
- 147.Tortoli, E., P. Kirschner, A. Bartoloni, C. Burrini, V. Manfrin, A. Mantella, M. Scagnelli, C. Scarparo, M. T. Simonetti, and E. C. Böttger. 1996. Isolation of an unusual mycobacterium from an AIDS patient. J. Clin. Microbiol. 34:2316-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tortoli, E., P. Kirschner, B. Springer, A. Bartoloni, C. Burrini, A. Mantella, M. Scagnelli, C. Scarparo, M. T. Simonetti, and E. C. Böttger. 1997. Cervical lymphadenitis due to an unusual mycobacterium. Eur. J. Clin. Microbiol. Infect. Dis. 16:308-311. [DOI] [PubMed] [Google Scholar]
- 149.Tortoli, E., R. M. Kroppenstedt, A. Bartoloni, G. Caroli, I. Jan, J. Pawlowsky, and S. Emler. 1999. Mycobacterium tusciae sp. nov. Int. J. Syst. Bacteriol. 49:1839-1844. [DOI] [PubMed] [Google Scholar]
- 150.Tortoli, E., A. Nanetti, C. Piersimoni, P. Cichero, C. Farina, G. Mucignat, C. Scarparo, L. Bartolini, R. Valentini, D. Nista, G. Gesu, C. Passerini Tosi, M. Crovatto, and G. Brusarosco. 2001. Performance assessment of new multiplex probe assay for identification of mycobacteria. J. Clin. Microbiol. 39:1079-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Tortoli, E., C. Piersimoni, D. Bacosi, A. Bartoloni, F. Betti, L. Bono, C. Burrini, G. De Sio, C. Lacchini, and A. Mantella. 1995. Isolation of the newly described species Mycobacterium celatum from AIDS patients. J. Clin. Microbiol. 33:137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tortoli, E., C. Piersimoni, P. Kirschner, A. Bartoloni, C. Burrini, C. Lacchini, A. Mantella, G. Muzzi, C. Passerini Tosi, V. Penati, C. Scarparo, M. T. Simonetti, and E. C. Böttger. 1997. Characterization of mycobacterial isolates phylogenetically related to, but different from, Mycobacterium simiae. J. Clin. Microbiol. 35:697-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tortoli, E., C. Piersimoni, R. M. Kroppenstedt, J. I. Montoya-Burgos, U. Reischl, A. Giacometti, and S. Emler. 2001. Mycobacterium doricum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2007-2012. [DOI] [PubMed] [Google Scholar]
- 154.Tortoli, E., U. Reischl, G. Besozzi, and S. Emler. 1998. Characterization of an isolate belonging to the newly described species Mycobacterium hassiacum. Diagn. Microbiol. Infect. Dis. 30:193-196. [DOI] [PubMed] [Google Scholar]
- 155.Tortoli, E., M. T. Simonetti, D. Dionisio, and M. Meli. 1994. Cultural studies on two isolates of “Mycobacterium genavense” from patients with acquired immunodeficiency syndrome. Diagn. Microbiol. Infect. Dis. 18:7-12. [DOI] [PubMed] [Google Scholar]
- 156.Turenne, C., P. Chedore, J. Wolfe, F. Jamieson, G. Broukhanski, K. May, and A. Kabani. 2002. Mycobacterium lacus sp. nov., a new slow-growing non-chromogenic clinical isolate. Int. J. Syst. Evol. Microbiol. 52:2135-2140. [DOI] [PubMed] [Google Scholar]
- 157.Turenne, C., P. Chedore, J. Wolfe, F. Jamieson, K. May, and A. Kabani. 2002. Phenotypic and molecular characterization of clinical isolates of Mycobacterium elephantis from human specimens. J. Clin. Microbiol. 40:1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Turenne, C. Y., L. Tschetter, J. Wolfe, and A. Kabani. 2001. Necessity of quality-controlled 16S rRNA gene sequence databases: identifying nontuberculous Mycobacterium species. J. Clin. Microbiol. 39:3637-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.van Soolingen, D., T. Hoogenboezem, P. E. W. de Haas, P. W. M. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. A. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 160.Vergnaud, M., C. Dauga, A. Dompmartin, B. Malbruny, C. Bazin, and P. A. D. Grimont. 1998. Genital infection due to Mycobacterium genavense in a patient with AIDS. Clin. Infect. Dis. 27:1531.. [DOI] [PubMed] [Google Scholar]
- 161.Vuorio, R., M. A. Andersson, F. A. Rainey, R. M. Kroppenstedt, P. Kämpfer, H. J. Busse, M. Viljanen, and M. Salkinoja Salonen. 1999. A new rapidly growing mycobacterial species, Mycobacterium murale sp. nov., isolated from the indoor walls of a children's day care centre. Int. J. Syst. Bacteriol. 49:25-35. [DOI] [PubMed] [Google Scholar]
- 162.Wallace, R. J., Jr. 1994. Recent changes in taxonomy and disease manifestations of the rapidly growing mycobacteria. Eur. J. Clin. Microbiol. Infect. Dis. 13:953-960. [DOI] [PubMed] [Google Scholar]
- 163.Wallace, R. J., Jr., B. A. Brown-Elliott, L. Hall, G. Roberts, R. W. Wilson, L. B. Mann, C. J. Crist, S. H. Chiu, R. Dunlap, M. J. Garcia, J. T. Bagwell, and K. C. Jost, Jr. 2002. Clinical and laboratory features of Mycobacterium mageritense. J. Clin. Microbiol. 40:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wallace, R. J., Jr., B. A. Brown, V. A. Silcox, M. Tsukamura, D. N. Nash, T. Udou, L. C. Steele, V. A. Steingrube, J. Smith, G. Sumter, Y. Zhang, and Z. Blacklock. 1991. Clinical disease, drug susceptibility, and biochemical patterns of the unnamed third biovariant complex of Mycobacterium fortuitum. J. Infect. Dis. 163:598-603. [DOI] [PubMed] [Google Scholar]
- 165.Wallace, R. J., Jr., R. O'Brien, J. Glassroth, J. Raleigh, and A. Dutt. 1990. Diagnosis and treatment of disease caused by nontuberculous mycobacteria. Statement of the American Thoracic Society; prepared by an ad hoc committee of the Scientific Assembly of Microbiology, Tuberculosis, and Pulmonary Infection. Am. Rev. Respir. Dis. 142:940-953. [DOI] [PubMed] [Google Scholar]
- 166.Wallace, R. J., Jr., V. A. Silcox, M. Tsukamura, B. A. Brown, J. O. Kilburn, W. R. Butler, and G. Onyi. 1993. Clinical significance, biochemical features, and susceptibility patterns of sporadic isolates of the Mycobacterium chelonae-like organism. J. Clin. Microbiol. 31:3231-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1995. Phylogenetic analysis of polycyclic aromatic hydrocarbon degrading mycobacteria by 16S rRNA sequencing. FEMS Microbiol. Lett. 130:75-80. [DOI] [PubMed] [Google Scholar]
- 168.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Trüper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 169.Wayne, L. G., and H. A. Sramek. 1992. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin. Microbiol. Rev. 5:1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Willumsen, P., U. Karlson, E. Stackebrandt, and R. M. Kroppenstedt. 2001. Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon-degrading Mycobacterium species. Int. J. Syst. Evol. Microbiol. 51:1715-1722. [DOI] [PubMed] [Google Scholar]
- 171.Wilson, R. W., V. A. Steingrube, E. C. Böttger, B. Springer, B. A. Brown-Elliott, V. Vincent, K. C. Jost, Jr., Y. Zhang, M. J. Garcia, S. H. Chiu, G. O. Onyi, H. Rossmoore, D. R. Nash, and R. J. Wallace, Jr. 2001. Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51:1751-1764. [DOI] [PubMed] [Google Scholar]
- 172.Wilson, R. W., V. A. Steingrube, B. A. Brown, and R. J. Wallace, Jr. 1998. Clinical application of PCR-restriction enzyme pattern analysis for rapid identification of aerobic actinomycete isolates. J. Clin. Microbiol. 36:148-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wilton, S., and D. Cousins. 1992. Detection and identification of multiple mycobacterial pathogens by DNA amplification in single tube. PCR Methods Appl. 1:269-273. [DOI] [PubMed] [Google Scholar]
- 174.Wolfe, J., C. Turenne, M. Alfa, G. Harding, L. Thibert, and A. Kabani. 2000. Mycobacterium branderi from both a hand infection and a case of pulmonary disease. J. Clin. Microbiol. 38:3896-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Wolinsky, E. 1979. Nontuberculous mycobacteria and associated diseases. Am. Rev. Respir. Dis. 119:107-159. [DOI] [PubMed] [Google Scholar]
- 176.Wolinsky, E. 1995. Mycobacterial lymphadenitis in children: a prospective study of 105 nontuberculous cases with long-term follow-up. Clin. Infect. Dis. 20:954-963. [DOI] [PubMed] [Google Scholar]
- 177.Yoshimura, H. H., and D. Y. Graham. 1988. Nucleic acid hybridization studies of mycobactin-dependent mycobacteria. J. Clin. Microbiol. 26:1309-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zurawski, C. A., G. D. Cage, D. Rimland, and H. M. Blumberg. 1997. Pneumonia and bacteremia due to Mycobacterium celatum masquerading as Mycobacterium xenopi in patient with AIDS: an underdiagnosed problem? Clin. Infect. Dis. 24:140-143. [DOI] [PubMed] [Google Scholar]